A High-Quality Genome Sequence of Model Legume Lotus japonicus (MG-20) Provides Insights into the Evolution of Root Nodule Symbiosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome Sequencing
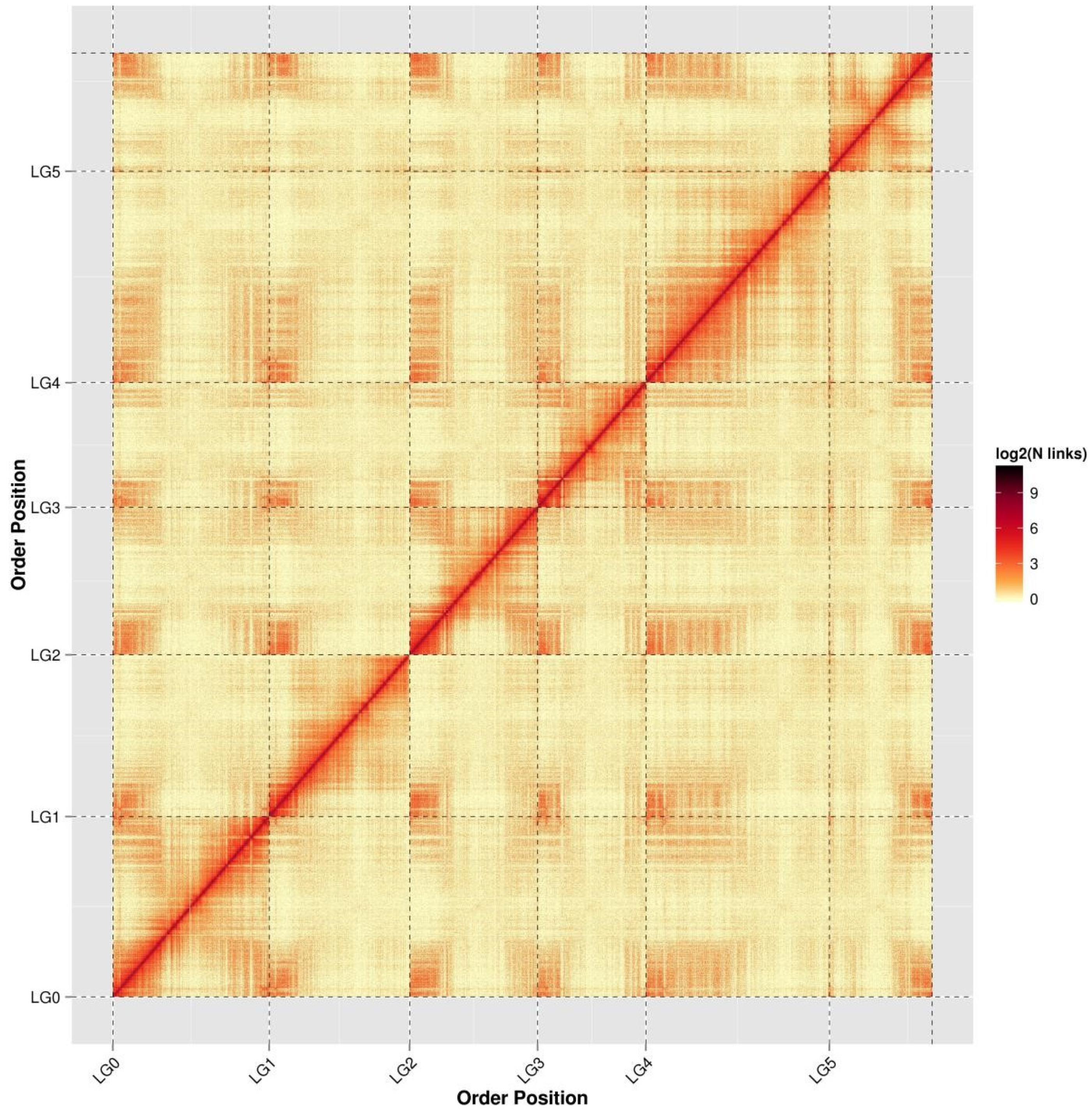
2.2. Hi-C Assembly
2.3. Evaluation of the Assembly
2.4. Genome Annotation
2.5. Genome Alignment and Gene Synteny Analysis
2.6. Phylogenetic Analysis
3. Results
3.1. Genome Sequencing and Assembly
3.2. Assessment of the Genomic Assembly
3.3. Genome Annotation
3.4. Synteny and Species Evolutionary Analysis
3.5. Orthogroup Analysis
4. Discussion
5. Conclusions
6. Patents
Availability of Supporting Data and Materials
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kawaguchi, M. Lotus japonicus ’Miyakojima’ MG-20: An early-flowering accession suitable for indoor handling. J. Plant Res. 2000, 113, 507–509. [Google Scholar] [CrossRef]
- Lto, M.; Miyamoto, J.; Mori, Y.; Fujimoto, S.; Uchiumi, T.; Abe, M.; Suzuki, A.; Tabata, S.; Fukui, K. Genome and Chromosome Dimensions of Lotus japonicus. J. Plant Res. 2000, 113, 435–442. [Google Scholar]
- Giovannetti, M.; Göschl, C.; Dietzen, C.; Andersen, S.U.; Kopriva, S.; Busch, W. Identification of novel genes involved in phosphate accumulation in Lotus japonicus through Genome Wide Association mapping of root system architecture and anion content. PLoS Genet. 2019, 15, e1008126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, N.; Wakabayashi, T.; Kawamura, Y.; Skovbjerg, C.K.; Wang, M.Z.; Mustamin, Y.; Isomura, Y.; Gupta, V.; Jin, H.; Mun, T.; et al. Extreme genetic signatures of local adaptation during Lotus japonicus colonization of Japan. Nat. Commun. 2020, 11, 253. [Google Scholar] [CrossRef] [Green Version]
- Sato, S.; Nakamura, Y.; Kaneko, T.; Asamizu, E.; Kato, T.; Nakao, M.; Sasamoto, S.; Watanabe, A.; Ono, A.; Kawashima, K.; et al. Genome Structure of the Legume, Lotus japonicus. Curr. Neuropharmacol. 2008, 15, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, S.; Murakami, Y. Genome Analysis of Lotus japonicus. J. Plant Res. 2000, 113, 497–506. [Google Scholar] [CrossRef]
- Kato, T.; Kaneko, T.; Sato, S.; Nakamura, Y.; Tabata, S. Complete Structure of the Chloroplast Genome of a Legume. Curr. Neuropharmacol. 2000, 7, 323–330. [Google Scholar]
- Pedrosa, A.; Sandal, N.; Stougaard, J.; Schweizer, D.; Bachmair, A. Chromosomal Map of the Model Legume Lotus japonicus. Genetics 2002, 161, 1661–1672. [Google Scholar]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Servant, N.; Varoquaux, N.; Lajoie, B.R.; Viara, E.; Chen, C.J.; Vert, J.P.; Heard, E.; Dekker, J.; Barillot, E. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015, 16, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burton, J.N.; Adey, A.; Patwardhan, R.P.; Qiu, R.; Kitzman, J.O.; Shendure, J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013, 31, 1119–1125. [Google Scholar] [CrossRef]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Parra, G.; Bradnam, K.; Korf, I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 2007, 23, 1061–1067. [Google Scholar] [CrossRef]
- Choi, V.; Farach-Colton, M. Barnacle: An assembly algorithm for clone-based sequences of whole genomes. Gene 2003, 320, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef] [Green Version]
- Burge, C.; Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef] [Green Version]
- Stanke, M.; Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 2003, 19 (Suppl. 2), ii215–ii225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef] [PubMed]
