Comparative Transcriptome Analysis Reveals Stem Secondary Growth of Grafted Rosa rugosa ‘Rosea’ Scion and R. multiflora ‘Innermis’ Rootstock
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
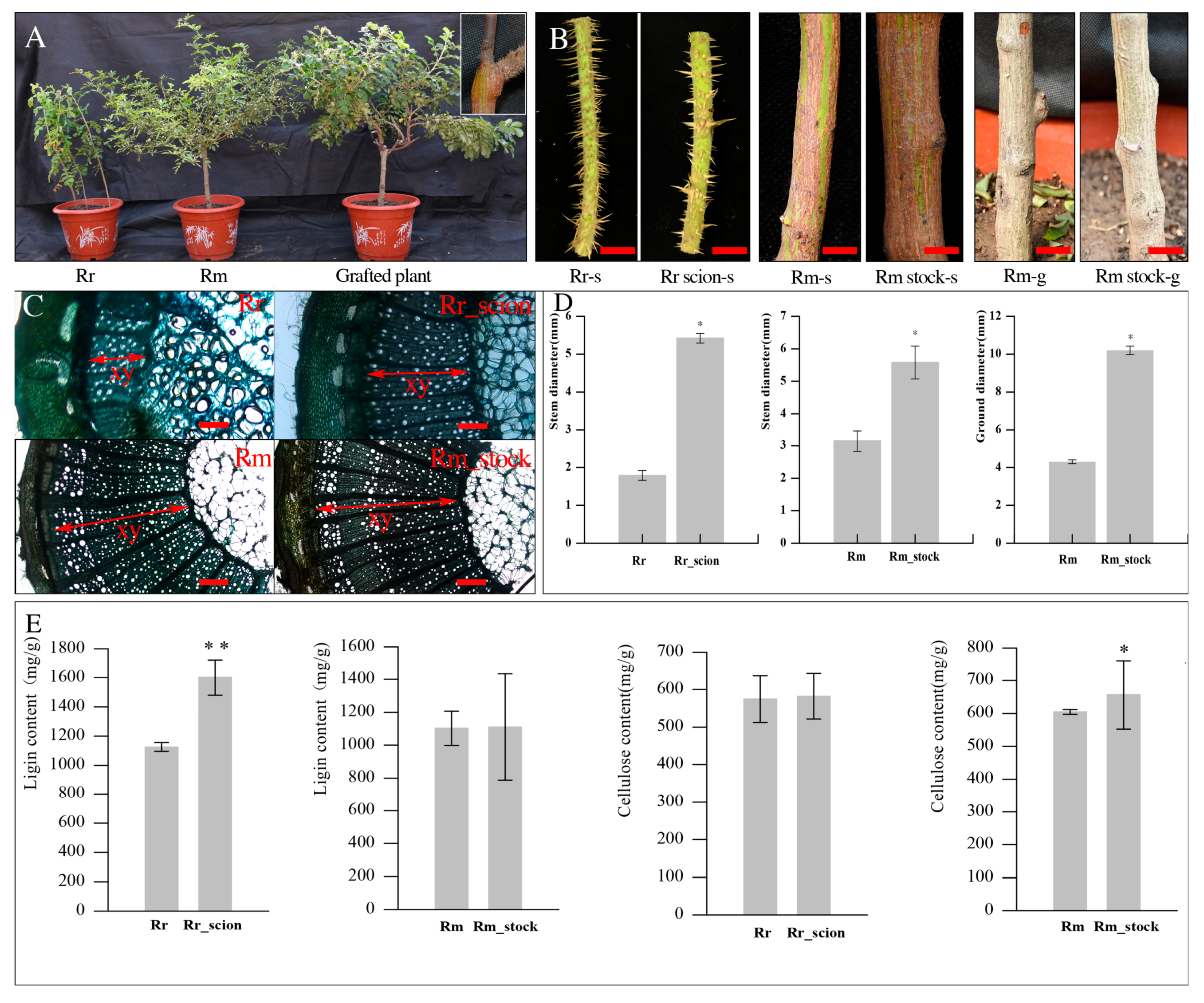
2.2. Histological Analysis
2.3. Construction of the cDNA Library and Solexa Sequencing for Transcriptomic Analysis
2.4. Sequencing, Assembly, and Functional Annotation of cDNA
2.5. Identification of DEGs and Functional Analysis
2.6. A Quantitative Reverse-Transcription PCR (qRT-PCR) for the Validation and Analysis of Expression Patterns
3. Results
3.1. Effects of Rosa Grafting on the Stem Growth of Scion and Stock
3.2. Illumina Sequencing and De Novo Assembly of the Grafted Rosa Transcriptome
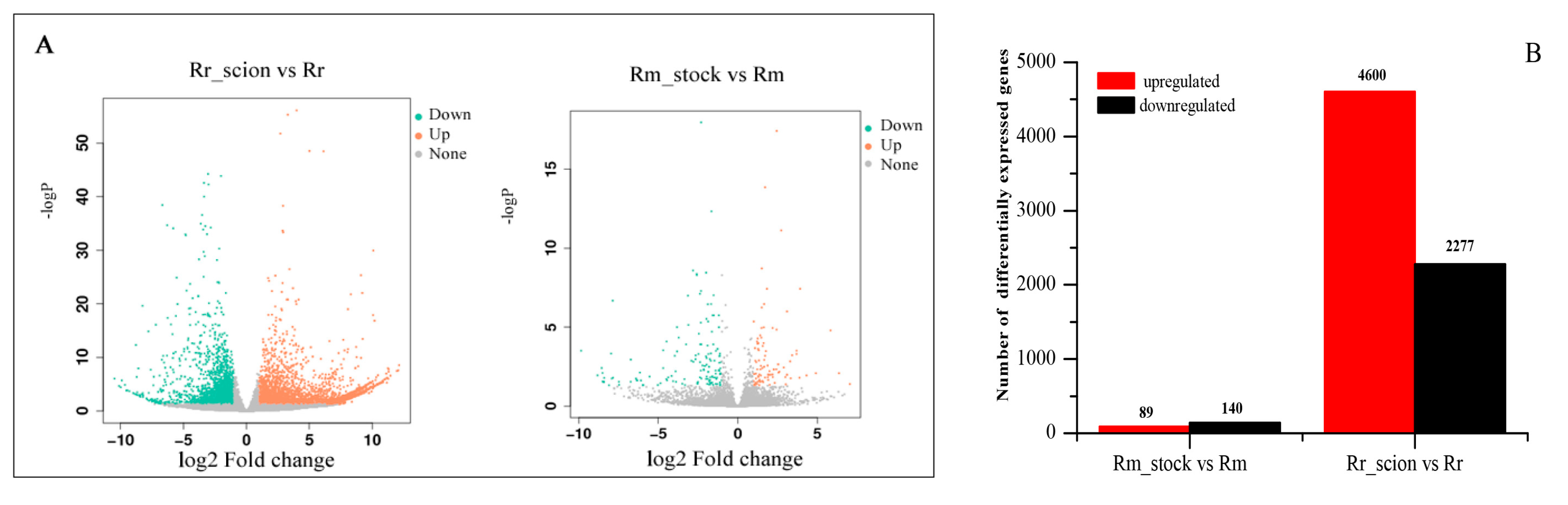
3.3. Analysis of DEGs of R. rugosa ‘Rosea’/R. multiflora ‘Innermis’ Grafts
3.4. GO Enrichment Analysis of DEGs
3.5. Pathway Analysis of DEGs in the Scion and Rootstock
3.6. Functional Genes in the Scion and Rootstock
3.6.1. Protein Kinases
3.6.2. Transcription Factors (TFs)
3.6.3. Transporter Genes
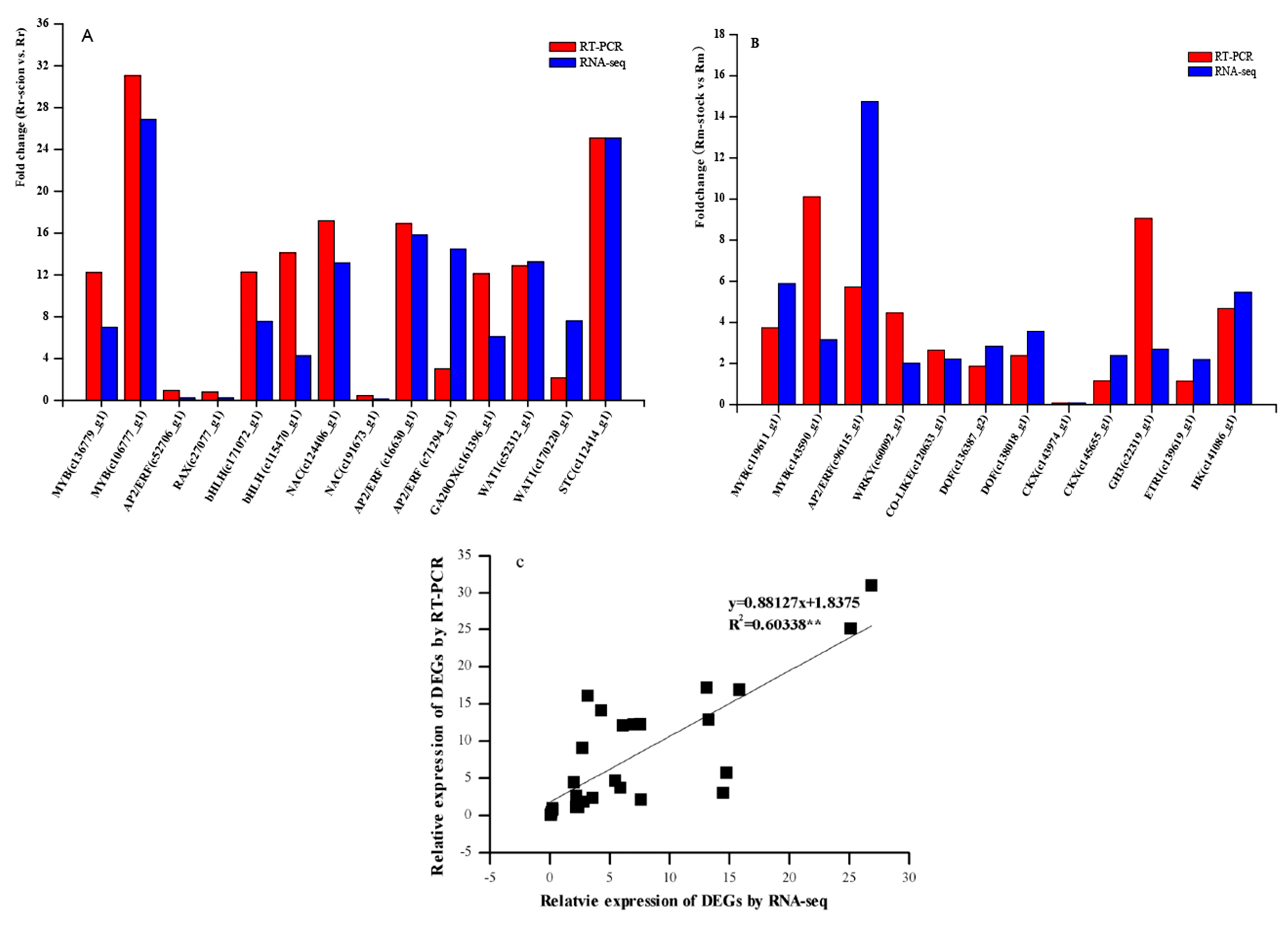
3.7. qRT-PCR Validation of Differentially Expressed Transcripts from RNA-seq
4. Discussion
4.1. Stem Secondary Growth Response for Grafted R. Multiflora ‘Innermis’/R. Rugosa ‘Rosea’ Plants
4.2. DEGs in Key Pathway about Lignin and Cellulose Biosynthesis in the Scion and Rootstock
4.3. DEGs in Response to Phytohormone Signal Transduction in the Scion and Rootstock
4.4. Protein Kinases Are Related to Stem Vascular Development in the Scion and Rootstock
4.5. Transcription Factors Involved in Stem Secondary Growth in the Scion and Rootstock
4.6. Transporter Proteins Are Important for Stem Secondary Growth in the Scion and Rootstock
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, S.G. Production of high quality vegetable seedling grafts. Acta Hortic. 2007. [Google Scholar] [CrossRef]
- Sakata, Y.; Ohara, T.; Sugiyama, M. The history and present state of the grafting of cucurbitaceous vegetables in Japan. Acta Hortic. 2007, 731, 159–170. [Google Scholar] [CrossRef]
- Tandonnet, J.P.; Cookson, S.J.; Vivin, P.; Ollat, N. Scion genotype controls biomass allocation and root development in grafted grapevine. Aust. J. Grape Wine Res. 2010, 16, 290–300. [Google Scholar] [CrossRef]
