Weighted Gene Correlation Network Meta-Analysis Reveals Functional Candidate Genes Associated with High- and Sub-Fertile Reproductive Performance in Beef Cattle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Approval and Consent to Participate
2.2. Data Collection
2.3. RNA-Sequencing Data Alignment and Variant Calling
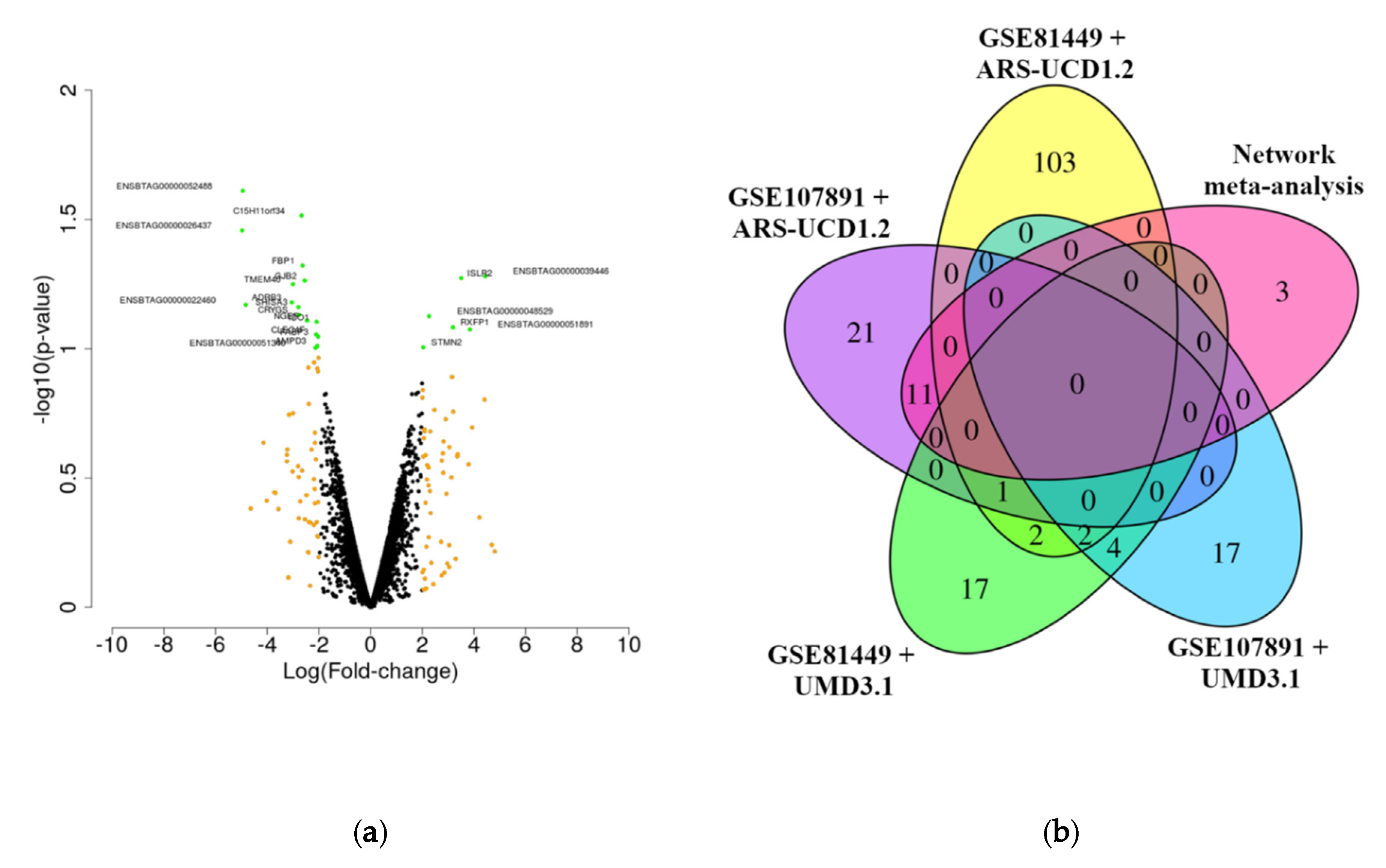
2.4. Identification of Genes with Expression Determined by the Study and Outliers
2.5. Meta-Analysis of Differentially Expressed Genes
2.6. Weighted Correlation Network Analysis
- -
- For each sample s in (HF and SF);
- -
- For each module m(s) in s;
- -
- Apply a Fisher’s exact test under the null hypothesis that there is no significant overlapping of m(HF) in SF and m(SF) in HF after a Bonferroni multiple test correction.
