Drosophila Interspecific Hybridization Causes a Deregulation of the piRNA Pathway Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Drosophila Stocks and Crosses
2.2. Sequencing of piRNA Pathway Genes in D. buzzatii and D. koepferae
2.3. Sequence Analysis
2.4. Quantification of Gene Transcripts by qRT-PCR
2.5. Fluorescent In Situ Hybridization in Ovaries
2.6. Statistical Methods
3. Results
3.1. piRNA Pathway Gene Characterization in D. buzzatii and D. koepferae
3.2. Gene Expression in Parental Species
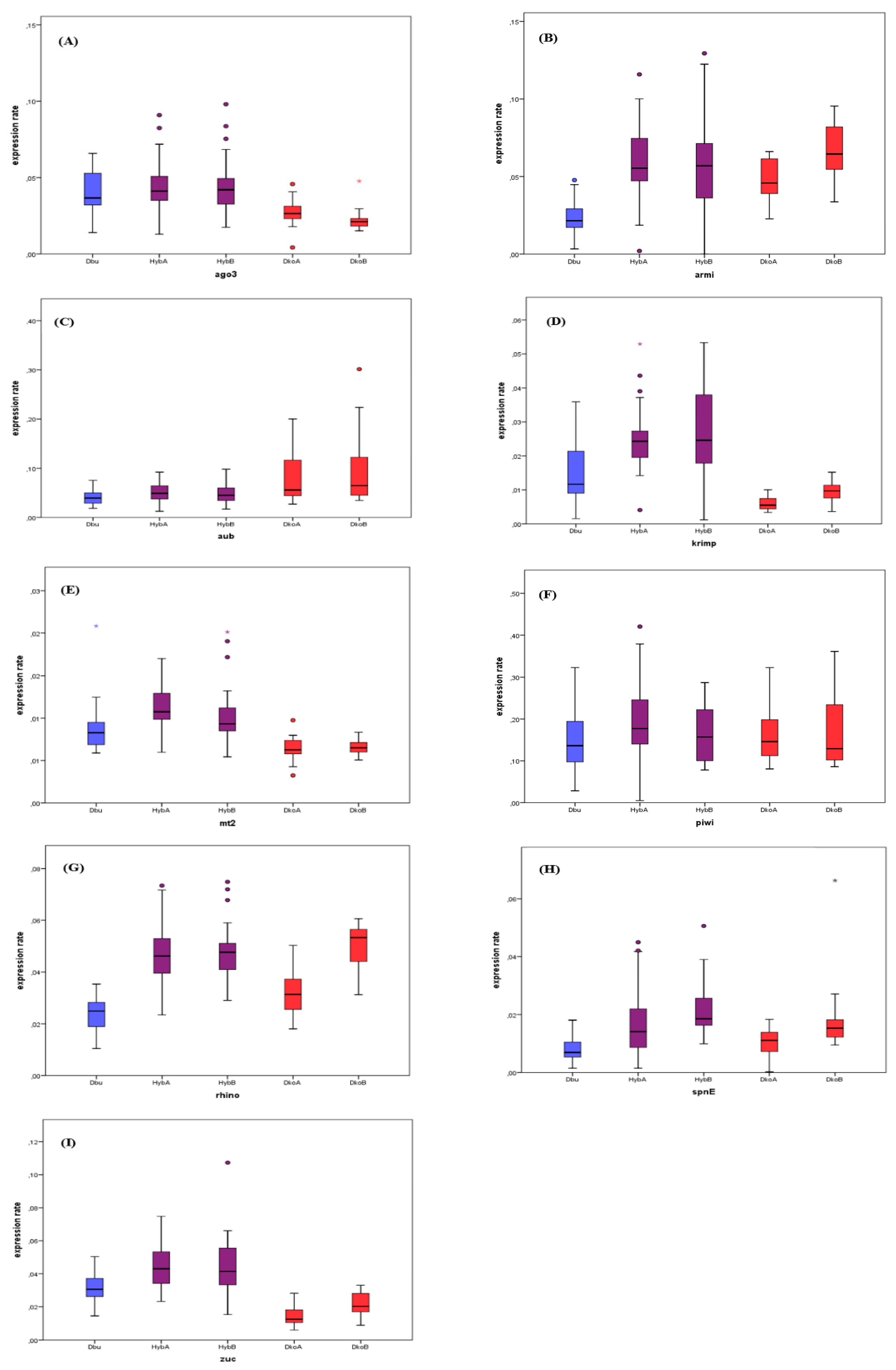
3.3. Gene Expression in Hybrids
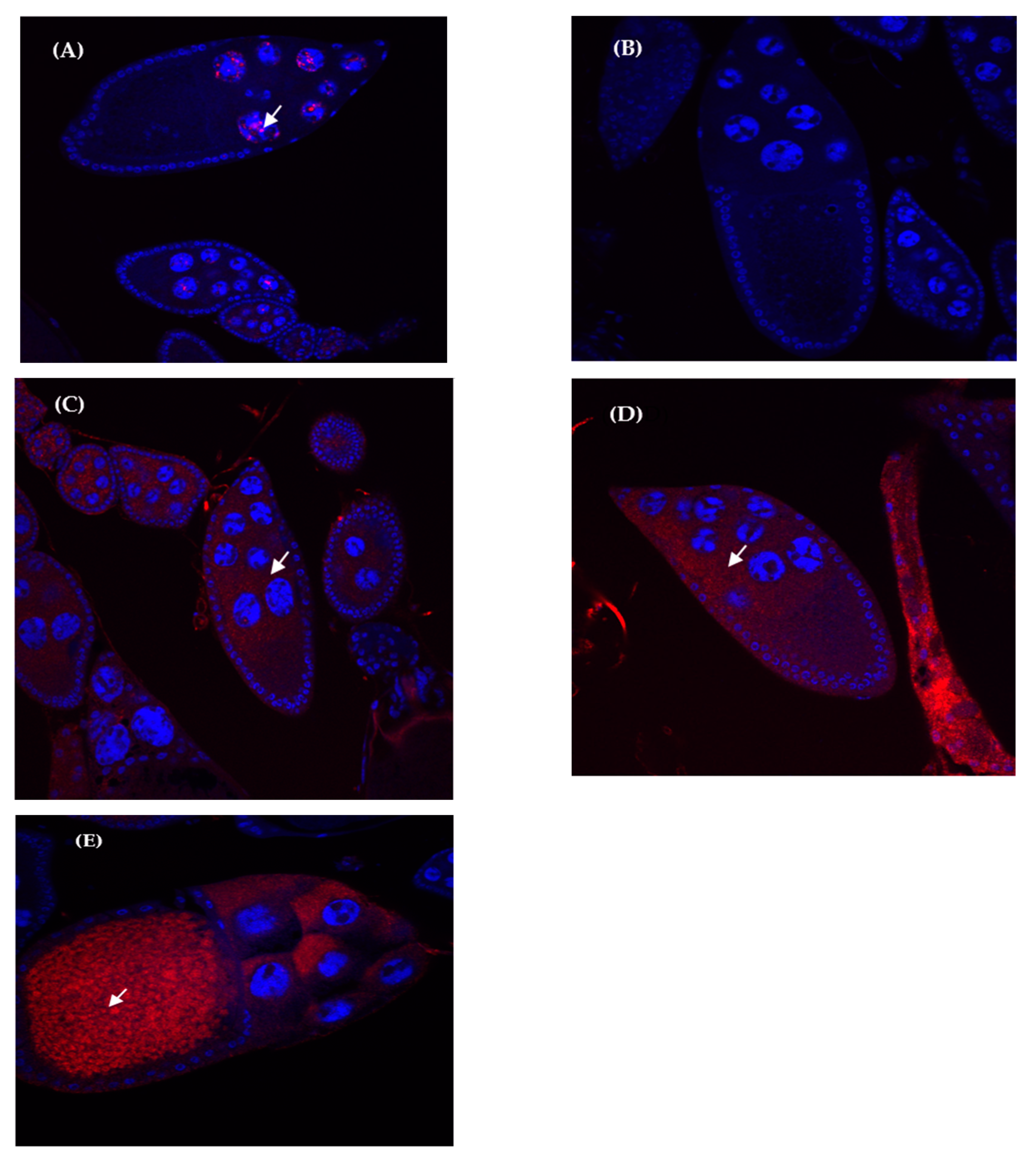
3.4. Expression Localization Patterns in Hybrid and Parental Species Ovaries
4. Discussion
4.1. piRNA Pathway Gene Structure is Conserved between D. buzzatii and D. koepferae
4.2. Armitage and Aubergine Bear Marks of Positive Selection
4.3. Misexpression of piRNA Pathway Genes in D. buzzatii–D. koepferae Hybrids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Iwasaki, A.; Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015, 16, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Hannon, G.J. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem. Sci. 2016, 41, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.; Theurkauf, W. Biogenesis and germline functions of piRNAs. Development 2007, 135, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Rozhkov, N.V.; Hammell, M.; Hannon, G.J. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013, 27, 400–412. [Google Scholar] [CrossRef]
- Gunawardane, L.S.; Saito, K.; Nishida, K.M.; Miyoshi, K.; Kawamura, Y.; Nagami, T.; Siomi, H.; Siomi, M.C. A Slicer-Mediated Mechanism for Repeat-Associated siRNA 5′ End Formation in Drosophila. Science (80-) 2007, 315, 1587–1590. [Google Scholar] [CrossRef]