- Rufato, A.; Tomaz, Z.; Gonçalves, C.; Silveira, S.; Rufato, L.; Gomes, F. The effect of intergrafting on fruit quality and productivity of peach tree ‘Jubileu’. Acta Hortic. 2010, 872, 349–352. [Google Scholar] [CrossRef]
- Marguerit, E.; Brendel, O.; Lebon, E.; Van Leeuwen, C.; Ollat, N. Rootstock control of scion transpiration and its acclimation to water deficit are controlled by different genes. New Phytol. 2012, 194, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Rouphael, Y.; Cardarelli, M.; Rea, E.; Colla, G. Improving melon and cucumber photosynthetic activity, mineral composition, and growth performance under salinity stress by grafting onto Cucurbita hybrid rootstocks. Photosynthetica 2012, 50, 180–188. [Google Scholar] [CrossRef]
- Sehr, E.M.; Agusti, J.; Lehner, R.; Farmer, E.E.; Schwarz, M.; Greb, T. Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 2010, 63, 811–822. [Google Scholar] [CrossRef]
- Cano-Delgado, A.; Lee, J.Y.; Demura, T. Regulatory mechanisms for specification and patterning of plant vascular tissues. Annu. Rev. Cell Dev. Biol. 2010, 26, 605–637. [Google Scholar] [CrossRef]
- Olmstead, M.A.; Lang, N.S.; Ewers, F.W.; Owens, S.A. Xylem vessel anatomy of sweet cherries grafted onto dwarfing and nondwarfing rootstocks. J. Am. Soc. Hortic. Sci. 2006, 131, 577–585. [Google Scholar] [CrossRef]
- Lauri, P.E.; Maguylo, K.; Trottier, C. Architecture and size relations: An essay on the apple (Malus X domestica, Rosaceae) tree. Am. J. Bot. 2006, 93, 357–368. [Google Scholar] [CrossRef]
- Cookson, S.J.; Ollat, N. Grafting with rootstocks induces extensive transcriptional re-programming in the shoot apical meristem of grapevine. BMC Plant Biol. 2013, 13, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.J.; Makalowska, I.; Altman, N.; Fazio, G.; Praul, C.; Maximova, S.N.; Crassweller, R.M.; Travis, J.W.; McNellis, T.W. Rootstock-regulated gene expression patterns in apple tree scions. Tree Genet. Genomes 2009, 6, 57–72. [Google Scholar] [CrossRef]
- Liu, X.Y.; Li, J.; Liu, M.M.; Yao, Q.; Chen, J.Z. Transcriptome profiling to understand the effect of citrus rootstocks on the growth of ‘Shatangju’ mandarin. PLoS ONE 2017, 12, e0169897. [Google Scholar] [CrossRef] [PubMed]
- Chitarra, W.; Perrone, I.; Avanzato, C.G.; Minio, A.; Boccacci, P.; Santini, D.; Gilardi, G.; Siciliano, I.; Gullino, M.L.; Delledonne, M.; et al. Grapevine grafting: Scion transcript profiling and defense-related metabolites induced by rootstocks. Front. Plant Sci. 2017, 8, 654–669. [Google Scholar] [CrossRef] [PubMed]