2.7. Functional Analysis and Annotation of Candidate Genes
3. Results
3.1. RNA-Sequencing and Variant Calling Statistics
3.2. Outlier Detection and Differential Expression Analysis between High-Fertile and Sub-Fertile Animals
3.3. Identification of Candidate Differentially Co-Expressed Gene Modules for High-Fertile and Sub-Fertile Animals
3.4. Functional Candidate Genes
4. Discussion
4.1. Network Meta-Analysis for Identification of Differentially Expressed Genes between High-Fertile and Sub-Fertile Animals
4.2. Differentially Co-Expressed Modules and the Identification of Functional Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thundathil, J.C.; Dance, A.L.; Kastelic, J.P. Fertility management of bulls to improve beef cattle productivity. Theriogenology 2016, 86, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.P.; Friggens, N.C.; Lucy, M.; Roche, J.R. Milk Production and Fertility in Cattle. Annu. Rev. Anim. Biosci. 2016, 4, 269–290. [Google Scholar] [CrossRef] [PubMed]
- De Souza Fonseca, P.A.; Id-Lahoucine, S.; Reverter, A.; Medrano, J.F.; Fortes, M.S.; Casellas, J.; Miglior, F.; Brito, L.; Carvalho, M.R.S.; Schenkel, F.S.; et al. Combining multi-OMICs information to identify key-regulator genes for pleiotropic effect on fertility and production traits in beef cattle. PLoS ONE 2018, 13, e0205295. [Google Scholar] [CrossRef]
- Kadri, N.K.; Sahana, G.; Charlier, C.; Iso-Touru, T.; Guldbrandtsen, B.; Karim, L.; Nielsen, U.S.; Panitz, F.; Aamand, G.P.; Schulman, N.; et al. A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock. PLoS Genet. 2014, 10, e1004049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saatchi, M.; Schnabel, R.D.; Taylor, J.F.; Garrick, D.J. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genom. 2014, 15, 442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuruta, S.; Lourenco, D.A.L.; Misztal, I.; Lawlor, T.J. Genomic analysis of cow mortality and milk production using a threshold-linear model. J. Dairy Sci. 2017, 100, 7295–7305. [Google Scholar] [CrossRef]
- De Camargo, G.M.F.; Porto-Neto, L.R.; Kelly, M.J.; Bunch, R.J.; McWilliam, S.M.; Tonhati, H.; Lehnert, S.A.; Fortes, M.R.S.; Moore, S.S. Non-synonymous mutations mapped to chromosome X associated with andrological and growth traits in beef cattle. BMC Genom. 2015, 16, 384. [Google Scholar] [CrossRef] [Green Version]
- Bellows, D.S.; Ott, S.L.; Bellows, R. a Review: Cost of reproductive diseases and conditions in cattle. Prof. Anim. Sci. 2002, 18, 26–32. [Google Scholar] [CrossRef]
- Shalloo, L.; Cromie, A.; McHugh, N. Effect of fertility on the economics of pasture-based dairy systems. Animal 2014, 8, 222–231. [Google Scholar] [CrossRef] [Green Version]
- Wiltbank, M.C.; Baez, G.M.; Garcia-Guerra, A.; Toledo, M.Z.; Monteiro, P.L.J.; Melo, L.F.; Ochoa, J.C.; Santos, J.E.P.; Sartori, R. Pivotal periods for pregnancy loss during the first trimester of gestation in lactating dairy cows. Theriogenology 2016, 86, 239–253. [Google Scholar] [CrossRef]
- Diskin, M.G.; Parr, M.H.; Morris, D.G. Embryo death in cattle: An update. Reprod. Fertil. Dev. 2012, 24, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Chebel, R.C.; Santos, J.E.P.; Reynolds, J.P.; Cerri, R.L.A.; Juchem, S.O.; Overton, M. Factors affecting conception rate after artificial insemination and pregnancy loss in lactating dairy cows. Anim. Reprod. Sci. 2004, 84, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Bormann, J.M.; Totir, L.R.; Kachman, S.D.; Fernando, R.L.; Wilson, D.E. Pregnancy rate and first-service conception rate in Angus heifers. J. Anim. Sci. 2006, 84, 2022–2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzam, S.M.; Kinder, J.E.; Nielsen, M.K. Conception rate at first insemination in beef cattle: Effects of season, age and previous reproductive performance. J. Anim. Sci. 1989, 67, 1405–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Id-Lahoucine, S.; Cánovas, A.; Jaton, C.; Miglior, F.; Fonseca, P.A.S.; Sargolzaei, M.; Miller, S.; Schenkel, F.S.; Medrano, J.F.; Casellas, J. Implementation of Bayesian methods to identify SNP and haplotype regions with transmission ratio distortion across the whole genome: TRDscan v.1.0. J. Dairy Sci. 2019, 102, 3175–3188. [Google Scholar] [CrossRef] [Green Version]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [Green Version]
- Butler, S.T.; Pryce, J.E.; Kemper, K.E.; Berry, D.P.; McCabe, M.; Lonergan, P.; Hayes, B.J.; Chamberlain, A.J.; Cormican, P.; Moore, S.G.; et al. Differentially Expressed Genes in Endometrium and Corpus Luteum of Holstein Cows Selected for High and Low Fertility Are Enriched for Sequence Variants Associated with Fertility1. Biol. Reprod. 2015, 94, 1–11. [Google Scholar]
- Moraes, J.G.N.; Behura, S.K.; Geary, T.W.; Hansen, P.J.; Neibergs, H.L.; Spencer, T.E. Uterine influences on conceptus development in fertility-classified animals. Proc. Natl. Acad. Sci. USA 2018, 115, E1749–E1758. [Google Scholar] [CrossRef] [Green Version]