- Mohn, F.; Handler, D.; Brennecke, J. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science (80-.) 2015, 348, 812–817. [Google Scholar] [CrossRef]
- Han, B.W.; Wang, W.; Li, C.; Weng, Z.; Zamore, P.D. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 2015, 348, 817–822. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.W.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science (80-) 2008, 320, 1077–1081. [Google Scholar] [CrossRef]
- García Guerreiro, M.P. What makes transposable elements move in the Drosophila genome? Heredity (Edinb.) 2012, 108, 461–468. [Google Scholar] [CrossRef]
- Horváth, V.; Merenciano, M.; González, J. Revisiting the Relationship between Transposable Elements and the Eukaryotic Stress Response. Trends Genet. 2017, 33, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Evgen’ev, M.B.; Yenikolopov, G.N.; Peunova, N.I.; Ilyin, Y.V. Transposition of mobile genetic elements in interspecific hybrids of Drosophila. Chromosoma 1982, 85, 375–386. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.J.W.; O’Neill, M.J.; Graves, J.A.M. Erratum: Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 1998, 393, 68–72. [Google Scholar] [CrossRef]
- Metcalfe, C.J.; Bulazel, K.V.; Ferreri, G.C.; Schroeder-Reiter, E.; Wanner, G.; Rens, W.; Obergfell, C.; Eldridge, M.D.B.; O’Neill, R.J. Genomic instability within centromeres of interspecific marsupial hybrids. Genetics 2007, 177, 2507–2517. [Google Scholar] [CrossRef] [PubMed]
- Vela, D. Estudio de la inestabilidad genómica inducida por transposición en los híbridos interespecíficos de Drosophila buzzatii y Drosophila koepferae; Universitat Autònoma de Barcelona: Barcelona, Spain, 2012. [Google Scholar]
