Human Nasal Epithelial Organoids for Therapeutic Development in Cystic Fibrosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Cell Culture and Expansion
2.3. Differentiated HNE Culture
2.4. Organoid Fixation and Immunoflurescence
2.5. Imaging and Analysis of Organoids
2.6. Short-Circuit Current Measurements of Monolayers
2.7. Statistical Analysis
3. Results
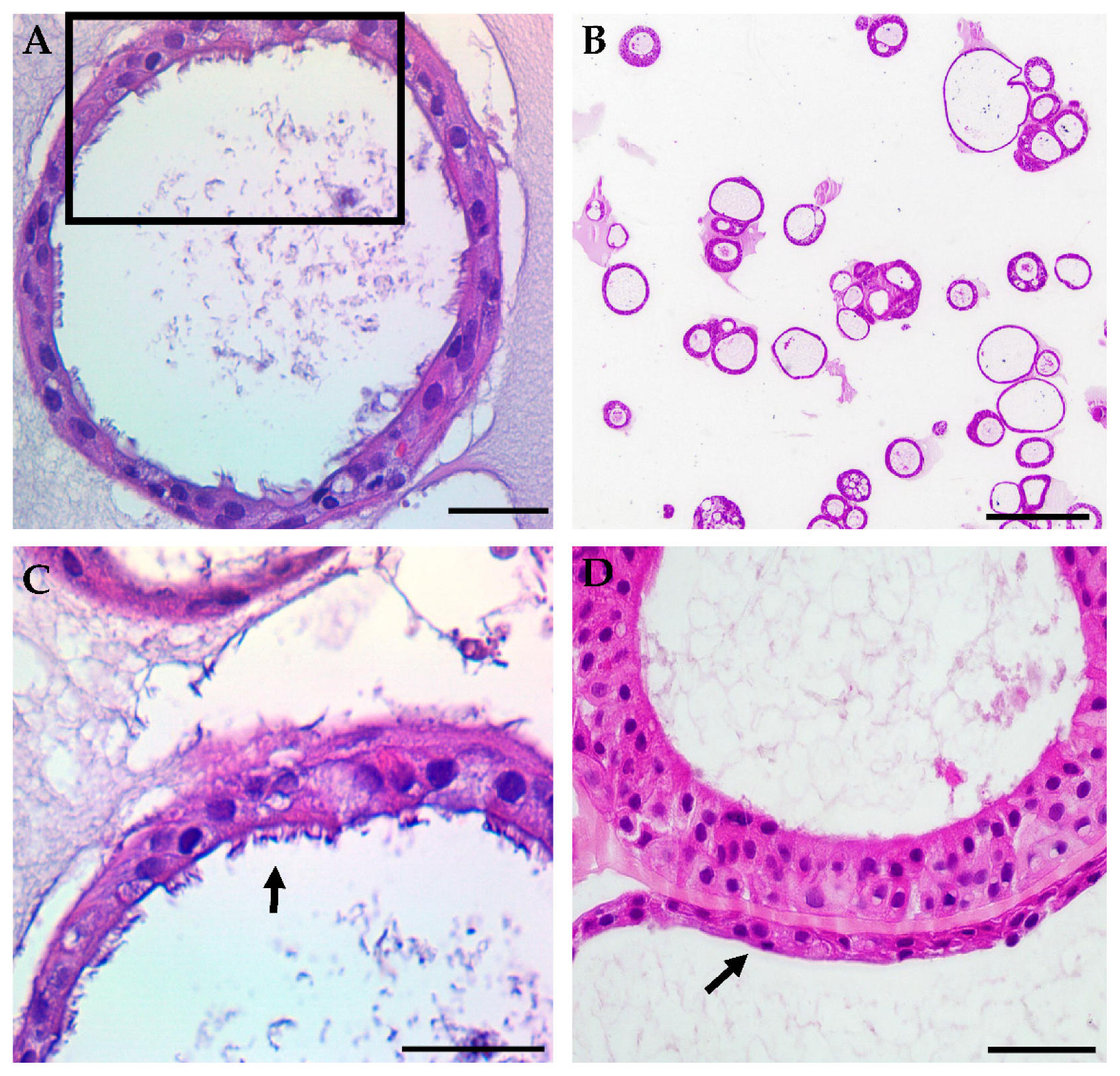
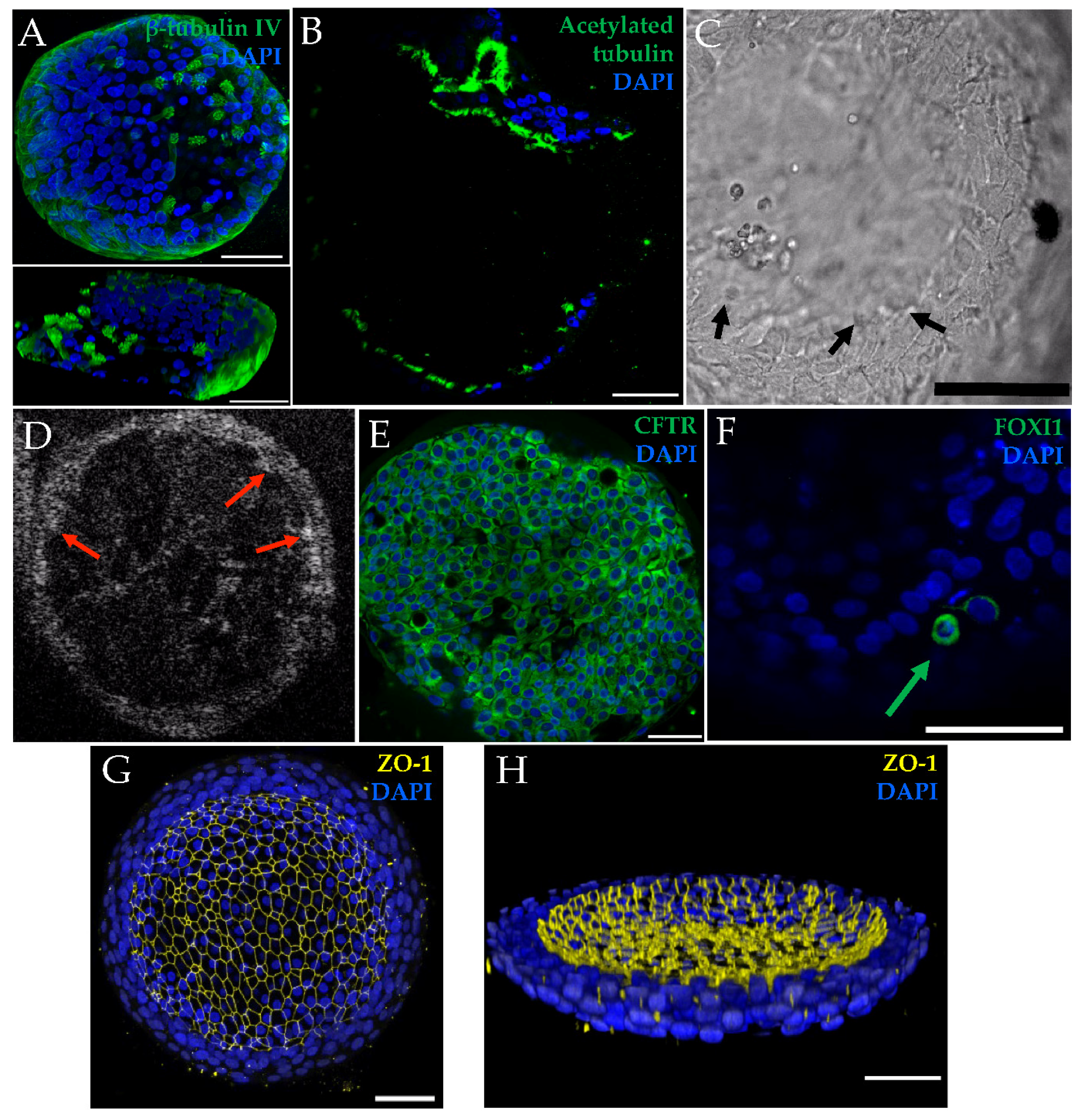
3.1. HNE Organoid Morphology Reflects That of Airway Epithelial Layers In Vivo
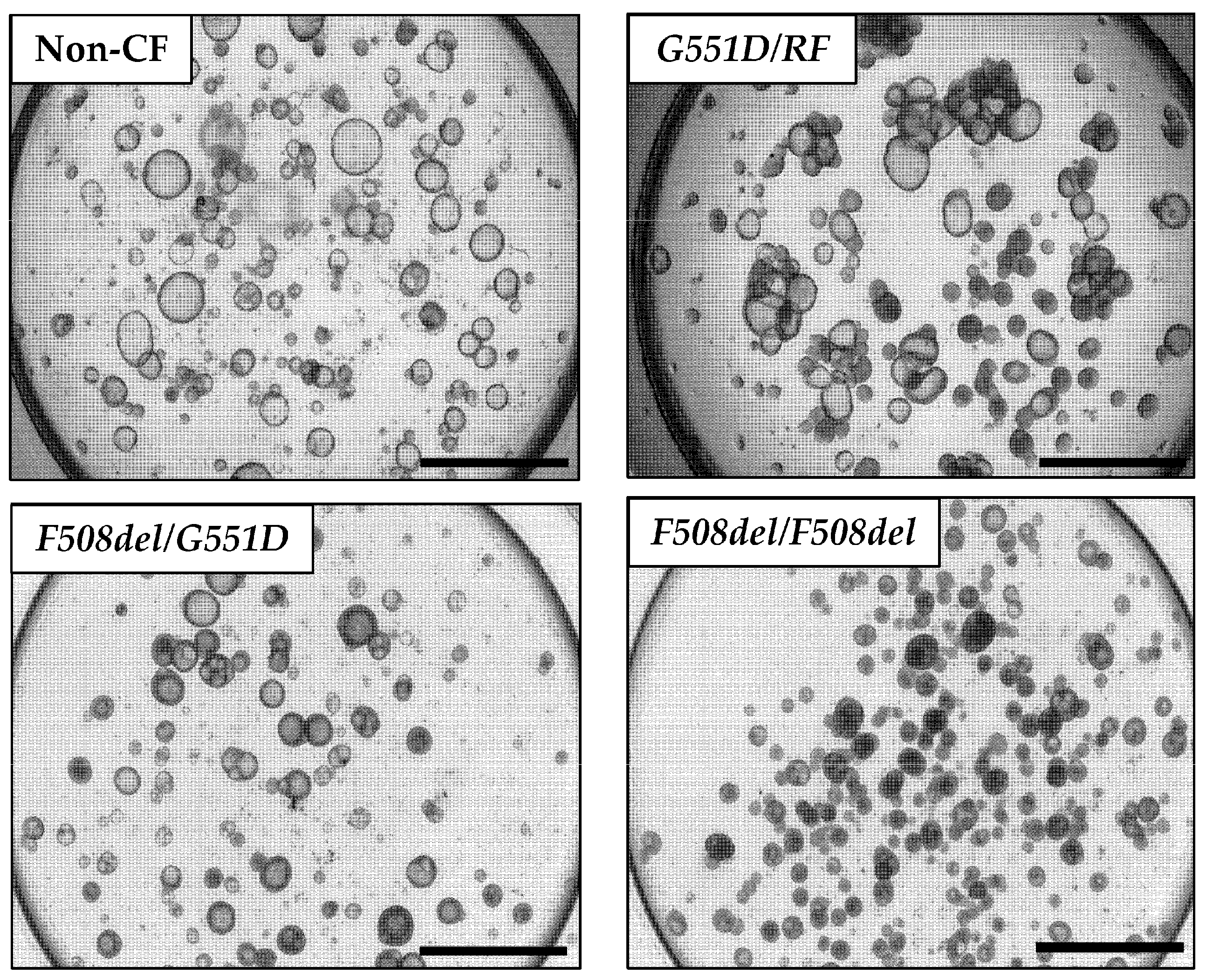
3.2. Organoid Lumen Size Differs between Different CFTR Genotypes
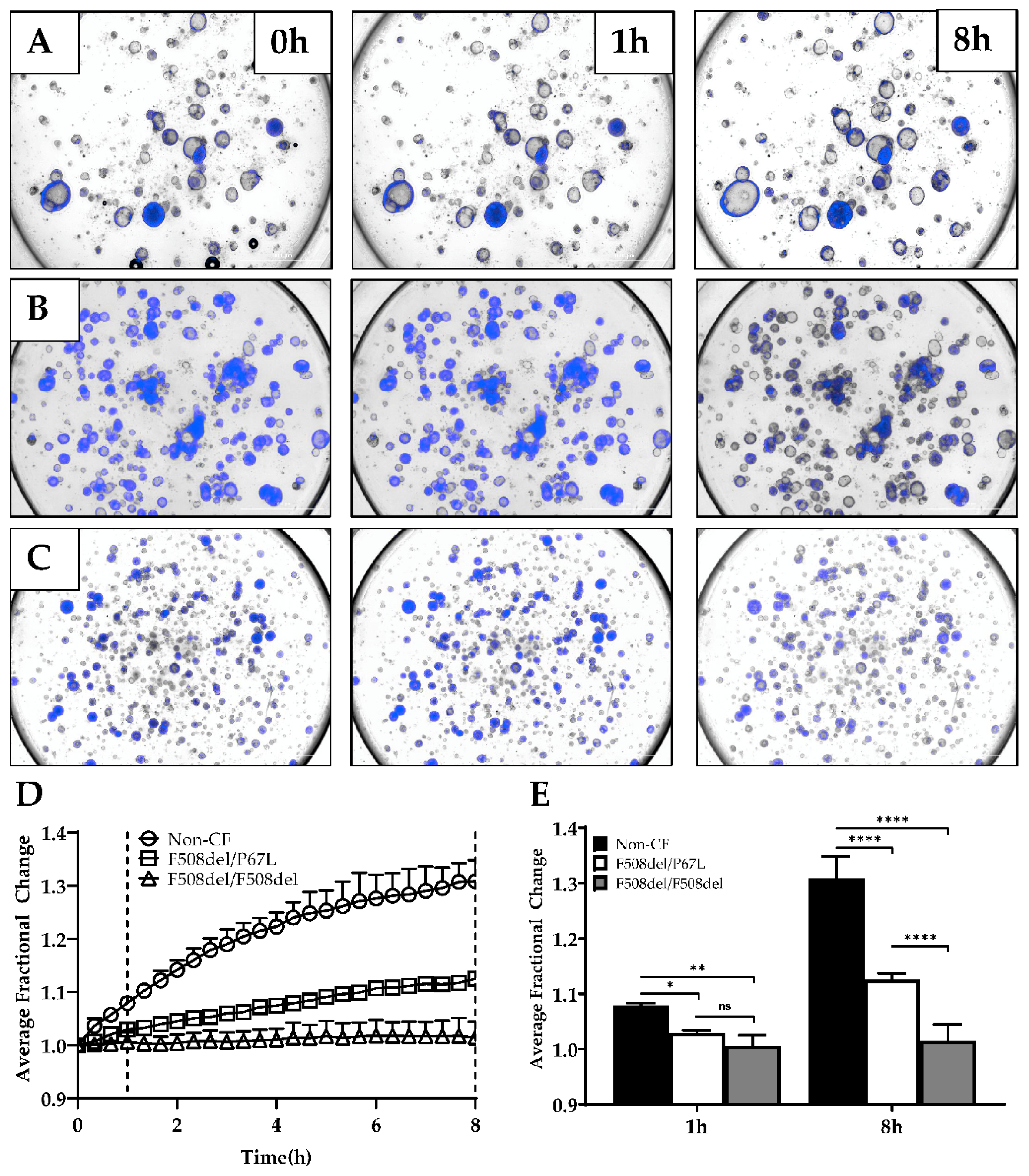
3.3. Optimization of Forskolin-Induced CFTR-Dependent Swelling Assay
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CF | cystic fibrosis |
| CFTR | Cystic Fibrosis Transmembrane conductance regulator |
References
- Bell, S.C.; De Boeck, K.; Amaral, M.D. New pharmacological approaches for cystic fibrosis: Promises, progress, pitfalls. Pharmacol. Ther. 2015, 145, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Bertoncini, E.; Colomb-Lippa, D. Pulmonology: CFTR modulators for cystic fibrosis. J. Am. Acad. Physician Assist. 2013, 26, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Derichs, N. Targeting a genetic defect: Cystic fibrosis transmembrane conductance regulator modulators in cystic fibrosis. Eur. Respir. Rev. 2013, 22, 58–65. [Google Scholar] [CrossRef]