- Moran, B.; Butler, S.T.; Moore, S.G.; Machugh, D.E.; Creevey, C.J. Differential gene expression in the endometrium reveals cytoskeletal and immunological genes in lactating dairy cows genetically divergent for fertility traits. Reprod. Fertil. Dev. 2017, 29, 274–282. [Google Scholar] [CrossRef]
- Geary, T.W.; Burns, G.W.; Moraes, J.G.N.; Moss, J.I.; Denicol, A.C.; Dobbs, K.B.; Ortega, M.S.; Hansen, P.J.; Wehrman, M.E.; Neibergs, H.; et al. Identification of Beef Heifers with Superior Uterine Capacity for Pregnancy. Biol. Reprod. 2016, 95, 47. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Reverter, A.; Cánovas, A.; Venus, B.; Islas-Trejo, A.; Porto-Neto, L.R.; Lehnert, S.A.; Medrano, J.F.; Moore, S.S.; Fortes, M.R.S. Global differential gene expression in the pituitary gland and the ovaries of Pre-And postpubertal brahman heifers. J. Anim. Sci. 2017, 95, 599–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortes, M.R.S.; Nguyen, L.T.; Weller, M.M.D.C.A.; Cánovas, A.; Islas-Trejo, A.; Porto-Neto, L.R.; Reverter, A.; Lehnert, S.A.; Boe-Hansen, G.B.; Thomas, M.G.; et al. Transcriptome analyses identify five transcription factors differentially expressed in the hypothalamus of post- versus prepubertal Brahman heifers. J. Anim. Sci. 2016, 94, 3693–3702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Reverter, A.; Cánovas, A.; Venus, B.; Anderson, S.T.; Islas-Trejo, A.; Dias, M.M.; Crawford, N.F.; Lehnert, S.A.; Medrano, J.F.; et al. STAT6, PBX2, and PBRM1 emerge as predicted regulators of 452 differentially expressed genes associated with puberty in Brahman heifers. Front. Genet. 2018, 9, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, M.M.; Cánovas, A.; Mantilla-Rojas, C.; Riley, D.G.; Luna-Nevarez, P.; Coleman, S.J.; Speidel, S.E.; Enns, R.M.; Islas-Trejo, A.; Medrano, J.F.; et al. SNP detection using RNA-sequences of candidate genes associated with puberty in cattle. Genet. Mol. Res. 2017, 16, 1–17. [Google Scholar] [CrossRef]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [Green Version]
- Oliver, S. Guilt-by-association goes global. Nature 2000, 403, 601–603. [Google Scholar] [CrossRef]
- Winter, C.; Kosch, R.; Ludlow, M.; Osterhaus, A.D.M.E.; Jung, K. Network meta-analysis correlates with analysis of merged independent transcriptome expression data. BMC Bioinform. 2019, 20, 144. [Google Scholar] [CrossRef]
- Wren, J.D. A global meta-analysis of microarray expression data to predict unknown gene functions and estimate the literature-data divide. Bioinformatics 2009, 25, 1694–1701. [Google Scholar] [CrossRef] [Green Version]
- Gillis, J.; Pavlidis, P. The impact of multifunctional genes on guilt “by association” analysis. PLoS ONE 2011, 6, e17258. [Google Scholar] [CrossRef] [Green Version]
- Blankenburg, H.; Pramstaller, P.P.; Domingues, F.S. A network-based meta-analysis for characterizing the genetic landscape of human aging. Biogerontology 2018, 19, 81–94. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Blom, U.M.; Wang, P.I.; Shim, J.E.; Marcotte, E.M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011, 21, 1109–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nygaard, V.; Rødland, E.A.; Hovig, E. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics 2016, 17, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Ayuso, M.; Fernández, A.; Núñez, Y.; Benítez, R.; Isabel, B.; Fernández, A.I.; Rey, A.I.; González-Bulnes, A.; Medrano, J.F.; Cánovas, Á.; et al. Developmental stage, muscle and genetic type modify muscle transcriptome in pigs: Effects on gene expression and regulatory factors involved in growth and metabolism. PLoS ONE 2016, 11, e0167858. [Google Scholar] [CrossRef] [Green Version]
- Cánovas, A.; Rincon, G.; Islas-Trejo, A.; Wickramasinghe, S.; Medrano, J.F. SNP discovery in the bovine milk transcriptome using RNA-Seq technology. Mamm. Genome 2010, 21, 592–598. [Google Scholar] [CrossRef] [Green Version]
- Cánovas, A.; Rincón, G.; Islas-Trejo, A.; Jimenez-Flores, R.; Laubscher, A.; Medrano, J.F. RNA sequencing to study gene expression and single nucleotide polymorphism variation associated with citrate content in cow milk. J. Dairy Sci. 2013, 96, 2637–2648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cánovas, A.; Rincón, G.; Bevilacqua, C.; Islas-Trejo, A.; Brenaut, P.; Hovey, R.C.; Boutinaud, M.; Morgenthaler, C.; Vanklompenberg, M.K.; Martin, P.; et al. Comparison of five different RNA sources to examine the lactating bovine mammary gland transcriptome using RNA-Sequencing. Sci. Rep. 2014, 4, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 2020, 9, giaa021. [Google Scholar] [CrossRef] [Green Version]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621. [Google Scholar] [CrossRef]
- Marioni, J.C.; Mason, C.E.; Mane, S.M.; Stephens, M.; Gilad, Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008, 18, 1509–1517. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Rücker, G.; Krahn, U.; Jochem, K.; Orestis, E.; Guido, S. Package ‘netmeta’. Network Meta-Analysis using Frequentist Methods (Version 0.7-0). 2015. Available online: http://kambing.ui.ac.id/cran/web/packages/netmeta/netmeta.pdf (accessed on 10 May 2020).