- Korf, I. Gene finding in novel genomes. BMC Bioinformatics 2004, 5, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keilwagen, J.; Wenk, M.; Erickson, J.L.; Schattat, M.H.; Grau, J.; Hartung, F. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016, 44, e89. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Tang, S.; Lomsadze, A.; Borodovsky, M. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res. 2015, 43, e78. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Campbell, M.A.; Haas, B.J.; Hamilton, J.P.; Mount, S.M.; Buell, C.R. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics 2006, 7, 327. [Google Scholar] [CrossRef] [Green Version]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- She, R.; Chu, J.S.; Wang, K.; Pei, J.; Chen, N. GenBlastA: Enabling BLAST to identify homologous gene sequences. Genome Res. 2009, 19, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Wang, H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007, 35, W265–W268. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Wessler, S.R. MITE-Hunter: A program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010, 38, e199. [Google Scholar] [CrossRef] [Green Version]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 2005, 21 (Suppl. 1), 351–358. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Myers, E.W. PILER: Identification and classification of genomic repeats. Bioinformatics 2005, 21 (Suppl. 1), 152–158. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using repeat masker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 25, 4–10. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Moxon, S.; Marshall, M.; Khanna, A.; Eddy, S.R.; Bateman, A. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005, 33, D121–D124. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [Green Version]
- Zdobnov, E.M.; Apweiler, R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef] [Green Version]
- Bairoch, A. PROSITE: A dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991, 19, 2241–2245. [Google Scholar] [CrossRef]
- Lima, T.; Auchincloss, A.H.; Coudert, E.; Keller, G.; Michoud, K.; Rivoire, C.; Bulliard, V.; de Castro, E.; Lachaize, C.; Baratin, D.; et al. HAMAP: A database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2009, 37, D471–D478. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Schuster-Bockler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006, 34, D247–D251. [Google Scholar] [CrossRef] [Green Version]
- Attwood, T.K.; Beck, M.E. PRINTS—A protein motif fingerprint database. Protein Eng. Des. Sel. 1994, 7, 841–848. [Google Scholar] [CrossRef]
- Bru, C.; Courcelle, E.; Carrere, S.; Beausse, Y.; Dalmar, S.; Kahn, D. The ProDom database of protein domain families: More emphasis on 3D. Nucleic Acids Res. 2005, 33, D212–D215. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Copley, R.R.; Schmidt, S.; Ciccarelli, F.D.; Doerks, T.; Schultz, J.; Ponting, C.P.; Bork, P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004, 32, D142–D144. [Google Scholar] [CrossRef] [Green Version]
- Haft, D.H.; Selengut, J.D.; White, O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003, 31, 371–373. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.H.; Nikolskaya, A.; Huang, H.; Yeh, L.S.; Natale, D.A.; Vinayaka, C.R.; Hu, Z.Z.; Mazumder, R.; Kumar, S.; Kourtesis, P.; et al. PIRSF: Family classification system at the Protein Information Resource. Nucleic Acids Res. 2004, 32, D112–D114. [Google Scholar] [CrossRef] [Green Version]