- Middleton, P.G.; Mall, M.A.; Drevinek, P.; Lands, L.C.; McKone, E.F.; Polineni, D.; Ramsey, B.W.; Taylor-Cousar, J.L.; Tullis, E.; Vermeulen, F.; et al. Elexacaftor-Tezacaftor-Ivacaftor for Cystic Fibrosis with a Single Phe508del Allele. N. Engl. J. Med. 2019, 381, 1809–1819. [Google Scholar] [CrossRef] [PubMed]
- Heijerman, H.G.M.; McKone, E.F.; Downey, D.G.; Van Braeckel, E.; Rowe, S.M.; Tullis, E.; Mall, M.A.; Welter, J.J.; Ramsey, B.W.; McKee, C.M.; et al. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: A double-blind, randomised, phase 3 trial. Lancet 2019. [Google Scholar] [CrossRef]
- Pedersen, P.S.; Braunstein, T.H.; Jorgensen, A.; Larsen, P.L.; Holstein-Rathlou, N.H.; Frederiksen, O. Stimulation of aquaporin-5 and transepithelial water permeability in human airway epithelium by hyperosmotic stress. Pflug. Arch. 2007, 453, 777–785. [Google Scholar] [CrossRef]
- Pedersen, P.S.; Frederiksen, O.; Holstein-Rathlou, N.H.; Larsen, P.L.; Qvortrup, K. Ion transport in epithelial spheroids derived from human airway cells. Am. J. Physiol. 1999, 276, L75–L80. [Google Scholar] [CrossRef]
- Pedersen, P.S.; Holstein-Rathlou, N.H.; Larsen, P.L.; Qvortrup, K.; Frederiksen, O. Fluid absorption related to ion transport in human airway epithelial spheroids. Am. J. Physiol. 1999, 277, L1096–L1103. [Google Scholar] [CrossRef]