- Agbaria, H.; Heuer, B.; Zieslin, N. Shoot-root interaction effects on nitrate reductase and glutamine synthetase activities in rose (Rosa × hybrida cvs. Ilseta and Mercedes) graftlings. J. Plant Physiol. 1996, 149, 559–563. [Google Scholar] [CrossRef]
- Dieleman, J.A.; Verstappen, F.W.A.; Kuiper, D. Bud break and cytokinin concentration in bleeding sap of Rosa hybrida as affected by the genotype of the rootstock. J. Plant Physiol. 1998, 152, 468–472. [Google Scholar] [CrossRef]
- Richer, C.; Rioux, J.; Lamy, M.; Drapeau, R. Evaluation of winter tolerance of 10 roses of Explorer Series (MC) propagated by micropropagation, cutting and grafting on Rosa multiflora Thunb. Can. J. Plant Sci. 2005, 85, 693–701. [Google Scholar] [CrossRef]
- Kucukoglu, M.; Nilsson, J.; Zheng, B.; Chaabouni, S.; Nilsson, O. WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 2017, 215, 642–657. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2009, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hu, N.; Qian, L.; Hu, Y.; Shou, J.-Z.; Wang, C.; Giffen, C.; Wang, Q.-H.; Wang, Y.; Goldstein, A.M.; Emmert-Buck, M.; et al. Quantitative real-time RT-PCR validation of differential mRNA expression of SPARC, FADD, Fascin, COL7A1, CK4, TGM3, ECM1, PPL and EVPLin esophageal squamous cell carcinoma. BMC Cancer 2006, 6. [Google Scholar] [CrossRef]
- Liu, N.; Yang, J.; Fu, X.; Zhang, L.; Tang, K.; Guy, K.M.; Hu, Z.; Guo, S.; Xu, Y.; Zhang, M. Genome-wide identification and comparative analysis of grafting-responsive mRNA in watermelon grafted onto bottle gourd and squash rootstocks by high-throughput sequencing. Mol. Genet. Genom. 2016, 291, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.J.; Rytter, J.; Detwiler, E.A.; Travis, J.W.; McNellis, T.W. Rootstock effects on gene expression patterns in apple tree scions. Plant Mol. Biol. 2003, 53, 493–511. [Google Scholar] [CrossRef]
- Zhao, C.; Craig, J.C.; Petzold, H.E.; Dickerman, A.W.; Beers, E.P. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 2005, 138, 803–818. [Google Scholar] [CrossRef]
- Shigeto, J.; Kiyonaga, Y.; Fujita, K.; Kondo, R.; Tsutsumi, Y. Putative cationic cell-wall-bound peroxidase homologues in Arabidopsis, AtPrx2, AtPrx25, and AtPrx71, are involved in lignification. J. Agric. Food Chem. 2013, 61, 3781–3788. [Google Scholar] [CrossRef]