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Zhang, B.; Horvath, S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics 2008, 24, 719–720. [Google Scholar] [CrossRef] [PubMed]
- Botía, J.A.; Vandrovcova, J.; Forabosco, P.; Guelfi, S.; D’Sa, K.; Hardy, J.; Lewis, C.M.; Ryten, M.; Weale, M.E.; Ramasamy, A.; et al. An additional k-means clustering step improves the biological features of WGCNA gene co-expression networks. BMC Syst. Biol. 2017, 11, 47. [Google Scholar] [CrossRef] [PubMed]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef] [Green Version]
- Guney, E.; Garcia-Garcia, J.; Oliva, B. GUILDify: A web server for phenotypic characterization of genes through biological data integration and network-based prioritization algorithms. Bioinformatics 2014, 30, 1789–1790. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, 305–311. [Google Scholar] [CrossRef]
- Fonseca, P.A.d.S.; dos Santos, F.C.; Lam, S.; Suárez-Vega, A.; Miglior, F.; Schenkel, F.S.; Diniz, L.d.A.F.; Id-Lahoucine, S.; Carvalho, M.R.S.; Cánovas, A. Genetic mechanisms underlying spermatic and testicular traits within and among cattle breeds: Systematic review and prioritization of GWAS results. J. Anim. Sci. 2018, 96, 4978–4999. [Google Scholar]
- Cánovas, A.; Reverter, A.; DeAtley, K.L.; Ashley, R.L.; Colgrave, M.L.; Fortes, M.R.S.; Islas-Trejo, A.; Lehnert, S.; Porto-Neto, L.; Rincón, G.; et al. Multi-tissue omics analyses reveal molecular regulatory networks for puberty in composite beef cattle. PLoS ONE 2014, 9, e102551. [Google Scholar] [CrossRef] [Green Version]
- Suárez-Vega, A.; Gutiérrez-Gil, B.; Benavides, J.; Perez, V.; Tosser-Klopp, G.; Klopp, C.; Keennel, S.J.; Arranz, J.J. Combining GWAS and RNA-Seq approaches for detection of the causal mutation for hereditary junctional epidermolysis bullosa in sheep. PLoS ONE 2015, 10, e0126416. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C. Defining cell types and states with single-cell genomics. Genome Res. 2015, 25, 1491–1498. [Google Scholar] [CrossRef] [Green Version]
- Petri, T.; Altmann, S.; Geistlinger, L.; Zimmer, R.; Küffner, R. Addressing false discoveries in network inference. Bioinformatics 2015, 31, 2836–2843. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.H.; Chu, L.J.; Wang, J.C.; Chen, T.W.; Tien, Y.J.; Lin, W.C.; Ng, W.V. Meta-analytical biomarker search of EST expression data reveals three differentially expressed candidates. BMC Genom. 2012, 13, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suravajhala, P.; Kogelman, L.J.A.; Kadarmideen, H.N. Multi-omic data integration and analysis using systems genomics approaches: Methods and applications In animal production, health and welfare. Genet. Sel. Evol. 2016, 48, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadarmideen, H.N. Genomics to systems biology in animal and veterinary sciences: Progress, lessons and opportunities. Livest. Sci. 2014, 166, 232–248. [Google Scholar] [CrossRef] [Green Version]
- Christians, E.; Davis, A.A.; Thomas, S.D.; Benjamin, I.J. Embryonic development: Maternal effect of hsf1 on reproductive success. Nature 2000, 407, 693–694. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Lv, H.; Wu, K.L.; Zhang, Y.S.; Luo, H.N.; Chen, Z.J. Discs large homologue 1 (Dlg1) coordinates mouse oocyte polarisation during maturation. Reprod. Fertil. Dev. 2017, 29, 1699–1707. [Google Scholar] [CrossRef]
- Tewes, A.C.; Rall, K.K.; Römer, T.; Hucke, J.; Kapczuk, K.; Brucker, S.; Wieacker, P.; Ledig, S. Variations in RBM8A and TBX6 are associated with disorders of the müllerian ducts. Fertil. Steril. 2015, 103, 1313–1318. [Google Scholar] [CrossRef]
- White, P.H.; Farkas, D.R.; McFadden, E.E.; Chapman, D.L. Defective somite patterning in mouse embryos with reduced levels of Tbx6. Development 2003, 130, 1681–1690. [Google Scholar] [CrossRef] [Green Version]
- Rosenbaum, M.; Leibel, R.L. Leptin: A molecule integrating somatic energy stores, energy expenditure and fertility. Trends Endocrinol. Metab. 1998, 9, 117–124. [Google Scholar] [CrossRef]
