DNA Helicase-SSB Interactions Critical to the Regression and Restart of Stalled DNA Replication Forks in Escherichia coli
Abstract
1. Introduction
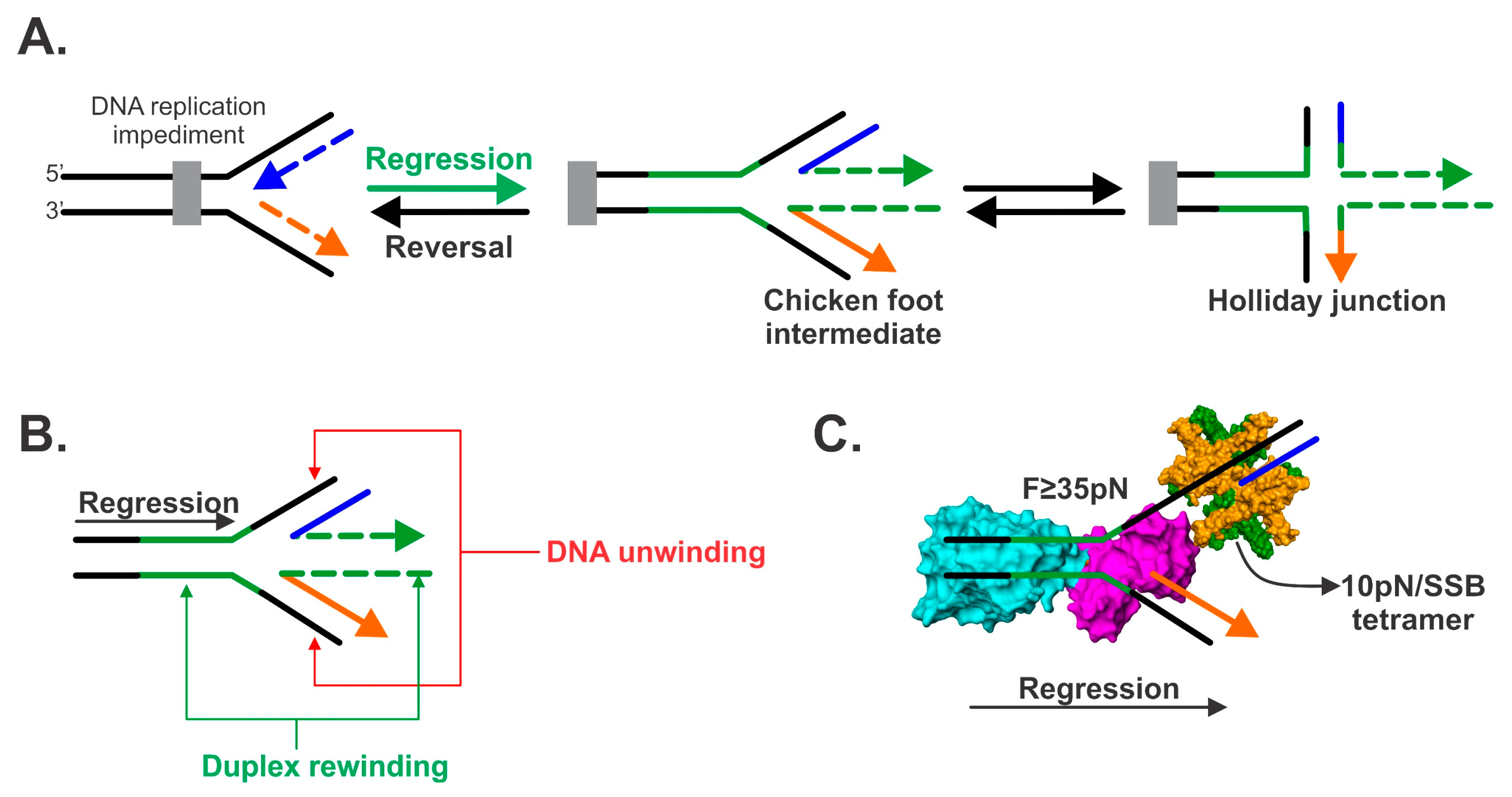
2. Fork Regression Defined
3. The Protein Players
3.1. SSB—The Mediator of DNA Transactions at Forks
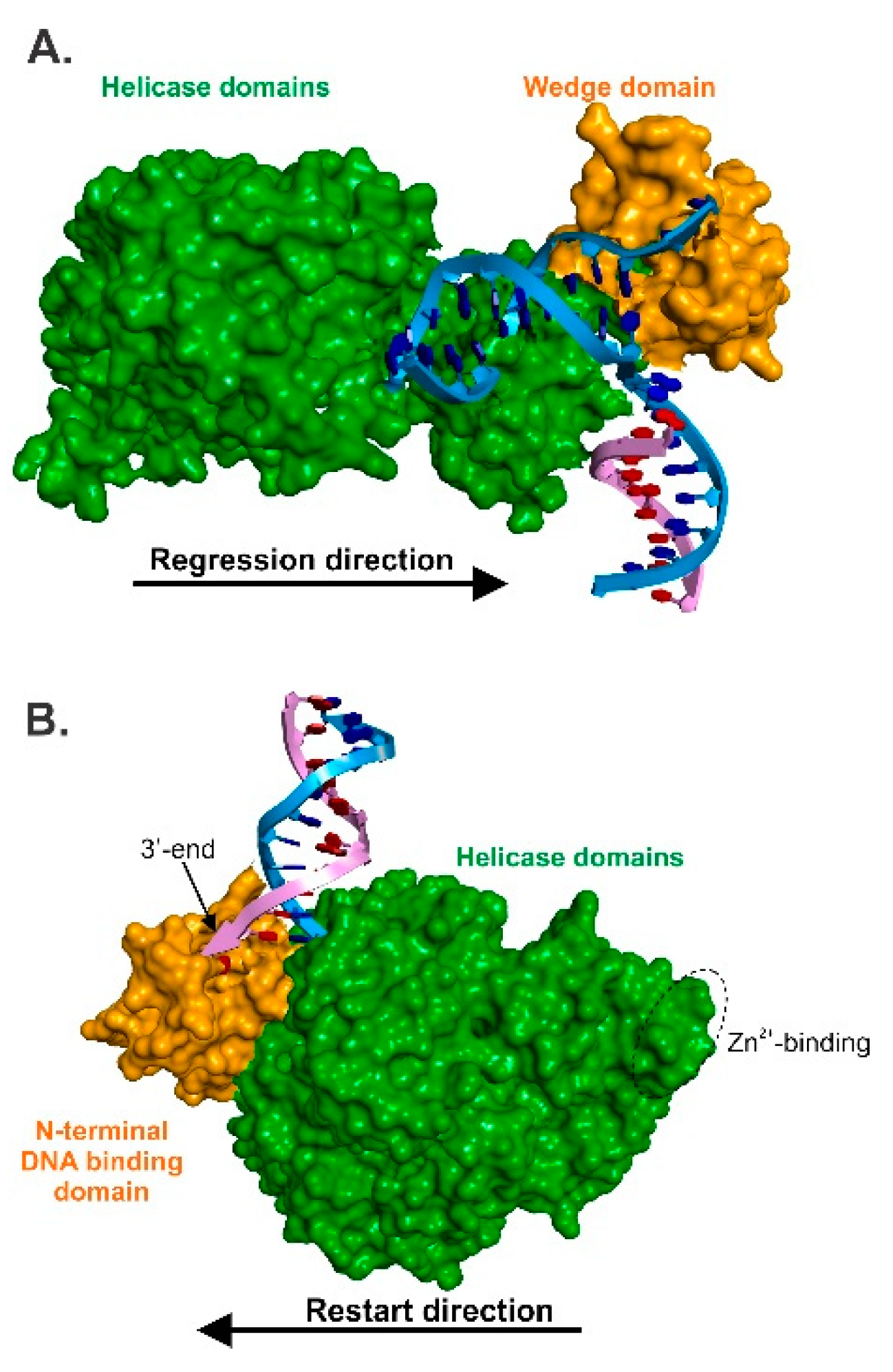
3.2. RecG—The Regression Beast
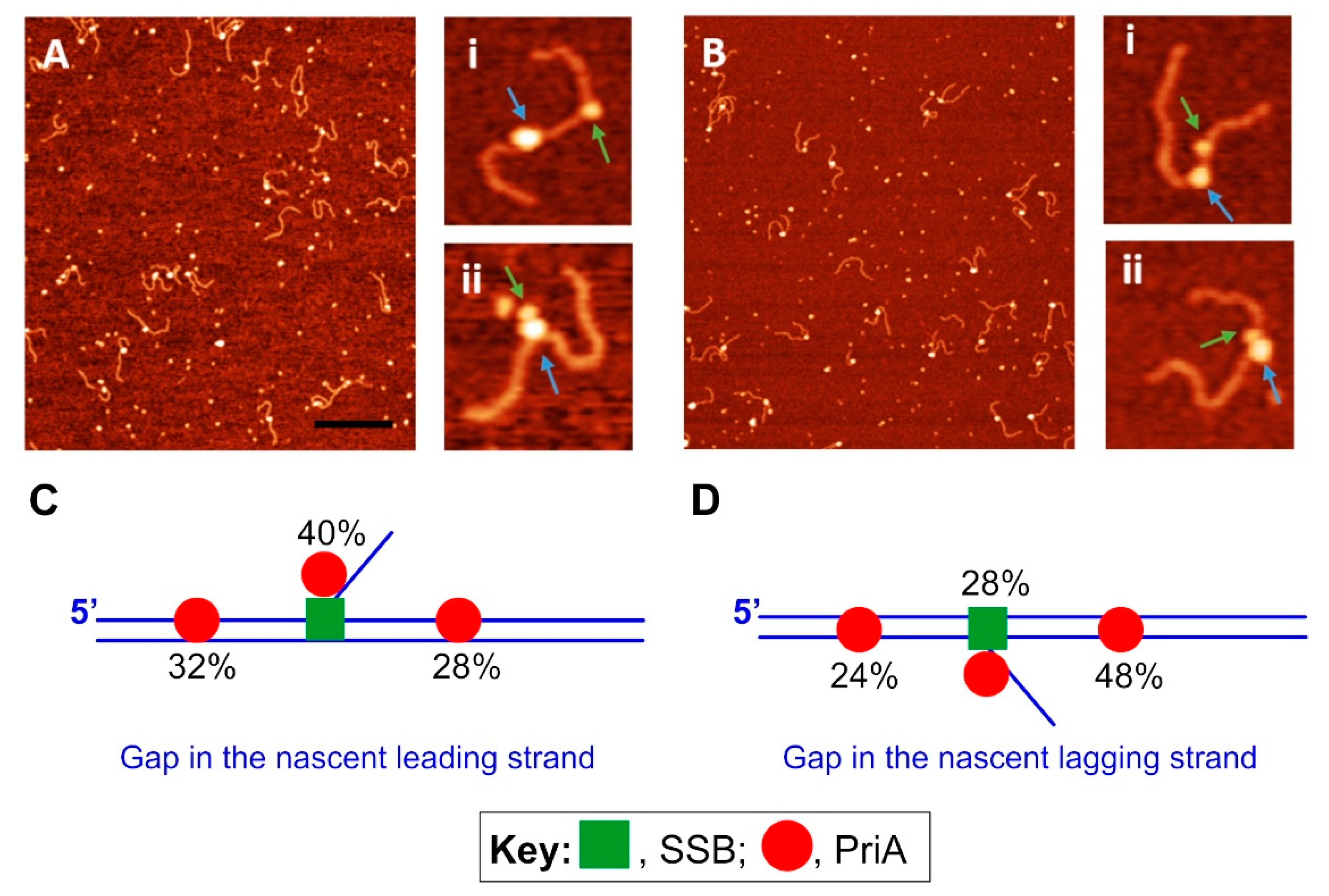
3.3. PriA—The Restart Specialist
4. SSB-DNA Helicase Interactions during Loading
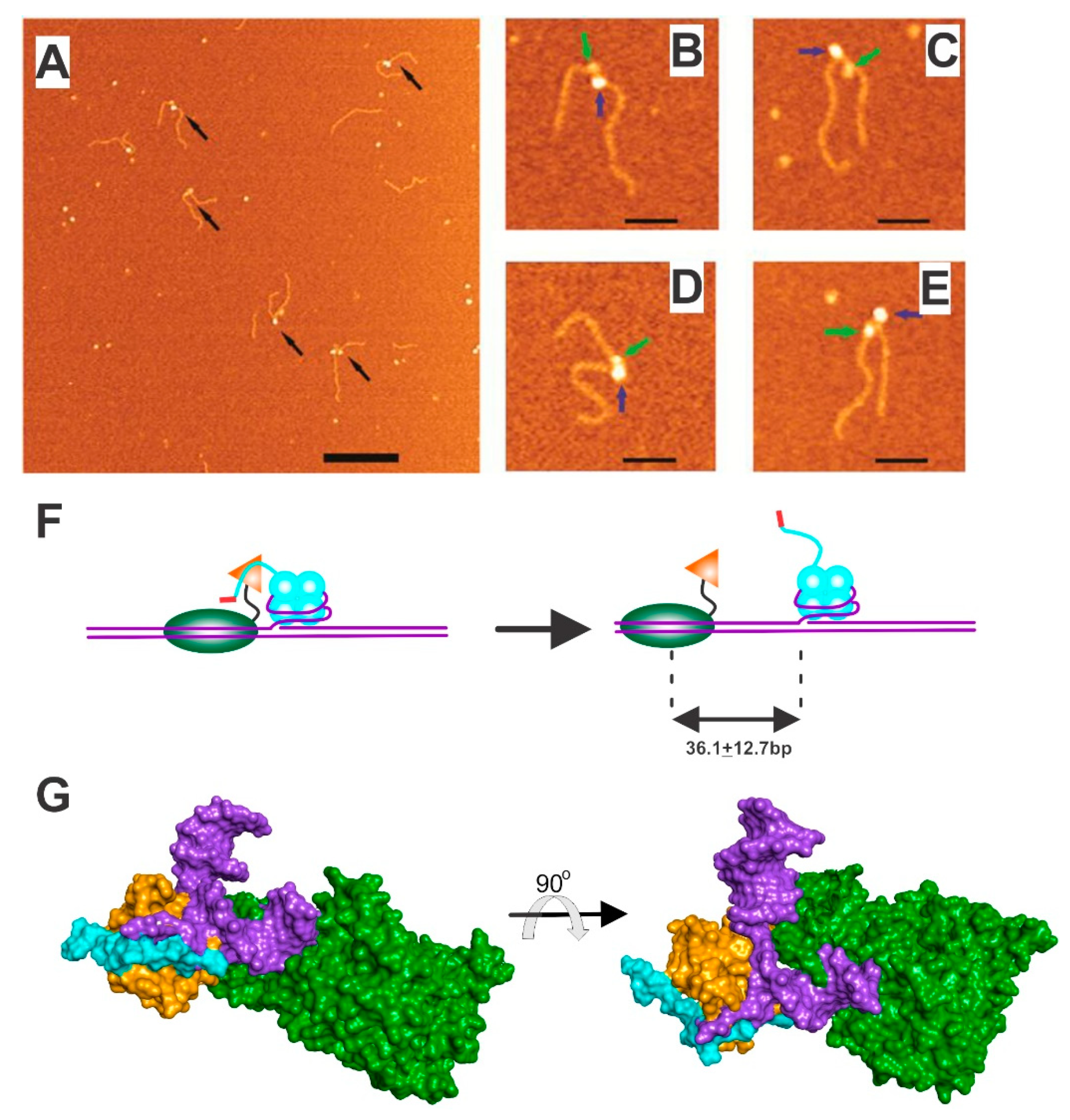
4.1. SSB-RecG
4.2. SSB-PriA
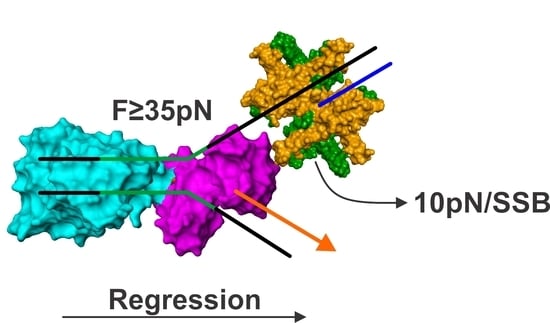
5. The Mechanics of Fork Regression by RecG
6. Summary
Funding
Conflicts of Interest
References
- Kogoma, T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Boil. Rev. 1997, 61, 212–238. [Google Scholar] [CrossRef]
- Kuzminov, A. Recombinational Repair of DNA Damage in Escherichia coli and Bacteriophage λ. Microbiol. Mol. Boil. Rev. 1999, 63, 751–813. [Google Scholar] [CrossRef]
- Kowalczykowski, S.C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000, 25, 156–165. [Google Scholar] [CrossRef]
- Cox, M. Recombinational DNA Repair of Damaged Replication Forks inEscherichia coli: Questions. Annu. Rev. Genet. 2001, 35, 53–82. [Google Scholar] [CrossRef]
- Kreuzer, K.N. Interplay between Dna Replication and Recombination in Prokaryotes. Annu. Rev. Microbiol. 2005, 59, 43–67. [Google Scholar] [CrossRef]
- McGlynn, P.; Lloyd, R.G. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Boil. 2002, 3, 859–870. [Google Scholar] [CrossRef]
- Cox, M.M.; Goodman, M.F.; Kreuzer, K.N.; Sherratt, D.J.; Sandler, S.J.; Marians, K.J. The importance of repairing stalled replication forks. Nature 2000, 404, 37–41. [Google Scholar] [CrossRef]
- McGlynn, P.; Lloyd, R.G. Replicating past lesions in DNA. Mol. Cell 2002, 10, 700–701. [Google Scholar] [CrossRef]
- Marians, K.J. Mechanisms of replication fork restart inEscherichia coli. Philos. Trans. R. Soc. B: Boil. Sci. 2004, 359, 71–77. [Google Scholar] [CrossRef][Green Version]
- Mirkin, E.V.; Mirkin, S.M. Replication Fork Stalling at Natural Impediments. Microbiol. Mol. Boil. Rev. 2007, 71, 13–35. [Google Scholar] [CrossRef]
- Voineagu, I.; Narayanan, V.; Lobachev, K.S.; Mirkin, S.M. Replication stalling at unstable inverted repeats: Interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 9936–9941. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.K.; Guy, C.P.; Yeeles, J.T.P.; Atkinson, J.; Bell, H.; Lloyd, R.G.; Marians, K.J.; McGlynn, P. Protein–DNA complexes are the primary sources of replication fork pausing in Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 7252–7257. [Google Scholar] [CrossRef]
- Michel, B.; Grompone, G.; Florès, M.-J.; Bidnenko, V. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. USA 2004, 101, 12783–12788. [Google Scholar] [CrossRef]
- McGlynn, P.; Lloyd, R.G. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 2002, 18, 413–419. [Google Scholar] [CrossRef]
- Marians, K.J. PriA-directed replication fork restart in Escherichia coli. Trends Biochem. Sci. 2000, 25, 185–189. [Google Scholar] [CrossRef]
- Marians, K.J. Replication and recombination intersect. Curr. Opin. Genet. Dev. 2000, 10, 151–156. [Google Scholar] [CrossRef]
- Courcelle, J.; Hanawalt, P.C. RecA-Dependent Recovery of Arrested DNA Replication Forks. Annu. Rev. Genet. 2003, 37, 611–646. [Google Scholar] [CrossRef]
- Henderson, M.L.; Kreuzer, K.N. Functions that Protect Escherichia coli from Tightly Bound DNA-Protein Complexes Created by Mutant EcoRII Methyltransferase. PLoS ONE 2015, 10, e0128092. [Google Scholar] [CrossRef]