- Goujon, T.; Sibout, R.; Eudes, A.; MacKay, J.; Jouanin, L. Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol. Biochem. 2003, 41, 677–687. [Google Scholar] [CrossRef]
- Thevenin, J.; Pollet, B.; Letarnec, B.; Saulnier, L.; Gissot, L.; Maia-Grondard, A.; Lapierre, C.; Jouanin, L. The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol. Plant 2011, 4, 70–82. [Google Scholar] [CrossRef]
- Van Acker, R.; Vanholme, R.; Storme, V.; Mortimer, J.C.; Dupree, P.; Boerjan, W. Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol. Biofuels 2013, 6, 46–63. [Google Scholar] [CrossRef]
- Taylor, N.G. Cellulose biosynthesis and deposition in higher plants. New Phytol. 2008, 178, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Herth, W. Arrays of plasma-membrane “rosettes” involved in cellulose microfibril formation of Spirogyra. Planta 1983, 159, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Van Sandt, V.S.; Suslov, D.; Verbelen, J.-P.; Vissenberg, K. Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. 2007, 100, 1467–1473. [Google Scholar] [CrossRef]
- Maris, A.; Suslov, D.; Fry, S.C.; Verbelen, J.-P.; Vissenberg, K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 2009, 60, 3959–3972. [Google Scholar] [CrossRef] [PubMed]
- Sorce, C.; Giovannelli, A.; Sebastiani, L.; Anfodillo, T. Hormonal signals involved in the regulation of cambial activity, xylogenesis and vessel patterning in trees. Plant Cell Rep. 2013, 32, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Cassan-Wang, H.; Goue, N.; Saidi, M.N.; Legay, S.; Sivadon, P.; Goffner, D.; Grima-Pettenati, J. Identification of novel transcription factors regulating secondary cell wall formation in Arabidopsis. Front. Plant Sci. 2013, 4, 189. [Google Scholar] [CrossRef] [PubMed]
- Hardtke, C.S.; Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998, 17, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Hamann, T.; Benkova, E.; Baurle, I.; Kientz, M.; Jurgens, G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002, 16, 1610–1615. [Google Scholar] [CrossRef]
- Miyashima, S.; Sebastian, J.; Lee, J.Y.; Helariutta, Y. Stem cell function during plant vascular development. EMBO J. 2013, 32, 178–193. [Google Scholar] [CrossRef]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef]
- Köllmer, I.; Novák, O.; Strnad, M.; Schmülling, T.; Werner, T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX 7) from A rabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 2014, 78, 359–371. [Google Scholar] [CrossRef]