- Lu, L.Y.; Yu, X. CHFR is important for the survival of male premeiotic germ cells. Cell Cycle 2015, 14, 3454–3460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Pan, H.; Zhu, L.; Deng, Y.; Pollard, J.W. Progesterone inhibits the estrogen-induced phosphoinositide 3-kinase→AKT→GSK-3β→cyclin D1→pRB pathway to block uterine epithelial cell proliferation. Mol. Endocrinol. 2005, 19, 1978–1990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, K.I.; Brummer, T.; O’Brien, P.M.; Daly, R.J. Dual-specificity phosphatases: Critical regulators with diverse cellular targets. Biochem. J. 2009, 418, 475–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Park, B.; Kim, G.; Kim, K.; Pak, J.; Kim, K.; Ye, M.B.; Park, S.G.; Park, D. Arhgef16, a novel Elmo1 binding partner, promotes clearance of apoptotic cells via RhoG-dependent Rac1 activation. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 2438–2447. [Google Scholar] [CrossRef] [Green Version]
- Elliott, M.R.; Zheng, S.; Park, D.; Woodson, R.I.; Reardon, M.A.; Juncadella, I.J.; Kinchen, J.M.; Zhang, J.; Lysiak, J.J.; Ravichandran, K.S. Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature 2010, 467, 333–337. [Google Scholar] [CrossRef]
- Gong, P.; Chen, S.; Zhang, L.; Hu, Y.; Gu, A.; Zhang, J.; Wang, Y. RhoG-ELMO1-RAC1 is involved in phagocytosis suppressed by mono-butyl phthalate in TM4 cells. Environ. Sci. Pollut. Res. 2018, 25, 35440–35450. [Google Scholar] [CrossRef]
- Yang, S.; Wang, C. The intraflagellar transport protein IFT80 is required for cilia formation and osteogenesis. Bone 2012, 51, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Yuan, X.; Yang, S. IFT80 is essential for chondrocyte differentiation by regulating Hedgehog and Wnt signaling pathways. Exp. Cell Res. 2013, 319, 623–632. [Google Scholar] [CrossRef] [Green Version]
- Beales, P.L.; Bland, E.; Tobin, J.L.; Bacchelli, C.; Tuysuz, B.; Hill, J.; Rix, S.; Pearson, C.G.; Kai, M.; Hartley, J.; et al. IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat. Genet. 2007, 39, 727–729. [Google Scholar] [CrossRef]
- Gaur, T.; Hussain, S.; Mudhasani, R.; Parulkar, I.; Colby, J.L.; Frederick, D.; Kream, B.E.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev. Biol. 2010, 340, 10–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, I.M.; Adams, C.S.; Freeman, T.; Srinivas, V. Fate of the hypertrophic chondrocyte: Microenvironmental perspectives on apoptosis and survival in the epiphyseal growth plate. Birth Defects Res. Part C Embryo Today Rev. 2005, 75, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Zelzer, E.; Mamluk, R.; Ferrara, N.; Johnson, R.S.; Schipani, E.; Olsen, B.R. VEGFA is necessary for chondrocyte survival during bone development. Development 2004, 131, 2161–2171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beagley, K.W.; Gockel, C.M. Regulation of innate and adaptive immunity by the female sex hormones oestradiol and progesterone. FEMS Immunol. Med. Microbiol. 2003, 38, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Kaetzel, C.S. The polymeric immunoglobulin receptor: Bridging innate and adaptive immune responses at mucosal surfaces. Immunol. Rev. 2005, 206, 83–99. [Google Scholar] [CrossRef]
- Richardson, J.M.; Kaushic, C.; Wira, C.R. Polymeric Immunoglobin (Ig) Receptor Production and IgA Transcytosis in Polarized Primary Cultures of Mature Rat Uterine Epithelial Cells. Biol. Reprod. 1995, 53, 488–498. [Google Scholar] [CrossRef]
- Rachman, F.; Casimiri, V.; Psychoyos, A.; Bernard, O. Immunoglobulins in the mouse uterus during the oestrous cycle. J. Reprod. Fertil. 1983, 69, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Kutteh, W.H.; Prince, S.J.; Hammond, K.R.; Kutteh, C.C.; Mestecky, J. Variations in immunoglobulins and IgA subclasses of human uterine cervical secretions around the time of ovulation. Clin. Exp. Immunol. 1996, 104, 538–542. [Google Scholar] [CrossRef]
- Riou, C.; Brionne, A.; Cordeiro, L.; Harichaux, G.; Gargaros, A.; Labas, V.; Gautron, J.; Gérard, N. Proteomic analysis of uterine fluid of fertile and subfertile hens before and after insemination. Reproduction 2019, 158, 335–356. [Google Scholar] [CrossRef]