- Liu, B.; Wendel, J.F. Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 2000, 43, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, M.C.; Strakosh, S.C.; Zhen, Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 2006, 16, R872–R873. [Google Scholar] [CrossRef]
- Obbard, D.J.; Gordon, K.H.; Buck, A.H.; Jiggins, F.M. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 99–115. [Google Scholar] [CrossRef]
- Simkin, A.; Wong, A.; Poh, Y.-P.; Theurkauf, W.E.; Jensen, J.D. Recurrent and recent selective sweeps in the piRNA pathway. Evolution (N. Y.) 2013, 67, 1081–1090. [Google Scholar] [CrossRef]
- Parhad, S.S.; Tu, S.; Weng, Z.; Theurkauf, W.E. Adaptive Evolution Leads to Cross-Species Incompatibility in the piRNA Transposon Silencing Machinery. Dev. Cell 2017, 43, 60–70.e5. [Google Scholar] [CrossRef]
- Fablet, M.; Akkouche, A.; Braman, V.; Vieira, C. Variable expression levels detected in the Drosophila effectors of piRNA biogenesis. Gene 2014, 537, 149–153. [Google Scholar] [CrossRef]
- Labrador, M.; Farré, M.; Utzet, F.; Fontdevila, A. Interspecific hybridization increases transposition rates of Osvaldo. Mol. Biol. Evol. 1999, 16, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Vela, D.; Fontdevila, A.; Vieira, C.; García Guerreiro, M.P. A genome-wide survey of genetic instability by transposition in Drosophila hybrids. PLoS ONE 2014, 9, e88992. [Google Scholar] [CrossRef] [PubMed]
- Romero-Soriano, V.; Burlet, N.; Vela, D.; Fontdevila, A.; Vieira, C.; García Guerreiro, M.P. Drosophila Females Undergo Genome Expansion after Interspecific Hybridization. Genome Biol. Evol. 2016, 8, 556–561. [Google Scholar] [CrossRef] [PubMed]