- Bishopp, A.; Help, H.; El-Showk, S.; Weijers, D.; Scheres, B.; Friml, J.; Benková, E.; Mähönen, A.P.; Helariutta, Y. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 2011, 21, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Bishopp, A.; Lehesranta, S.; Vatén, A.; Help, H.; El-Showk, S.; Scheres, B.; Helariutta, K.; Mähönen, A.P.; Sakakibara, H.; Helariutta, Y. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 2011, 21, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Gou, X.; He, K.; Yang, H.; Yuan, T.; Lin, H.; Clouse, S.D.; Li, J. Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genom. 2010, 11, 19. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xun, Q.; Guo, Y.; Zhang, J.; Cheng, K.; Shi, T.; He, K.; Hou, S.; Gou, X.; Li, J. Genome-wide expression pattern analyses of the arabidopsis leucine-rich repeat receptor-like kinases. Mol. Plant 2016, 9, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Bryan, A.C.; Obaidi, A.; Wierzba, M.; Tax, F.E. XYLEM INTERMIXED WITH PHLOEM1, a leucine-rich repeat receptor-like kinase required for stem growth and vascular development in Arabidopsis thaliana. Planta 2012, 235, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar] [CrossRef]
- Li, W.; Tian, Z.; Yu, D. WRKY13 acts in stem development in Arabidopsis thaliana. Plant Sci. 2015, 236, 205–213. [Google Scholar] [CrossRef]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef]
- Legay, S.; Sivadon, P.; Blervacq, A.S.; Pavy, N.; Baghdady, A.; Tremblay, L.; Levasseur, C.; Ladouce, N.; Lapierre, C.; Seguin, A.; et al. EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol. 2010, 188, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Ye, Z.H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010, 15, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, A.; Mansfield, S.D.; Hall, H.C.; Douglas, C.J.; Ellis, B.E. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol. 2010, 154, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288–306. [Google Scholar] [CrossRef]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef]
- Larsen, B.; Xu, D.; Halkier, B.A.; Nour-Eldin, H.H. Advances in methods for identification and characterization of plant transporter function. J. Exp. Bot. 2017, 68, 4045–4056. [Google Scholar] [CrossRef]