- Wetendorf, M.; DeMayo, F.J. The progesterone receptor regulates implantation, decidualization, and glandular development via a complex paracrine signaling network. Mol. Cell. Endocrinol. 2012, 357, 108–118. [Google Scholar] [CrossRef] [Green Version]
- Lessey, B.A.; Yeh, I.; Castelbaum, A.J.; Fritz, M.A.; Ilesanmi, A.O.; Korzeniowski, P.; Sun, J.; Chwalisż, K. Endometrial progesterone receptors and markers of uterine receptivity in the window of implantation. Fertil. Steril. 1996, 65, 477–483. [Google Scholar] [CrossRef]
- Hsieh, M.; Lee, D.; Panigone, S.; Horner, K.; Chen, R.; Theologis, A.; Lee, D.C.; Threadgill, D.W.; Conti, M. Luteinizing Hormone-Dependent Activation of the Epidermal Growth Factor Network Is Essential for Ovulation. Mol. Cell. Biol. 2007, 27, 1914–1924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martini, M.; De Santis, M.C.; Braccini, L.; Gulluni, F.; Hirsch, E. PI3K/AKT signaling pathway and cancer: An updated review. Ann. Med. 2014, 46, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Rajareddy, S.; Liu, L.; Jagarlamudi, K.; Boman, K.; Selstam, G.; Reddy, P. Control of mammalian oocyte growth and early follicular development by the oocyte PI3 kinase pathway: New roles for an old timer. Dev. Biol. 2006, 299, 1–11. [Google Scholar] [CrossRef]
- Sansregret, L.; Nepveu, A. The multiple roles of CUX1: Insights from mouse models and cell-based assays. Gene 2008, 412, 84–94. [Google Scholar] [CrossRef]
- Abdollahi-arpanahi, R.; Carvalho, M.R.; Ribeiro, E.S.; Peñagaricano, F. Association of lipid-related genes implicated in conceptus elongation with female fertility traits in dairy cattle. J. Dairy Sci. 2019, 102, 10020–10029. [Google Scholar] [CrossRef]
- Chen, C.; Ouyang, W.; Grigura, V.; Zhou, Q.; Carnes, K.; Lim, H.; Zhao, G.Q.; Arber, S.; Kurpios, N.; Murphy, T.L.; et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 2005, 436, 1030–1034. [Google Scholar] [CrossRef] [Green Version]
- Oatley, J.M.; Avarbock, M.R.; Brinster, R.L. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J. Biol. Chem. 2007, 282, 25842–25851. [Google Scholar] [CrossRef] [Green Version]
- Alankarage, D.; Lavery, R.; Svingen, T.; Kelly, S.; Ludbrook, L.; Bagheri-Fam, S.; Koopman, P.; Harley, V. SOX9 regulates expression of the male fertility gene Ets variant factor 5 (ETV5) during mammalian sex development. Int. J. Biochem. Cell Biol. 2016, 79, 41–51. [Google Scholar] [CrossRef]
- Eo, J.; Shin, H.; Kwon, S.; Song, H.; Murphy, K.M.; Lim, J.H. Complex ovarian defects lead to infertility in Etv5-/- female mice. Mol. Hum. Reprod. 2011, 17, 568–576. [Google Scholar] [CrossRef] [Green Version]
- Guo, G.; Yang, J.; Nichols, J.; Hall, J.S.; Eyres, I.; Mansfield, W.; Smith, A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 2009, 136, 1063–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, Y.; Takeuchi, T.; Mita, S.; Notsu, T.; Mizuguchi, K.; Kyo, S. Krüppel-like factor 4 mediates anti-proliferative effects of progesterone with G0/G1arrest in human endometrial epithelial cells. J. Endocrinol. Investig. 2010, 33, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.C.; Jiang, M.; Beaudet, A.L.; Wu, M.Y. ARID4A and ARID4B regulate male fertility, a functional link to the AR and RB pathways. Proc. Natl. Acad. Sci. USA 2013, 110, 4616–4621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, T.; Banno, K.; Okawa, R.; Yanokura, M.; Iijima, M.; Iriekunitomi, H.; Nakamura, K.; Iida, M.; Adachi, M.; Umene, K.; et al. ARID1A gene mutation in ovarian and endometrial cancers (Review). Oncol. Rep. 2016, 35, 607–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, C.; Han, H.J.; Fan, L.J.; Guan, J.; Zheng, X.B.; Chen, X.; Liang, R.; Zhang, X.W.; Sun, K.K.; Cui, Q.H.; et al. Diverse endometrial mRNA signatures during the window of implantation in patients with repeated implantation failure. Hum. Fertil. 2018, 21, 183–194. [Google Scholar] [CrossRef]
- Harris, T.P.; Schimenti, K.J.; Munroe, R.J.; Schimenti, J.C. IQ motif-containing G (Iqcg) is required for mouse spermiogenesis. G3 Genes Genomes Genet. 2014, 4, 367–372. [Google Scholar] [CrossRef] [Green Version]