- Heller, R.C.; Marians, K.J. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 2006, 439, 557–562. [Google Scholar] [CrossRef]
- Heller, R.C.; Marians, K.J. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Boil. 2006, 7, 932–943. [Google Scholar] [CrossRef]
- Lusetti, S.L.; Cox, M. The Bacterial RecA Protein and the Recombinational DNA Repair of Stalled Replication Forks. Annu. Rev. Biochem. 2002, 71, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Heller, R.C.; Marians, K.J. Unwinding of the Nascent Lagging Strand by Rep and PriA Enables the Direct Restart of Stalled Replication Forks. J. Boil. Chem. 2005, 280, 34143–34151. [Google Scholar] [CrossRef] [PubMed]
- Kuzminov, A. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 1995, 16, 373–384. [Google Scholar] [CrossRef]
- Manosas, M.; Perumal, S.K.; Bianco, P.R.; Ritort, F.; Benkovic, S.J.; Croquette, V. RecG and UvsW catalyse robust DNA rewinding critical for stalled DNA replication fork rescue. Nat. Commun. 2013, 4, 2368. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.R. Stalled replication fork rescue requires a novel DNA helicase. Methods 2016, 108, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Masai, H.; Asai, T.; Kubota, Y.; Arai, K.; Kogoma, T. Escherichia coli PriA protein is essential for inducible and constitutive stable DNA replication. EMBO J. 1994, 13, 5338–5345. [Google Scholar] [CrossRef] [PubMed]
- Gabbai, C.B.; Marians, K.J. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair 2010, 9, 202–209. [Google Scholar] [CrossRef]
- Sandler, S.J.; Marians, K.J. Role of PriA in Replication Fork Reactivation inEscherichia coli. J. Bacteriol. 2000, 182, 9–13. [Google Scholar] [CrossRef]
- Lewis, J.; Spenkelink, L.M.; Jergic, S.; Wood, E.A.; Monachino, E.; Horan, N.P.; Duderstadt, K.E.; Cox, M.M.; Robinson, A.; Dixon, N.E.; et al. Single-molecule visualization of fast polymerase turnover in the bacterial replisome. eLife 2017, 6, 6. [Google Scholar] [CrossRef]
- Jeiranian, H.A.; Schalow, B.J.; Courcelle, C.T.; Courcelle, J. Fate of the replisome following arrest by UV-induced DNA damage in Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 11421–11426. [Google Scholar] [CrossRef]
- Yeeles, J.T.P.; Marians, K.J. The Escherichia coli Replisome Is Inherently DNA Damage Tolerant. Science 2011, 334, 235–238. [Google Scholar] [CrossRef]
- Postow, L.; Crisona, N.J.; Peter, B.J.; Hardy, C.D.; Cozzarelli, N.R. Topological challenges to DNA replication: Conformations at the fork. Proc. Natl. Acad. Sci. USA 2001, 98, 8219–8226. [Google Scholar] [CrossRef] [PubMed]
- Postow, L.; Ullsperger, C.; Keller, R.W.; Bustamante, C.; Vologodskii, A.V.; Cozzarelli, N.R. Positive Torsional Strain Causes the Formation of a Four-way Junction at Replication Forks. J. Boil. Chem. 2000, 276, 2790–2796. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; Lloyd, R.G. Modulation of RNA Polymerase by (p)ppGpp Reveals a RecG-Dependent Mechanism for Replication Fork Progression. Cell 2000, 101, 35–45. [Google Scholar] [CrossRef]
- Robu, M.E.; Inman, R.B.; Cox, M.M. Situational repair of replication forks: Roles of RecG and RecA proteins. J. Biol. Chem. 2004, 279, 10973–10981. [Google Scholar] [CrossRef]
- Robu, M.E.; Inman, R.B.; Cox, M. RecA protein promotes the regression of stalled replication forks in vitro. Proc. Natl. Acad. Sci. USA 2001, 98, 8211–8218. [Google Scholar] [CrossRef]