- García Guerreiro, M.P. Changes of Osvaldo expression patterns in germline of male hybrids between the species Drosophila buzzatii and Drosophila koepferae. Mol. Genet. Genom. 2015, 290, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Romero-Soriano, V.; Garcia Guerreiro, M.P. Expression of the Retrotransposon Helena Reveals a Complex Pattern of TE Deregulation in Drosophila Hybrids. PLoS ONE 2016, 11, e0147903. [Google Scholar] [CrossRef] [PubMed]
- Romero-Soriano, V.; Modolo, L.; Lopez-Maestre, H.; Mugat, B.; Pessia, E.; Chambeyron, S.; Vieira, C.; Garcia Guerreiro, M.P. Transposable Element Misregulation Is Linked to the Divergence between Parental piRNA Pathways in Drosophila Hybrids. Genome Biol. Evol. 2017, 9, 1450–1470. [Google Scholar] [CrossRef]
- Marin, I.; Ruiz, A.; Pla, C.; Fontdevila, A. Reproductive relationships among ten species of the Drosophila repleta group from South America and the West Indies. Evolution (N. Y.) 1993, 47, 1616–1624. [Google Scholar]
- Gramates, L.S.; Marygold, S.J.; dos Santos, G.; Urbano, J.-M.; Antonazzo, G.; Matthews, B.B.; Rey, A.J.; Tabone, C.J.; Crosby, M.A.; Emmert, D.B.; et al. FlyBase at 25: Looking to the future. Nucleic Acids Res. 2017, 45, D663–D671. [Google Scholar] [CrossRef]
- Guillén, Y.; Rius, N.; Delprat, A.; Williford, A.; Muyas, F.; Puig, M.; Casillas, S.; Ràmia, M.; Egea, R.; Negre, B.; et al. Genomics of Ecological Adaptation in Cactophilic Drosophila. Genome Biol. Evol. 2015, 7, 349–366. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Zheng, C.; Chitsaz, F.; Derbyshire, M.K.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Lanczycki, C.J.; et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2012, 41, D348–D352. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-3.0. Available online: http://www.repeatmasker.org (accessed on 1 July 2010).