- Li, R.K.; Tan, J.L.; Chen, L.T.; Feng, J.S.; Liang, W.X.; Guo, X.J.; Liu, P.; Chen, Z.; Sha, J.H.; Wang, Y.F.; et al. Iqcg is essential for sperm flagellum formation in mice. PLoS ONE 2014, 9, e98053. [Google Scholar] [CrossRef] [Green Version]
- Murakami, M.; Ichisaka, T.; Maeda, M.; Oshiro, N.; Hara, K.; Edenhofer, F.; Kiyama, H.; Yonezawa, K.; Yamanaka, S. mTOR Is Essential for Growth and Proliferation in Early Mouse Embryos and Embryonic Stem Cells. Mol. Cell. Biol. 2004, 24, 6710–6718. [Google Scholar] [CrossRef] [Green Version]
- Gansmo, L.B.; Bjørnslett, M.; Halle, M.K.; Salvesen, H.B.; Dørum, A.; Birkeland, E.; Hveem, K.; Romundstad, P.; Vatten, L.; Lønning, P.E.; et al. The MDM4 SNP34091 (rs4245739) C-allele is associated with increased risk of ovarian—but not endometrial cancer. Tumor Biol. 2016, 24, 6710–6718. [Google Scholar] [CrossRef] [Green Version]
- Forde, N.; Duffy, G.B.; McGettigan, P.A.; Browne, J.A.; Mehta, J.P.; Kelly, A.K.; Mansouri-Attia, N.; Sandra, O.; Loftus, B.J.; Crowe, M.A.; et al. Evidence for an early endometrial response to pregnancy in cattle: Both dependent upon and independent of interferon tau. Physiol. Genom. 2012, 44, 799–810. [Google Scholar] [CrossRef]
- Beis, A.; Zammit, V.A.; Newsholme, E.A. Activities of 3-Hydroxybutyrate Dehydrogenase, 3-Oxoacid CoA-Transferase and Acetoacetyl-CoA Thiolase in Relation to Ketone-Body Utilisation in Muscles from Vertebrates and Invertebrates. Eur. J. Biochem. 1980, 104, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Koundakjian, P.P.; Snoswell, A.M. Ketone body and fatty acid metabolism in sheep tissues. 3-Hydroxybutyrate dehydrogenase, a cytoplasmic enzyme in sheep liver and kidney. Biochem. J. 1970, 119, 49–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cochran, S.D.; Cole, J.B.; Null, D.J.; Hansen, P.J. Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genet. 2013, 14, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Module | Number of Genes | Top Hub-Genes |
|---|---|---|
| Cyan HF | 204 | ANKRD65, TIMM17A, ENSBTAG00000046047, ENSBTAG00000051586, RRP1, PWP2, PSMB5, LTBP2, C11H2orf81, ENSBTAG00000050675 |
| Darkgreen HF | 147 | SREBF2, DACT2, SDHA, PPFIA4, ENSBTAG00000047824, ENSBTAG00000052047, ELF3, ENSBTAG00000051421, NPTN, DHRS4 |
| Grey60 HF | 177 | IARS2, ENSBTAG00000053801, ENSBTAG00000033740, ENSBTAG00000048975, WRB, ENSBTAG00000052845, FAM214A, EIF2AK3, MPV17, MAPKAP1 |
| Lightgreen HF | 136 | CEP104, PKP1, PPP1R12B, ENSBTAG00000051541, ENSBTAG00000049133, ARF6, NUMB, SLC25A15, EEF1AKMT1, PARP4 |
| Purple HF | 227 | TIRAP, PYCR2, FMO2, MIIP, ENSBTAG00000049485, ENSBTAG00000054279, ENSBTAG00000052750, EAF1, SDR39U1, TINF2 |
| Red HF | 237 | STRADA, ENSBTAG00000054600, MDM4, MARK1, KLHL20, CACYBP, ABL2, RABIF, ENSBTAG00000051120, TMEM50B |
| Saddlebrown HF | 114 | NME7, CCDC181, ENSBTAG00000054228, TCTEX1D2, IL20RB, ITGB2, ENSBTAG00000023186, F2RL2, OIP5, DUT |
| Tan HF | 172 | ZMAT2, CPOX, IQCG, WDR53, ENSBTAG00000042475, HACL1, INTS14, ENSBTAG00000043377, REC8, RPS27L |
| Turquoise HF | 544 | SLC45A3, IKBKE, PIGR, PRRX1, TNFRSF1B, SPSB1, CAMTA1, NOL9, TNFRSF25, ARHGEF16 |
| Green HF | 240 | CTCF, MIA3, PSEN2, CASZ1, SDF4, COLGALT2, ENSBTAG00000054874, ENSBTAG00000051836, ENSBTAG00000051084, GOLGB1 |
| Lightgreen SF | 244 | SOX13, TMEM81, COQ8A, ZBTB48, VWA1, EFHD2, PWP2, PLEKHO2, NRDE2, MALL |
| Paleturqouise SF | 130 | MTHFR, ENSBTAG00000031572, JDP2, CCDC142, CD8A, DNAJC27, ENSBTAG00000053045, ENSBTAG00000048432, CABLES2, NECAB3 |
| Gene ID | Mapped Candidate Modules | Top Hub-Gene Candidate Modules | Overall Adjusted p-Value (FDR 5%) |
|---|---|---|---|
| ENSBTAG00000046047 | Cyan HF | Cyan HF | NA * |
| PWP2 | Cyan HF, Lightgreen SF | Cyan HF, Lightgreen SF | 0.112 |
| DACT2 | Darkgreen HF | Darkgreen HF | 0.028 ** |
| MIA3 | Green HF | Green HF | 0.032 ** |
| COLGALT2 | Green HF | Green HF | 0.109 |
| SKA2 | Grey60 HF | Grey60 HF | 0.05 |
| MAPKAP1 | Grey60 HF | Grey60 HF | 0.018 ** |
| PPP1R12B | Lightgreen HF | Lightgreen HF | 0.026 ** |
| SLC25A15 | Lightgreen HF | Lightgreen HF | 0.09 |
| EEF1AKMT1 | Lightgreen HF | Lightgreen HF | 0.1 |