- Kaneda, M.; Schuetz, M.; Lin, B.S.; Chanis, C.; Hamberger, B.; Western, T.L.; Ehlting, J.; Samuels, A.L. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J. Exp. Bot. 2011, 62, 2063–2077. [Google Scholar] [CrossRef]
- Ko, D.; Kang, J.; Kiba, T.; Park, J.; Kojima, M.; Do, J.; Kim, K.Y.; Kwon, M.; Endler, A.; Song, W.Y.; et al. Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc. Natl. Acad. Sci. USA 2014, 111, 7150–7155. [Google Scholar] [CrossRef]
- Casu, R.E.; Grof, C.P.; Rae, A.L.; McIntyre, C.L.; Dimmock, C.M.; Manners, J.M. Identification of a novel sugar transporter homologue strongly expressed in maturing stem vascular tissues of sugarcane by expressed sequence tag and microarray analysis. Plant Mol. Biol. 2003, 52, 371–386. [Google Scholar] [CrossRef]
- Le Hir, R.; Spinner, L.; Klemens, P.A.; Chakraborti, D.; de Marco, F.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Gery, C.; et al. Disruption of the Sugar Transporters AtSWEET11 and AtSWEET12 Affects Vascular Development and Freezing Tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Gong, Z.; Zhu, J.K. Abscisic acid-mediated epigenetic processes in plant development and stress responses. J. Integr. Plant Biol. 2008, 50, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Singla-Pareek, S.L.; Pareek, A. Engineering abiotic stress response in plants for biomass production. J. Biol. Chem. 2018, 293, 5035–5043. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hu, Q.; Luo, S.; Li, Q.; Yang, X.; Wang, X.; Wang, S. Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front. Plant Sci. 2015, 6, 388. [Google Scholar] [CrossRef]
- Shafi, A.; Chauhan, R.; Gill, T.; Swarnkar, M.K.; Sreenivasulu, Y.; Kumar, S.; Kumar, N.; Shankar, R.; Ahuja, P.S.; Singh, A.K. Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol. Biol. 2015, 87, 615–631. [Google Scholar] [CrossRef]
- Moura, J.C.; Bonine, C.A.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]




| Sample | Total Reads | Clean Reads | Clean Base | Error (%) | Q30 |
|---|---|---|---|---|---|
| Rr | 199,710,618 | 184,953,434 | 27,743,015,100 | 0.02 | 96.82 |
| Rr_scion | 198,497,050 | 184,574,214 | 27,686,132,100 | 0.02 | 96.75 |
| Rm | 199,381,448 | 184,304, 392 | 27,645,658,800 | 0.02 | 96.32 |
| Rm_stock | 196,825,416 | 180,846,594 | 27,126,989,100 | 0.02 | 96.70 |
| Sample | Transcripts | Unigene | Aligned Reads | GC (%) | Min (nt) | Max (nt) | Mean (nt) | N50 | N90 |
|---|---|---|---|---|---|---|---|---|---|