- Seigneur, M.; Bidnenko, V.; Ehrlich, S.; Michel, B. RuvAB Acts at Arrested Replication Forks. Cell 1998, 95, 419–430. [Google Scholar] [CrossRef]
- Gupta, S.; Yeeles, J.T.P.; Marians, K.J. Regression of Replication Forks Stalled by Leading-strand Template Damage. J. Boil. Chem. 2014, 289, 28388–28398. [Google Scholar] [CrossRef]
- Gupta, S.; Yeeles, J.T.P.; Marians, K.J. Regression of Replication Forks Stalled by Leading-strand Template Damage I-Both Recg and RuvAB Catalyze Regression, but RuvC Cleaves the Holliday Junctions FORMED BY RecG Preferentially. J. Biol. Chem. 2014, 289, 28376–28387. [Google Scholar] [CrossRef]
- Rosenberg, M.; Echols, H. Differential recognition of ultraviolet lesions by RecA protein. Possible mechanism for preferential targeting of SOS mutagenesis to (6-4) dipyrimidine sites. J. Boil. Chem. 1990, 265, 20641–20645. [Google Scholar]
- Wahab, S.A.; Choi, M.; Bianco, P.R. Characterization of the ATPase Activity of RecG and RuvAB Proteins on Model Fork Structures Reveals Insight into Stalled DNA Replication Fork Repair*. J. Boil. Chem. 2013, 288, 26397–26409. [Google Scholar] [CrossRef] [PubMed]
- Buss, J.A.; Kimura, Y.; Bianco, P.R. RecG interacts directly with SSB: Implications for stalled replication fork regression. Nucleic Acids Res. 2008, 36, 7029–7042. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Tan, H.Y.; Bianco, P.R.; Lyubchenko, Y.L. Remodeling of RecG Helicase at the DNA Replication Fork by SSB Protein. Sci. Rep. 2015, 5, 9625. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.R.; Lyubchenko, Y.L. SSB and the RecG DNA helicase: An intimate association to rescue a stalled replication fork. Protein Sci. 2017, 26, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Marians, K.J. Purification and Characterization of DnaC810, a Primosomal Protein Capable of Bypassing PriA Function. J. Boil. Chem. 2000, 275, 8196–8205. [Google Scholar] [CrossRef]
- Yu, C.; Tan, H.Y.; Choi, M.; Stanenas, A.J.; Byrd, A.; Raney, K.D.; Cohan, C.S.; Bianco, P.R. SSB binds to the RecG and PriA helicases in vivo in the absence of DNA. Genes Cells 2016, 21, 163–184. [Google Scholar] [CrossRef]
- Cadman, C.J.; McGlynn, P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004, 32, 6378–6387. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Z.; Bianco, P.R.; Lyubchenko, Y.L. AFM characterization of the interaction of PriA helicase with stalled DNA replication forks. J. Biol. Chem. 2020, in press. [Google Scholar] [CrossRef]
- West, S. Processing of recombination intermediates by the ruvabc proteins. Annu. Rev. Genet. 1997, 31, 213–244. [Google Scholar] [CrossRef]
- Shereda, R.D.; Kozlov, A.G.; Lohman, T.M.; Cox, M.M.; Keck, J.L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Boil. 2008, 43, 289–318. [Google Scholar] [CrossRef]
- Chase, J.W.; Williams, K.R. Single-stranded DNA binding proteins required for DNA replication. Annu. Rev. Biochem. 1986, 55, 103–136. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.R.; Laine, P.S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990, 54, 342–380. [Google Scholar] [CrossRef] [PubMed]
- Kowalczykowski, S.C.; A Dixon, D.; Eggleston, A.; Lauder, S.D.; Rehrauer, W.M. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994, 58, 401–465. [Google Scholar] [CrossRef] [PubMed]
- Lohman, T.M.; E Ferrari, M. Escherichia coli Single-Stranded DNA-Binding Protein: Multiple DNA-Binding Modes and Cooperativities. Annu. Rev. Biochem. 1994, 63, 527–570. [Google Scholar] [CrossRef] [PubMed]