- Gough, J.; Chothia, C. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 2002, 30, 268–272. [Google Scholar] [CrossRef]
- Lees, J.; Yeats, C.; Perkins, J.; Sillitoe, I.; Rentzsch, R.; Dessailly, B.H.; Orengo, C. Gene3D: A domain-based resource for comparative genomics, functional annotation and protein network analysis. Nucleic Acids Res. 2012, 40, D465–D471. [Google Scholar] [CrossRef]
- Thomas, P.D.; Kejariwal, A.; Campbell, M.J.; Mi, H.; Diemer, K.; Guo, N.; Ladunga, I.; Ulitsky-Lazareva, B.; Muruganujan, A.; Rabkin, S.; et al. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003, 31, 334–341. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Verdier, J.; Torres-Jerez, I.; Wang, M.; Andriankaja, A.; Allen, S.N.; He, J.; Tang, Y.; Murray, J.D.; Udvardi, M.K. Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 2013, 74, 351–362. [Google Scholar] [CrossRef]
- Mun, T.; Bachmann, A.; Gupta, V.; Stougaard, J.; Andersen, S.U. Lotus Base: An integrated information portal for the model legume Lotus japonicus. Sci. Rep. 2016, 6, 39447. [Google Scholar] [CrossRef] [Green Version]
- Małolepszy, A.; Mun, T.; Sandal, N.; Gupta, V.; Dubin, M.; Urbański, D.; Shah, N.; Bachmann, A.; Fukai, E.; Hirakawa, H.; et al. The LORE1 insertion mutant resource. Plant J. 2016, 88, 306–317. [Google Scholar] [CrossRef] [Green Version]
- Urban´ski, D.F.; Małolepszy, A.; Stougaard, J.; Andersen, S.U. Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J. 2012, 69, 731–741. [Google Scholar] [CrossRef] [Green Version]
- Fukai, E.; Soyano, T.; Umehara, Y.; Nakayama, S.; Hirakawa, H.; Tabata, S.; Sato, S.; Hayashi, M. Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. Plant J. 2012, 69, 720–730. [Google Scholar] [CrossRef]







| Genomic Feature | LjPB_ver1.0 | L. japonicus (MG-20) v3.0 |
|---|---|---|
| Total length of contigs | 544,144,611 | NA |
| Total sequences length of clustered | 517,538,720 | NA |
| Total sequences ordered and oriented | 499,061,371 | 394,454,697 |
| Percentage of total sequences clustered | 95.11% | NA |
| Percentage of total sequences ordered and oriented | 96.43% | NA |
| Number of contigs | 616 | 44,464 |
| Contig N50(bp) | 2,515,607 | 25,054 |
| Contig N90 (bp) | 675,000 | NA |
| Contig max (bp) | 11,738,399 | NA |
| GC content | 37.99% | 36.6% |
| Genome coverage | 60× | 35× |
| Percentage of repeat sequences | 65.61% | NA |
| Percentage of 458 CEGs present in assemblies | 97.82% | NA |
| Percentage of 248 highly conserved CEGs present | 91.53% | NA |
| Number of genes | 28,251 | 10,951 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Jiang, F.; Wu, P.; Wang, K.; Cao, Y. A High-Quality Genome Sequence of Model Legume Lotus japonicus (MG-20) Provides Insights into the Evolution of Root Nodule Symbiosis. Genes 2020, 11, 483. https://doi.org/10.3390/genes11050483
Li H, Jiang F, Wu P, Wang K, Cao Y. A High-Quality Genome Sequence of Model Legume Lotus japonicus (MG-20) Provides Insights into the Evolution of Root Nodule Symbiosis. Genes. 2020; 11(5):483. https://doi.org/10.3390/genes11050483
Chicago/Turabian StyleLi, Haoxing, Fan Jiang, Ping Wu, Ke Wang, and Yangrong Cao. 2020. "A High-Quality Genome Sequence of Model Legume Lotus japonicus (MG-20) Provides Insights into the Evolution of Root Nodule Symbiosis" Genes 11, no. 5: 483. https://doi.org/10.3390/genes11050483