| Rr | 154,572 | 136,293 | 174,417,164 | 43.80 | 201 | 93521 | 816 | 1069 | 286 |
| Rm | 184,747 | 108,651 | 172,594,386 | 43.24 | 201 | 100409 | 859 | 1220 | 290 |
| Rr_scion vs. Rr | |||||
|---|---|---|---|---|---|
| Contig | Grafting | Non-Grafting | Fold Change | Log2 Fold Change | Gene Description |
| c161396_g1 | 168.94 | 27.72 | 6.09 | 2.619 | gibberellin 20 oxidase |
| c150592_g1 | 25.44 | 6.36 | 3.99 | 2.00 | gibberellin 20 oxidase |
| c137188_g1 | 10.864 | 42.50 | 0.26534 | −1.97 | gibberellin 20 oxidase |
| c139468_g3 | 29.924 | 4.65 | 6.43 | 2.69 | gibberellin 20 oxidase |
| c169466_g1 | 1202.19 | 329 | 0.36 | −1.45 | Gasa4-like protein |
| c145719_g1 | 29.71 | 7.28 | 4.083 | 2.03 | gibberellin 2-β-dioxygenase |
| c144986_g1 | 2.30 | 9.58 | 0.24 | −2.06 | auxin transporter-like protein |
| c80584_g1 | 1075.22 | 6844.72 | 0.16 | −2.67 | auxin transporter-like protein |
| c159798_g7 | 932.27 | 399.73 | 2.33 | 1.22 | auxin binding protein |
| c111040_g1 | 213.30 | 43.45 | 4.91 | 2.30 | auxin binding protein |
| c170220_g1 | 315.98 | 41.63 | 7.59 | 2.92 | wall are thin1-related protein |
| c191576_g1 | 32.29 | 7.46 | 4.33 | 2.11 | wall are thin1-related protein |
| c152340_g1 | 69.88 | 30.49 | 2.29 | 1.197 | wall are thin1-related protein |
| c81300_g1 | 747.41 | 296.19 | 2.52 | 1.34 | wall are thin1-related protein |
| c52312_g1 | 12.708 | 0.96 | 13.25 | 3.73 | wall are thin1-related protein |
| c114045_g1 | 99.80 | 40.19 | 2.48 | 1.31 | auxin-induced protein |
| c122006_g1 | 3.03 | 27.71 | 0.11 | −3.19 | auxin-induced protein |
| c142151_g1 | 104.22 | 35.71 | 2.92 | 1.55 | auxin-induced in root cultures protein |
| c142151_g2 | 36.64 | 13.81 | 2.65 | 1.41 | auxin-induced in root cultures protein |
| c162170_g1 | 24.58 | 76.66 | 0.32 | −1.64 | auxin-induced in root cultures protein |
| c136754_g1 | 4.49 | 0.33 | 13.757 | 3.78 | indole-3-acetic acid-induced protein |
| c134374_g1 | 13.33 | 30.53 | 0.44 | −1.195 | auxin-responsive protein |
| c21225_g1 | 69.86 | 209.07 | 0.33 | −1.58 | auxin-responsive protein |
| c146441_g2 | 4.44 | 16.22 | 0.27 | −1.87 | auxin efflux carrier component |
| c140539_g1 | 105.48 | 332.63 | 0.32 | −1.66 | abscisic acid 8′-hydroxylase |
| c158586_g1 | 312.40 | 56.48 | 5.53 | 2.47 | abscisic acid 8′-hydroxylase |
| c122806_g1 | 35.01 | 135.65 | 0.26 | −1.95 | abscisic acid 8′-hydroxylase |
| c150342_g1 | 330.36 | 24.20 | 13.65 | 3.77 | abscisic acid receptor |