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Akkouche, A.; Rebollo, R.; Burlet, N.; Esnault, C.; Martinez, S.; Viginier, B.; Terzian, C.; Vieira, C.; Fablet, M. tirant, a Newly Discovered Active Endogenous Retrovirus in Drosophila simulans. J. Virol. 2012, 86, 3675–3681. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a Test of Whether one of Two Random Variables is Stochastically Larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- McDonald, J.H.; Kreitman, M. Adaptive protein evolution at the Adh locus in Drosophila. Nature 1991, 351, 652–654. [Google Scholar] [CrossRef]
- Putnam, N.H.; Srivastava, M.; Hellsten, U.; Dirks, B.; Chapman, J.; Salamov, A.; Terry, A.; Shapiro, H.; Lindquist, E.; Kapitonov, V.V.; et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317, 86–94. [Google Scholar] [CrossRef]
- Kolaczkowski, B.; Hupalo, D.N.; Kern, A.D. Recurrent Adaptation in RNA Interference Genes Across the Drosophila Phylogeny. Mol. Biol. Evol. 2011, 28, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Coulombe-Huntington, J.; Majewski, J. Intron Loss and Gain in Drosophila. Mol. Biol. Evol. 2007, 24, 2842–2850. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 2009, 10, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Barker, J.S.F. Effective population size of natural populations of Drosophila buzzatii, with a comparative evaluation of nine methods of estimation. Mol. Ecol. 2011, 20, 4452–4471. [Google Scholar] [CrossRef]
- Gaffney, D.J.; Keightley, P.D. Genomic Selective Constraints in Murid Noncoding DNA. PLoS Genet. 2006, 2, e204. [Google Scholar] [CrossRef]
- Gomez, G.A.; Hasson, E. Transpecific Polymorphisms in an Inversion Linked Esterase Locus in Drosophila buzzatii. Mol. Biol. Evol. 2003, 20, 410–423. [Google Scholar] [CrossRef]
- Palmer, W.H.; Hadfield, J.D.; Obbard, D.J. RNA-Interference Pathways Display High Rates of Adaptive Protein Evolution in Multiple Invertebrates. Genetics 2018, 208, 1585–1599. [Google Scholar] [CrossRef]
- Alvarez-Ponce, D.; Aguadé, M.; Rozas, J. Network-level molecular evolutionary analysis of the insulin/TOR signal transduction pathway across 12 Drosophila genomes. Genome Res. 2009, 19, 234–242. [Google Scholar] [CrossRef]
- Blumenstiel, J.P.; Erwin, A.A.; Hemmer, L.W. What Drives Positive Selection in the Drosophila piRNA Machinery? The Genomic Autoimmunity Hypothesis. Yale J. Biol. Med. 2016, 89, 499–512. [Google Scholar]
- Duggal, N.K.; Emerman, M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 2012, 12, 687–695. [Google Scholar] [CrossRef]
- Kelleher, E.S.; Edelman, N.B.; Barbash, D.A. Drosophila Interspecific Hybrids Phenocopy piRNA-Pathway Mutants. PLoS Biol. 2012, 10, e1001428. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sun, Y.; Shen, K.; Sun, S.; Wang, J.; Jiang, T.; Cao, S.; Josiah, S.M.; Pang, J.; Lin, X.; et al. Immediate Genetic and Epigenetic Changes in F1 Hybrids Parented by Species with Divergent Genomes in the Rice Genus (Oryza). PLoS ONE 2015, 10, e0132911. [Google Scholar] [CrossRef] [PubMed]