- Costes, A.; Lecointe, F.; McGovern, S.; Quevillon-Cheruel, S.; Polard, P. The C-terminal Domain of the Bacterial SSB Protein Acts as a DNA Maintenance Hub at Active Chromosome Replication Forks. PLoS Genet. 2010, 6, e1001238. [Google Scholar] [CrossRef]
- Sancar, A.; Williams, K.; Chase, J.W.; Rupp, W.D. Sequences of the ssb gene and protein. Proc. Natl. Acad. Sci. USA 1981, 78, 4274–4278. [Google Scholar] [CrossRef]
- Curth, U.; Genschel, J.; Urbanke, C.; Greipel, J. In Vitro and in Vivo Function of the C-Terminus of Escherichia coli Single-Stranded DNA Binding Protein. Nucleic Acids Res. 1996, 24, 2706–2711. [Google Scholar] [CrossRef]
- Raghunathan, S.; Kozlov, A.G.; Lohman, T.M.; Waksman, G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Genet. 2000, 7, 648–652. [Google Scholar] [CrossRef]
- Kuznetsov, S.V.; Kozlov, A.G.; Lohman, T.M.; Ansari, A. Microsecond Dynamics of Protein–DNA Interactions: Direct Observation of the Wrapping/Unwrapping Kinetics of Single-stranded DNA around the E. coli SSB Tetramer. J. Mol. Boil. 2006, 359, 55–65. [Google Scholar] [CrossRef]
- Ding, W.; Tan, H.Y.; Zhang, J.X.; Wilczek, L.A.; Hsieh, K.R.; Mulkin, J.A.; Bianco, P.R. The mechanism of SSB-RecG binding: Implications for SSB interactome function. Protein Sci. 2020, in press. [Google Scholar] [CrossRef]
- Simossis, V.A.; Heringa, J. PRALINE: A multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005, 33, W289–W294. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.-C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Ther. Antib. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Kozlov, A.G.; Weiland, E.; Mittal, A.; Waldman, V.; Pappu, R.V.; Timothy, L.M. The Intrinsically Disordered C-terminal Tails of E. coli Single-Stranded DNA Binding Protein Regulate Cooperative Binding to Single-Stranded DNA. Biophys. J. 2015, 108, 389a. [Google Scholar] [CrossRef]
- Bianco, P.R. The tale of SSB. Prog. Biophys. Mol. Boil. 2016, 127, 111–118. [Google Scholar] [CrossRef]
- Kozlov, A.G.; Jezewska, M.J.; Bujalowski, W.; Lohman, T.M. Binding specificity of Escherichia coli single-stranded DNA binding protein for the chi subunit of DNA pol III holoenzyme and PriA helicase. Biochemistry 2010, 49, 3555–3566. [Google Scholar] [CrossRef]
- Sandigursky, M.; Mendez, F.; Bases, R.E.; Matsumoto, T.; Franklin, W.A. Protein-Protein Interactions between the Escherichia coli Single-Stranded DNA-Binding Protein and Exonuclease I. Radiat. Res. 1996, 145, 619. [Google Scholar] [CrossRef]
- Bianco, P.R.; Pottinger, S.; Tan, H.Y.; Nguyenduc, T.; Rex, K.; Varshney, U. The IDL of E. coli SSB links ssDNA and protein binding by mediating protein–protein interactions. Protein Sci. 2017, 26, 227–241. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Huang, C.-Y. The glycine-rich flexible region in SSB is crucial for PriA stimulation. RSC Adv. 2018, 8, 35280–35288. [Google Scholar] [CrossRef]
- Nigam, R.; Mohan, M.; Shivange, G.; Dewangan, P.K.; Anindya, R. Escherichia coli AlkB interacts with single-stranded DNA binding protein SSB by an intrinsically disordered region of SSB. Mol. Boil. Rep. 2018, 45, 865–870. [Google Scholar] [CrossRef]
- Su, X.-C.; Wang, Y.; Yagi, H.; Shishmarev, D.; Mason, C.E.; Smith, P.J.; Vandevenne, M.; Dixon, N.E.; Otting, G. Bound or Free: Interaction of the C-terminal Domain of Escherichia coli Single-Stranded DNA-Binding Protein (SSB) with the Tetrameric Core of SSB. Biochemistry 2014, 53, 1925–1934. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.R.; Spicer, E.K.; Lopresti, M.B.; A Guggenheimer, R.; Chase, J.W. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J. Boil. Chem. 1983, 258, 3346–3355. [Google Scholar]