- Guimbellot, J.S.; Leach, J.M.; Chaudhry, I.G.; Quinney, N.L.; Boyles, S.E.; Chua, M.; Aban, I.; Jaspers, I.; Gentzsch, M. Nasospheroids permit measurements of CFTR-dependent fluid transport. JCI Insight 2017, 2. [Google Scholar] [CrossRef]
- Brewington, J.J.; Filbrandt, E.T.; LaRosa, F.J., 3rd; Ostmann, A.J.; Strecker, L.M.; Szczesniak, R.D.; Clancy, J.P. Detection of CFTR function and modulation in primary human nasal cell spheroids. J. Cyst. Fibros 2017. [Google Scholar] [CrossRef]
- Birket, S.E.; Chu, K.K.; Houser, G.H.; Liu, L.; Fernandez, C.M.; Solomon, G.M.; Lin, V.; Shastry, S.; Mazur, M.; Sloane, P.A.; et al. Combination therapy with cystic fibrosis transmembrane conductance regulator modulators augment the airway functional microanatomy. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 310, L928–L939. [Google Scholar] [CrossRef]
- Birket, S.E.; Chu, K.K.; Liu, L.; Houser, G.H.; Diephuis, B.J.; Wilsterman, E.J.; Dierksen, G.; Mazur, M.; Shastry, S.; Li, Y.; et al. A functional anatomic defect of the cystic fibrosis airway. Am. J. Respir. Crit. Care Med. 2014, 190, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Chu, K.K.; Mojahed, D.; Fernandez, C.M.; Li, Y.; Liu, L.; Wilsterman, E.J.; Diephuis, B.; Birket, S.E.; Bowers, H.; Martin Solomon, G.; et al. Particle-Tracking