- Becalska, A.N.; Gavis, E.R. Lighting up mRNA localization in Drosophila oogenesis. Development 2009, 136, 2493–2503. [Google Scholar] [CrossRef]
- Dion-Côté, A.-M.; Renaut, S.; Normandeau, E.; Bernatchez, L. RNA-seq Reveals Transcriptomic Shock Involving Transposable Elements Reactivation in Hybrids of Young Lake Whitefish Species. Mol. Biol. Evol. 2014, 31, 1188–1199. [Google Scholar] [CrossRef] [PubMed]
- Renaut, S.; Rowe, H.C.; Ungerer, M.C.; Rieseberg, L.H. Genomics of homoploid hybrid speciation: Diversity and transcriptional activity of long terminal repeat retrotransposons in hybrid sunflowers. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Carnelossi, E.A.G.; Lerat, E.; Henri, H.; Martinez, S.; Carareto, C.M.A.; Vieira, C. Specific Activation of an I-Like Element in Drosophila Interspecific Hybrids. Genome Biol. Evol. 2014, 6, 1806–1817. [Google Scholar] [CrossRef] [PubMed]
- Josefsson, C.; Dilkes, B.; Comai, L. Parent-dependent loss of gene silencing during interspecies hybridization. Curr. Biol. 2006, 16, 1322–1328. [Google Scholar] [CrossRef]

| D. buzzatii | D. koepferae | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genes a | Length | CDS | Protein | Exons | Length | CDS | Protein | Exons | NI (%) | PS (%) |
| ago3 | 2009 | 1688 | 562 | 2 | 3961 | 1686 | 562 | 2 | 95.6 | 93.2 |
| armi | 3701 | 3432 | 1141 | 5 | 3688 | 3424 | 1122 | 5 | 93.2 | 92.5 |
| aub | 4250 | 2562 | 852 | 9 | 4474 | 2466 | 821 | 8b | 94 | 92.9 |
| krimp | 1929 | 1852 | 617 | 2 | 1920 | 1843 | 615 | 2 | 92.5 | 90.9 |
| mt2 | 812 | 810 | 270 | 1 b | 1064 | 1005 | 335 | 2 | 94 | 94.4 |
| piwi | 3924 | 2625 | 874 | 8 | 2484 | 2097 | 699 | 7b | 94.5 | 95.6 |
| rhino | 1865 | 1749 | 602 | 2 | 1944 | 1885 | 628 | 2 | 78 | 69 |
| spnE | 6052 | 4012 | 1337 | 11 | 6245 | 4125 | 1375 | 11 | 94 | 91.1 |
| zuc | 651 | 651 | 217 | 1 | 654 | 654 | 218 | 1 | 86.5 | 72.7 |
| Genes | Region/Domain | Pn/Ps | Dn/Ds | NI | α | P |
|---|---|---|---|---|---|---|
| ago3 | CDS | - | 0.911 | - | - | - |
| armi | CDS | 8.33 × 10−2 | 6.35 × 10-1 | 1.31 × 10-1 | 8.69 × 10-1 | 1.00 × 10-3 ** |
| aub | CDS | 5.00 × 10-1 | 6.37 × 10-1 | 7.84 × 10-1 | 3.27 × 10-1 | 2.15 × 10-1 |
| krimp | CDS | 0.00 | 8.25 × 10-1 | 0.00 | 1.00 | 6.00 × 10-2 |
| mt2 | CDS | 1.67 × 10-1 | 4.38 × 10-1 | 3.80 × 10-1 | 6.19 × 10-1 | 3.75 × 10-1 |
| piwi | CDS | 3.08 × 10-1 | 4.33 × 10-1 | 7.11 × 10-1 | 2.88 × 10-1 | 5.78 × 10-1 |
| rhino | CDS | 7.50 × 10-1 | 4.94 × 10-1 | 6.58 × 10-1 | 3.41 × 10-1 | 5.89 × 10-1 |
| spnE | CDS | 5.00 × 10-1 | 9.31 × 10-1 | 4.65 × 10-1 | 5.34 × 10-1 | 1.65 × 10-1 |
| zuc | CDS | 5.00 × 10-1 | 3.34 × 10-1 | 6.67 × 10-1 | 3.32 × 10-1 | 7.44 × 10-1 |
| aub | PAZ | 1.60 | 6.30 × 10-1 | 9.94 × 10-1 | 5.00 × 10-3 | 7.5 × 10-1 *** |
| D. koepferae vs. D. buzzatii | ||
|---|---|---|
| Gene | W | p-Value |
| ago3 | 790 | 1.46 × 10-6 *** |
| armi | 36 | 1.24 × 10-7 *** |
| aub | 190 | 1.31 × 10-3 ** |
| krimp | 582 | 4.98 × 10-3 ** |