- Brown, A.M.; Zondlo, N.J. A Propensity Scale for Type II Polyproline Helices (PPII): Aromatic Amino Acids in Proline-Rich Sequences Strongly Disfavor PPII Due to Proline–Aromatic Interactions. Biochemistry 2012, 51, 5041–5051. [Google Scholar] [CrossRef] [PubMed]
- Meirson, T.; Bomze, D.; Kahlon, L.; Gil-Henn, H.; Samson, A.O. A helical lock and key model of polyproline II conformation with SH3. Bioinformatics 2019, 36, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Kurochkina, N.; Guha, U. SH3 domains: Modules of protein–protein interactions. Biophys. Rev. 2012, 5, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Saksela, K.; Permi, P. SH3 domain ligand binding: What’s the consensus and where’s the specificity? FEBS Lett. 2012, 586, 2609–2614. [Google Scholar] [CrossRef] [PubMed]
- Kay, B.K. SH3 domains come of age. FEBS Lett. 2012, 586, 2606–2608. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, V.; Kishan, K.V.R. Functional evolution of two subtly different (similar) folds. BMC Struct. Boil. 2001, 1, 5. [Google Scholar]
- Cheng, K.; Xu, H.; Chen, X.; Wang, L.; Tian, B.; Zhao, Y.; Hua, Y. Structural basis for DNA 5′-end resection by RecJ. eLife 2016, 5, 14294. [Google Scholar] [CrossRef]
- Wakamatsu, T.; Kitamura, Y.; Kotera, Y.; Nakagawa, N.; Kuramitsu, S.; Masui, R. Structure of RecJ Exonuclease Defines Its Specificity for Single-stranded DNA*. J. Boil. Chem. 2010, 285, 9762–9769. [Google Scholar] [CrossRef]
- Singleton, M.; Scaife, S.; Wigley, D.B. Structural Analysis of DNA Replication Fork Reversal by RecG. Cell 2001, 107, 79–89. [Google Scholar] [CrossRef]
- Ryzhikov, M.; Koroleva, O.; Postnov, D.; Tran, A.; Korolev, S. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 2011, 39, 6305–6314. [Google Scholar] [CrossRef] [PubMed]
- Windgassen, T.A.; Leroux, M.; Sandler, S.J.; Keck, J.L. Function of a strand-separation pin element in the PriA DNA replication restart helicase. J. Boil. Chem. 2018, 294, 2801–2814. [Google Scholar] [CrossRef] [PubMed]
- Bharti, S.K.; Rex, K.; Sreedhar, P.; Krishnan, N.; Varshney, U. Chimeras of Escherichia coli and Mycobacterium tuberculosis Single-Stranded DNA Binding Proteins: Characterization and Function in Escherichia coli. PLoS ONE 2011, 6, 27216. [Google Scholar] [CrossRef] [PubMed]
- Storm, P.; Hoekstra, W.; De Haan, P.; Verhoef, C. Genetic recombination in Escherichia coliIV. Isolation and characterization of recombinaion-deficient mutants of Escherichia coli K12. Mutat. Res. Mol. Mech. Mutagen. 1971, 13, 9–17. [Google Scholar] [CrossRef][Green Version]
- Benson, F.E.; Collier, S.; Lloyd, R.G. Evidence of abortive recombination in ruv mutants of Escherichia coli K12. Mol. Genet. Genom. 1991, 225, 266–272. [Google Scholar] [CrossRef]
- Lloyd, R.G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J. Bacteriol. 1991, 173, 5414–5418. [Google Scholar] [CrossRef]
- Kalman, M.; Murphy, H.; Cashel, M. The nucleotide sequence of recG, the distal spo operon gene in Escherichia coli K-12. Gene 1992, 110, 95–99. [Google Scholar] [CrossRef]
- Singleton, M.; Dillingham, M.; Wigley, D.B. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef]
- Vincent, S.D.; Mahdi, A.A.; Lloyd, R.G. The RecG Branch Migration Protein of Escherichia coli Dissociates R-loops. J. Mol. Boil. 1996, 264, 713–721. [Google Scholar] [CrossRef]