| PARP4 | Lightgreen HF | Lightgreen HF | 0.052 |
| FMO2 | Purple HF | Purple HF | 0.061 |
| MDM4 | Red HF | Red HF | 0.018 ** |
| RABIF | Red HF | Red HF | 0.109 |
| CCDC181 | Saddlebrown HF | Saddlebrown HF | 0.085 |
| F2RL2 | Saddlebrown HF | Saddlebrown HF | 0.028 ** |
| IQCG | Tan HF | Tan HF | 0.022 ** |
| HACL1 | Tan HF | Tan HF | 0.09 |
| PIGR | Turquoise HF | Turquoise HF | 0.026 ** |
| ARHGEF16 | Turquoise HF | Turquoise HF | 0.028 ** |
| NRDE2 | Lightgreen SF | Lightgreen SF | 0.278 |
| IFT80 | Turquoise HF, Paleturquoise SF | Paleturquoise SF | 0.028 ** |
| Gene ID | Mapped Candidate Modules | Regulated Module | Target Genes | Adjusted p-Value (FDR 5%) Upstream Regulation | Overall Adjusted p-Value (FDR 5%) for Prioritization |
|---|---|---|---|---|---|
| EGFR | Darkgreen HF | Turquoise HF | BIRC5, CCNA2, CXCL5, E2F1, EXOSC5, FKBP11, FOXP3, GFAP, HMGB3, HNRNPA1, IGBP1, ITGA6, MYBL2, PDK1, PROM1, PSEN1, RANBP1, SEMA7A, SKP2, TUBA4A, TUBB4A, VEGFA | 0.003 | 0.002 * |
| EGFR | Darkgreen HF | Cyan HF | CCT5, EIF5A, EPS15, GADD45A, NUTF2, ODC1, PPIA, PSMB5, STAT3, TPST1 | 0.003 | 0.002 * |
| ETV5 | - | Turquoise HF | AQP5, CHSY1, KRT19, KRT7, MYB, RAB27A, TJP3, VEGFA | 0.016 | 0.009 * |
| KLF4 | - | Turquoise HF | CCND2, CRABP2, DUSP5, E2F1, HES1, KRT14, KRT19, KRT7, MSX2, PAX2, PROM1, VEGFA, WNT5A | 0.032 | 0.006 * |
| TCHP | - | Turquoise HF | VEGFA | 0.039 | 0.028 * |
| COX7A2 | - | Turquoise HF | STAR | 0.039 | 0.147 |
| PIK3C2A | - | Turquoise HF | VEGFA | 0.039 | 0.009 * |
| ARID4A | - | Turquoise HF | E2F1, FOXP3 | 0.039 | 0.046 * |
| ARID4A | - | Saddlebrown HF | HOXB6 | 0.039 | 0.046 * |
| ARID4A | - | Grey60 HF | HOXB3, HOXB5 | 0.039 | 0.046 * |
| CUX1 | Cyan HF | Turquoise HF | CCNA2, LTF, RAB36, WNT5A | 0.047 | 0.006 * |
| PGR | Turquoise HF | Turquoise HF | AK3, HES1, HPGD, ITGA6, MSX2, NPC1, PDGFA, PGR, PPM1H, PRRX1, TAT | 0.047 | 0.006 * |
| PGR | Turquoise HF | Cyan HF | LIG1, MAP2K3, STAT3, TSC22D3, UCK2, URB2 | 0.047 | 0.006 * |
| IPO9 | - | Turquoise HF | PTK2B | 0.047 | 0.09 |
| DLG1 | Red HF | Tan HF | KCNJ2 | 0.045 | 0.006 * |
| AGER | - | Saddlebrown HF | CCL4, TJP1 | 0.046 | 0.006 * |
| SORT1 | - | Saddlebrown HF | UBE2I | 0.047 | 0.009 * |
| HNRNPAB | Purple HF | Saddlebrown HF | TJP1 | 0.047 | 0.031 * |
| TBX6 | Tan HF | Red HF | HES7 | 0.049 | 0.024 * |
| HSF1 | - | Purple HF | CCT4, FKBP4, HSF2, HSP90AA1, HSPA8, HSPH1, KNTC1, RELA, SPHK2, STIP1 | 0.003 | 0.009 * |
| HSF1 | - | Darkgreen HF | CSRP2, EFEMP1, INHBB, RPL22 | 0.003 | 0.009 * |
| NUB1 | - | Red HF | NEDD8 | 0.046 | 0.088 |
| DPH5 | - | Red HF | NFKBIA, RELA | 0.046 | 0.077 |
| LEPR | Darkgreen HF | Lightgreen HF | ANGPTL4, CDK2, MMP7, PLP1, SOCS2 | 0.044 | 0.006 * |
| DYSF | Tan HF | Lightgreen HF | CD48, DNAJB1, FCGR2B | 0.044 | 0.012 * |
| API5 | Purple HF | Lightgreen HF | CDK2 | 0.044 | 0.077 |
| BCKDK | - | Lightgreen HF | PLP1 | 0.047 | 0.041 * |
| DUSP16 | Turquoise HF | Lightgreen HF | VCAM1 | 0.047 | 0.014 * |
| CHFR | Cyan HF | Grey60 HF | PLK1 | 0.046 | 0.032 * |
| PROM1 | Turquoise HF | Green HF | DSG2 | 0.044 | 0.006 * |
| ERN2 | Grey60 HF | Green HF | XBP1 | 0.046 | 0.031 * |
| UTP3 | - | Darkgreen HF | IGLL1/IGLL5 | 0.043 | 0.112 |
| RDH10 | - | Darkgreen HF | RDH5 | 0.045 | 0.032 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca, P.A.S.; Suárez-Vega, A.; Cánovas, A. Weighted Gene Correlation Network Meta-Analysis Reveals Functional Candidate Genes Associated with High- and Sub-Fertile Reproductive Performance in Beef Cattle. Genes 2020, 11, 543. https://doi.org/10.3390/genes11050543
Fonseca PAS, Suárez-Vega A, Cánovas A. Weighted Gene Correlation Network Meta-Analysis Reveals Functional Candidate Genes Associated with High- and Sub-Fertile Reproductive Performance in Beef Cattle. Genes. 2020; 11(5):543. https://doi.org/10.3390/genes11050543
Chicago/Turabian StyleFonseca, Pablo A. S., Aroa Suárez-Vega, and Angela Cánovas. 2020. "Weighted Gene Correlation Network Meta-Analysis Reveals Functional Candidate Genes Associated with High- and Sub-Fertile Reproductive Performance in Beef Cattle" Genes 11, no. 5: 543. https://doi.org/10.3390/genes11050543