Microrheology Using Micro-Optical Coherence Tomography. Biophys. J. 2016, 111, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Chu, K.K.; Unglert, C.; Ford, T.N.; Cui, D.; Carruth, R.W.; Singh, K.; Liu, L.; Birket, S.E.; Solomon, G.M.; Rowe, S.M.; et al. In vivo imaging of airway cilia and mucus clearance with micro-optical coherence tomography. Biomed. Opt. Express 2016, 7, 2494–2505. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chu, K.K.; Houser, G.H.; Diephuis, B.J.; Li, Y.; Wilsterman, E.J.; Shastry, S.; Dierksen, G.; Birket, S.E.; Mazur, M.; et al. Method for quantitative study of airway functional microanatomy using micro-optical coherence tomography. PLoS ONE 2013, 8, e54473. [Google Scholar] [CrossRef]
- Tuggle, K.L.; Birket, S.E.; Cui, X.; Hong, J.; Warren, J.; Reid, L.; Chambers, A.; Ji, D.; Gamber, K.; Chu, K.K.; et al. Characterization of defects in ion transport and tissue development in cystic fibrosis transmembrane conductance regulator (CFTR)-knockout rats. PLoS ONE 2014, 9, e91253. [Google Scholar] [CrossRef]
- Chapman, S.; Liu, X.; Meyers, C.; Schlegel, R.; McBride, A.A. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Investig. 2010, 120, 2619–2626. [Google Scholar] [CrossRef]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef]