- Sharples, G.J.; Ingleston, S.M.; Lloyd, R.G. Holliday Junction Processing in Bacteria: Insights from the Evolutionary Conservation of RuvABC, RecG, and RusA. J. Bacteriol. 1999, 181, 5543–5550. [Google Scholar] [CrossRef]
- Whitby, M.C.; Vincent, S.; Lloyd, R. Branch migration of Holliday junctions: Identification of RecG protein as a junction specific DNA helicase. EMBO J. 1994, 13, 5220–5228. [Google Scholar] [CrossRef]
- Peter, M.; Lloyd, R.G. RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 1999, 27, 3049–3056. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McGlynn, P. Characterisation of the catalytically active form of RecG helicase. Nucleic Acids Res. 2000, 28, 2324–2332. [Google Scholar] [CrossRef] [PubMed]
- Slocum, S.L.; Buss, J.A.; Kimura, Y.; Bianco, P.R. Characterization of the ATPase Activity of the Escherichia coli RecG Protein Reveals that the Preferred Cofactor is Negatively Supercoiled DNA. J. Mol. Boil. 2007, 367, 647–664. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peter, B.J.; Ullsperger, C.; Hiasa, H.; Marians, K.J.; Cozzarelli, N.R. The Structure of Supercoiled Intermediates in DNA Replication. Cell 1998, 94, 819–827. [Google Scholar] [CrossRef]
- McGlynn, P.; Lloyd, R.G.; Marians, K.J. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. USA 2001, 98, 8235–8240. [Google Scholar] [CrossRef]
- Lecointe, F.; Sérèna, C.; Velten, M.; Costes, A.; McGovern, S.; Meile, J.-C.; Errington, J.; Ehrlich, S.D.; Noirot, P.; Polard, P. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J. 2007, 26, 4239–4251. [Google Scholar] [CrossRef]
- Wen, Q.; Mahdi, A.A.; Briggs, G.S.; Sharples, G.J.; Lloyd, R.G. Conservation of RecG activity from pathogens to hyperthermophiles. DNA Repair 2005, 4, 23–31. [Google Scholar] [CrossRef]
- Mahdi, A.A.; Briggs, G.S.; Sharples, G.J.; Wen, Q.; Lloyd, R.G. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 2003, 22, 724–734. [Google Scholar] [CrossRef]
- Sun, Z.; Hashemi, M.; Warren, G.; Bianco, P.R.; Lyubchenko, Y.L. Dynamics of the Interaction of RecG Protein with Stalled Replication Forks. Biochemistry 2018, 57, 1967–1976. [Google Scholar] [CrossRef]
- Briggs, G.S.; Mahdi, A.A.; Wen, Q.; Lloyd, R.G. DNA Binding by the Substrate Specificity (Wedge) Domain of RecG Helicase Suggests a Role in Processivity. J. Boil. Chem. 2005, 280, 13921–13927. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, A.A.; McGlynn, P.; Levett, S.D.; Lloyd, R.G. DNA Binding and Helicase Domains of the Escherichia coli Recombination Protein RecG. Nucleic Acids Res. 1997, 25, 3875–3880. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marians, K.J. PriA: At the crossroads of DNA replication and recombination. In Progress in Nucleic Acid Research and Molecular Biology; Academic Press: Cambridge, MA, USA, 1999; Volume 63, pp. 39–67. [Google Scholar] [CrossRef]
- Masai, H. DnaA- and PriA-dependent primosomes Two distinct replication complexes for replication of Escherichia coli chromosome. Front. Biosci. 1996, 1, d48–d58. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.H.; Masai, H.; Allen, G.C.; Kornberg, A. The priA gene encoding the primosomal replicative n’ protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 4620–4624. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.M.; Nakai, H. PriA and phage T4 gp59: Factors that promote DNA replication on forked DNA substrates microreview. Mol. Microbiol. 2000, 36, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P.; Zavitz, K.H.; Marians, K.J. Inactivation of the Escherichia coli priA DNA replication protein induces the SOS response. J. Bacteriol. 1991, 173, 6686–6693. [Google Scholar] [CrossRef]