| mt2 | 773 | 9.00 × 10-5 *** |
| piwi | 349 | 4.28 × 10-1 |
| rhino | 118 | 3.00 × 10-6 *** |
| spnE | 232 | 9.21 × 10-4 *** |
| zuc | 691 | 1.25 × 10-5 *** |
| Gene | Cross | N | Median | SD | vs. D. buzzatii | vs. D. koepferae A | vs. D. koepferae B | |||
|---|---|---|---|---|---|---|---|---|---|---|
| W | p-Value | W | p-Value | W | p-Value | |||||
| ago3 | A | 45 | 4.11 × 10-2 | 1.28 × 10-2 | 143 | 8.41 × 10-1 | 31 | 1.02 × 10-2 * | ||
| B | 26 | 4.20 × 10-2 | 1.88 × 10-2 | 136 | 7.51 × 10-1 | 0 | 0.00 *** | |||
| armi | A | 36 | 5.53 × 10-2 | 2.33 × 10-2 | 207 | 0.00 *** | 152 | 1.05 × 10-1 | ||
| B | 34 | 5.69 × 10-2 | 2.74 × 10-2 | 210 | 0.00 *** | 156 | 6.20 × 10-2 | |||
| aub | A | 36 | 4.90 × 10-2 | 1.87 × 10-2 | 75 | 4.67 × 10-1 | 52 | 7.20 × 10-2 | ||
| B | 34 | 4.49 × 10-2 | 2.05 × 10-2 | 90 | 6.90 × 10-1 | 32 | 9.00 x 10-3 ** | |||
| krimp | A | 36 | 2.43 × 10-2 | 8.80 × 10-3 | 38 | 2.00 × 10-2 * | 0 | 0.00 *** | ||
| B | 34 | 2.46 × 10-2 | 1.27 × 10-2 | 37 | 1.80 × 10-2 * | 1 | 0.00 *** | |||
| mt2 | A | 45 | 1.07 × 10-2 | 2.30 × 10-3 | 58 | 1.80 × 10-2 * | 1 | 0.00 *** | ||
| B | 26 | 9.32 × 10-3 | 4.50 × 10-3 | 86 | 1.01 × 10-1 | 1 | 0.00 *** | |||
| piwi | A | 36 | 1.77 × 10-1 | 8.81 × 10-2 | 66 | 1.81 × 10-1 | 91 | 6.01 × 10-1 | ||
| B | 34 | 1.57 × 10-1 | 6.85 × 10-2 | 102 | 9.11 × 10-1 | 84 | 4.33 × 10-1 | |||
| rhino | A | 45 | 1.31 × 10-2 | 1.01 × 10-2 | 246 | 1.70 × 10-2 * | 149 | 1.09 × 10-1 | ||
| B | 26 | 1.86 × 10-2 | 9.00 × 10-3 | 295 | 0.00 *** | 156 | 6.20 × 10-2 | |||
| spnE | A | 45 | 1.31 × 10-2 | 1.01 × 10-2 | 55 | 1.70 × 10-2 * | 61 | 1.09 × 10-1 | ||
| B | 26 | 1.85 × 10-2 | 9.00 × 10-3 | 5 | 0.00 *** | 54 | 6.20 × 10-2 | |||
| zuc | A | 36 | 4.30 × 10-2 | 1.31 × 10-2 | 48 | 4.70 × 10-2 * | 1 | 0.00 *** | ||
| B | 34 | 4.14 × 10-2 | 1.78 × 10-2 | 59 | 1.96 × 10-1 | 12 | 2.00 × 10-3 ** | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gámez-Visairas, V.; Romero-Soriano, V.; Martí-Carreras, J.; Segarra-Carrillo, E.; García Guerreiro, M.P. Drosophila Interspecific Hybridization Causes a Deregulation of the piRNA Pathway Genes. Genes 2020, 11, 215. https://doi.org/10.3390/genes11020215
Gámez-Visairas V, Romero-Soriano V, Martí-Carreras J, Segarra-Carrillo E, García Guerreiro MP. Drosophila Interspecific Hybridization Causes a Deregulation of the piRNA Pathway Genes. Genes. 2020; 11(2):215. https://doi.org/10.3390/genes11020215
Chicago/Turabian StyleGámez-Visairas, Víctor, Valèria Romero-Soriano, Joan Martí-Carreras, Eila Segarra-Carrillo, and Maria Pilar García Guerreiro. 2020. "Drosophila Interspecific Hybridization Causes a Deregulation of the piRNA Pathway Genes" Genes 11, no. 2: 215. https://doi.org/10.3390/genes11020215
APA StyleGámez-Visairas, V., Romero-Soriano, V., Martí-Carreras, J., Segarra-Carrillo, E., & García Guerreiro, M. P. (2020). Drosophila Interspecific Hybridization Causes a Deregulation of the piRNA Pathway Genes. Genes, 11(2), 215. https://doi.org/10.3390/genes11020215