- Avramescu, R.G.; Kai, Y.; Xu, H.; Bidaud-Meynard, A.; Schnur, A.; Frenkiel, S.; Matouk, E.; Veit, G.; Lukacs, G.L. Mutation-specific downregulation of CFTR2 variants by gating potentiators. Hum. Mol. Genet. 2017, 26, 4873–4885. [Google Scholar] [CrossRef]
- De Courcey, F.; Zholos, A.V.; Atherton-Watson, H.; Williams, M.T.; Canning, P.; Danahay, H.L.; Elborn, J.S.; Ennis, M. Development of primary human nasal epithelial cell cultures for the study of cystic fibrosis pathophysiology. Am. J. Physiol. Cell Physiol. 2012, 303, C1173–C1179. [Google Scholar] [CrossRef]
- Mosler, K.; Coraux, C.; Fragaki, K.; Zahm, J.M.; Bajolet, O.; Bessaci-Kabouya, K.; Puchelle, E.; Abely, M.; Mauran, P. Feasibility of nasal epithelial brushing for the study of airway epithelial functions in CF infants. J. Cyst. Fibros 2008, 7, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Martinovich, K.M.; Iosifidis, T.; Buckley, A.G.; Looi, K.; Ling, K.M.; Sutanto, E.N.; Kicic-Starcevich, E.; Garratt, L.W.; Shaw, N.C.; Montgomery, S.; et al. Conditionally reprogrammed primary airway epithelial cells maintain morphology, lineage and disease specific functional characteristics. Sci. Rep. 2017, 7, 17971. [Google Scholar] [CrossRef]
- Neuberger, T.; Burton, B.; Clark, H.; Van Goor, F. Use of primary cultures of human bronchial epithelial cells isolated from cystic fibrosis patients for the pre-clinical testing of CFTR modulators. In Cystic Fibrosis; Springer: Berlin/Heidelberg, Germany, 2011; pp. 39–54. [Google Scholar]