- Lee, E.H.; Kornberg, A. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n’ protein. Proc. Natl. Acad. Sci. USA 1991, 88, 3029–3032. [Google Scholar] [CrossRef]
- Sandler, S.J.; Samra, H.S.; Clark, A.J. Differential Suppression of Pria2::Kan Phenotypes in Escherichia coli K-12 by Mutations in Pria, Lexa, and Dnac. Genetics 1996, 143, 5–13. [Google Scholar]
- Kogoma, T.; Cadwell, G.W.; Barnard, K.G.; Asai, T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J. Bacteriol. 1996, 178, 1258–1264. [Google Scholar] [CrossRef]
- Heller, R.C.; Marians, K.J. Non-replicative helicases at the replication fork. DNA Repair 2007, 6, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Masai, H. Stabilization of a Stalled Replication Fork by Concerted Actions of Two Helicases. J. Boil. Chem. 2005, 281, 3484–3493. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Mizukoshi, T.; Taniyama, C.; Kohda, D.; Arai, K.-I.; Masai, H. DNA Binding of PriA Protein Requires Cooperation of the N-terminal D-loop/Arrested-fork Binding and C-terminal Helicase Domains. J. Boil. Chem. 2002, 277, 38062–38071. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-W.; North, S.H.; Nakai, H. Properties of the PriA Helicase Domain and Its Role in Binding PriA to Specific DNA Structures. J. Boil. Chem. 2004, 279, 38503–38512. [Google Scholar] [CrossRef]
- Windgassen, T.; Leroux, M.; Satyshur, K.; Sandler, S.J.; Keck, J.L. Structure-specific DNA replication-fork recognition directs helicase and replication restart activities of the PriA helicase. Proc. Natl. Acad. Sci. USA 2018, 115, E9075–E9084. [Google Scholar] [CrossRef]
- Zavitz, K.H.; Marians, K.J. Helicase-deficient cysteine to glycine substitution mutants of Escherichia coli replication protein PriA retain single-stranded DNA-dependent ATPase activity. Zn2+ stimulation of mutant PriA helicase and primosome assembly activities. J. Boil. Chem. 1993, 268, 4337–4346. [Google Scholar]
- Masai, H.; Deneke, J.; Furui, Y.; Tanaka, T.; Arai, K.I. Escherichia coli and Bacillus subtilis PriA proteins essential for recombination-dependent DNA replication: Involvement of ATPase/helicase activity of PriA for inducible stable DNA replication. Biochimie 1999, 81, 847–857. [Google Scholar] [CrossRef]
- Liu, J.; Nurse, P.; Marians, K.J. The ordered assembly of the phiX174-type primosome. III. PriB facilitates complex formation between PriA and DnaT. J. Boil. Chem. 1996, 271, 15656–15661. [Google Scholar] [CrossRef]
- Nurse, P.; Liu, J.; Marians, K.J. Two modes of PriA binding to DNA. J. Boil. Chem. 1999, 274, 25026–25032. [Google Scholar] [CrossRef]
- Jones, J.M.; Nakai, H. Duplex opening by primosome protein PriA for replisome assembly on a recombination intermediate. J. Mol. Boil. 1999, 289, 503–515. [Google Scholar] [CrossRef]
- McGlynn, P.; A Al-Deib, A.; Liu, J.; Marians, K.J.; Lloyd, R.G. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J. Mol. Boil. 1997, 270, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Mizukoshi, T.; Tanaka, T.; Arai, K.-I.; Kohda, D.; Masai, H. A Critical Role of the 3′ Terminus of Nascent DNA Chains in Recognition of Stalled Replication Forks. J. Boil. Chem. 2003, 278, 42234–42239. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Marians, K.J. Escherichia coli replication factor Y, a component of the primosome, can act as a DNA helicase. Proc. Natl. Acad. Sci. USA 1987, 84, 8345–8349. [Google Scholar] [CrossRef] [PubMed]