| c192915_g1 | 29.7 | 7.89 | 3.77 | 1.91 | abscisic acid receptor |
| c161435_g1 | 326.73 | 52.34 | 6.24 | 2.64 | abscisic acid receptor |
| c140884_g1 | 1942.26 | 69017 | 0.28 | −1.83 | abscisic acid insentive protein |
| c120991_g2 | 8.082 | 25.33 | 0.32 | −1.65 | abscisic acid insentive protein |
| c132075_g1 | 170.47 | 83.47 | 2.04 | 1.03 | cytokinin dehydrogenase |
| c158300_g2 | 160.80 | 59.70 | 2.69 | 1.4 | two-component response regulator |
| c152947_g1 | 215.33 | 106.11 | 2.03 | 1.02 | ethylene receptor |
| c145980_g1 | 267.47 | 130.48 | 2.05 | 1.04 | ethylene receptor |
| c150029_g1 | 76.21 | 17.624 | 4.32 | 2.11 | reversion-to-ethylene sensitivity |
| Rm_stock vs. Rm | |||||
| c143974_g1 | 9.60 | 135.88 | 0.071 | −3.82 | cytokinin dehydrogenase |
| c145655_g1 | 276.45 | 116.26 | 2.38 | 1.25 | cytokinin dehydrogenase |
| c22319_g1 | 27.75 | 10.31 | 2.69 | 1.43 | auxin-responsive GH3 gene family |
| c139619_g1 | 953.89 | 435.35 | 2.19 | 1.13 | ethylene receptor |
| C140747_g1 | 375.42 | 147.53 | 2.54 | 1.35 | EIN3-binding F-box protein |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.-s.; Hu, R.-y.; Lv, F.-l.; Yang, Y.-f.; Tang, Z.-m.; Zheng, G.-s.; Li, J.-b.; Tian, H.; Xu, Y.; Li, S.-f. Comparative Transcriptome Analysis Reveals Stem Secondary Growth of Grafted Rosa rugosa ‘Rosea’ Scion and R. multiflora ‘Innermis’ Rootstock. Genes 2020, 11, 228. https://doi.org/10.3390/genes11020228
Sun J-s, Hu R-y, Lv F-l, Yang Y-f, Tang Z-m, Zheng G-s, Li J-b, Tian H, Xu Y, Li S-f. Comparative Transcriptome Analysis Reveals Stem Secondary Growth of Grafted Rosa rugosa ‘Rosea’ Scion and R. multiflora ‘Innermis’ Rootstock. Genes. 2020; 11(2):228. https://doi.org/10.3390/genes11020228
Chicago/Turabian StyleSun, Jing-shuang, Rui-yang Hu, Fu-ling Lv, Yan-fang Yang, Zhi-min Tang, Guang-shun Zheng, Jian-bo Li, Hua Tian, Yan Xu, and Shao-feng Li. 2020. "Comparative Transcriptome Analysis Reveals Stem Secondary Growth of Grafted Rosa rugosa ‘Rosea’ Scion and R. multiflora ‘Innermis’ Rootstock" Genes 11, no. 2: 228. https://doi.org/10.3390/genes11020228
APA StyleSun, J.-s., Hu, R.-y., Lv, F.-l., Yang, Y.-f., Tang, Z.-m., Zheng, G.-s., Li, J.-b., Tian, H., Xu, Y., & Li, S.-f. (2020). Comparative Transcriptome Analysis Reveals Stem Secondary Growth of Grafted Rosa rugosa ‘Rosea’ Scion and R. multiflora ‘Innermis’ Rootstock. Genes, 11(2), 228. https://doi.org/10.3390/genes11020228