- Lasken, R.S.; Kornberg, A. The primosomal protein n’ of Escherichia coli is a DNA helicase. J. Boil. Chem. 1988, 263, 5512–5518. [Google Scholar]
- Tanaka, T.; Taniyama, C.; Arai, K.-I.; Masai, H. ATPase/helicase motif mutants of Escherichia coli PriA protein essential for recombination-dependent DNA replication. Genes Cells 2003, 8, 251–261. [Google Scholar] [CrossRef]
- Liu, J.; Marians, K.J. PriA-directed Assembly of a Primosome on D Loop DNA. J. Boil. Chem. 1999, 274, 25033–25041. [Google Scholar] [CrossRef]
- McGlynn, P.; Lloyd, R.G. Rescue of stalled replication forks by RecG: Simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA 2001, 98, 8227–8234. [Google Scholar] [CrossRef]
- Gregg, A.V.; McGlynn, P.; Jaktaji, R.P.; Lloyd, R.G. Direct Rescue of Stalled DNA Replication Forks via the Combined Action of PriA and RecG Helicase Activities. Mol. Cell 2002, 9, 241–251. [Google Scholar] [CrossRef]
- Sasaki, K.; Ose, T.; Okamoto, N.; Maenaka, K.; Tanaka, T.; Masai, H.; Saito, M.; Shirai, T.; Kohda, D. Structural basis of the 3′-end recognition of a leading strand in stalled replication forks by PriA. EMBO J. 2007, 26, 2584–2593. [Google Scholar] [CrossRef]
- Tanaka, T.; Mizukoshi, T.; Sasaki, K.; Kohda, D.; Masai, H. Escherichia coli PriA Protein, Two Modes of DNA Binding and Activation of ATP Hydrolysis. J. Boil. Chem. 2007, 282, 19917–19927. [Google Scholar] [CrossRef]
- Bhattacharyya, B.; George, N.P.; Thurmes, T.M.; Zhou, R.; Jani, N.; Wessel, S.R.; Sandler, S.J.; Ha, T.; Keck, J.L. Structural mechanisms of PriA-mediated DNA replication restart. Proc. Natl. Acad. Sci. USA 2013, 111, 1373–1378. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.M.; Nakai, H. Escherichia coli PriA helicase: Fork binding orients the helicase to unwind the lagging strand side of arrested replication forks11Edited by M. Gottesman. J. Mol. Boil. 2001, 312, 935–947. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Senac, M.D.M.; Webb, M. Mechanism of Translocation and Kinetics of DNA Unwinding by the Helicase RecG†. Biochemistry 2005, 44, 16967–16976. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Kozlov, A.G.; Roy, R.; Zhang, J.; Korolev, S.; Lohman, T.M.; Ha, T. SSB Functions as a Sliding Platform that Migrates on DNA via Reptation. Cell 2011, 146, 485. [Google Scholar] [CrossRef]
- Tan, H.Y.; Wilczek, L.A.; Pottinger, S.; Manosas, M.; Yu, C.; Nguyenduc, T.; Bianco, P.R. The intrinsically disordered linker of E. coli SSB is critical for the release from single-stranded DNA. Protein Sci. 2017, 26, 700–717. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianco, P.R. DNA Helicase-SSB Interactions Critical to the Regression and Restart of Stalled DNA Replication Forks in Escherichia coli. Genes 2020, 11, 471. https://doi.org/10.3390/genes11050471
Bianco PR. DNA Helicase-SSB Interactions Critical to the Regression and Restart of Stalled DNA Replication Forks in Escherichia coli. Genes. 2020; 11(5):471. https://doi.org/10.3390/genes11050471
Chicago/Turabian StyleBianco, Piero R. 2020. "DNA Helicase-SSB Interactions Critical to the Regression and Restart of Stalled DNA Replication Forks in Escherichia coli" Genes 11, no. 5: 471. https://doi.org/10.3390/genes11050471
APA StyleBianco, P. R. (2020). DNA Helicase-SSB Interactions Critical to the Regression and Restart of Stalled DNA Replication Forks in Escherichia coli. Genes, 11(5), 471. https://doi.org/10.3390/genes11050471