- Brewington, J.J.; Filbrandt, E.T.; LaRosa, F.J., 3rd; Moncivaiz, J.D.; Ostmann, A.J.; Strecker, L.M.; Clancy, J.P. Brushed nasal epithelial cells are a surrogate for bronchial epithelial CFTR studies. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhao, H.; Dong, H.; Wu, Y.; Yao, L.; Zou, F.; Cai, S. High-mobility group box 1 impairs airway epithelial barrier function through the activation of the RAGE/ERK pathway. Int. J. Mol. Med. 2016, 37, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- McCray, T.; Richards, Z.; Marsili, J.; Prins, G.S.; Nonn, L. Handling and Assessment of Human Primary Prostate Organoid Culture. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [PubMed]
- Moonwiriyakit, A.; Koval, M.; Muanprasat, C. Pharmacological stimulation of G-protein coupled receptor 40 alleviates cytokine-induced epithelial barrier disruption in airway epithelial Calu-3 cells. Int. Immunopharmacol. 2019, 73, 353–361. [Google Scholar] [CrossRef]
- Park, H.R.; O’Sullivan, M.; Vallarino, J.; Shumyatcher, M.; Himes, B.E.; Park, J.A.; Christiani, D.C.; Allen, J.; Lu, Q. Transcriptomic response of primary human airway epithelial cells to flavoring chemicals in electronic cigarettes. Sci. Rep. 2019, 9, 1400. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, J.F.; Wiegerinck, C.L.; de Jonge, H.R.; Bronsveld, I.; Janssens, H.M.; de Winter-de Groot, K.M.; Brandsma, A.M.; de Jong, N.W.; Bijvelds, M.J.; Scholte, B.J.; et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013, 19, 939–945. [Google Scholar] [CrossRef]
- Wojtkowski, M.; Srinivasan, V.; Ko, T.; Fujimoto, J.; Kowalczyk, A.; Duker, J. Ultrahigh-resolution, high-speed, Fourier domain optical coherence tomography and methods for dispersion compensation. Opt. Express 2004, 12, 2404–2422. [Google Scholar] [CrossRef]
- Liu, Z.; Mackay, S.; Gordon, D.M.; Anderson, J.D.; Haithcock, D.W.; Garson, C.J.; Tearney, G.J.; Solomon, G.M.; Pant, K.; Prabhakarpandian, B.; et al. Co-cultured microfluidic model of the airway optimized for microscopy and micro-optical coherence tomography imaging. Biomed. Opt. Express 2019, 10, 5414–5430. [Google Scholar] [CrossRef]
- Rowe, S.M.; Pyle, L.C.; Jurkevante, A.; Varga, K.; Collawn, J.; Sloane, P.A.; Woodworth, B.; Mazur, M.; Fulton, J.; Fan, L.; et al. DeltaF508 CFTR processing correction and activity in polarized airway and non-airway cell monolayers. Pulm. Pharmacol. Ther. 2010, 23, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Ermund, A.; Meiss, L.N.; Rodriguez-Pineiro, A.M.; Bahr, A.; Nilsson, H.E.; Trillo-Muyo, S.; Ridley, C.; Thornton, D.J.; Wine, J.J.; Hebert, H.; et al. The normal trachea is cleaned by MUC5B mucin bundles from the submucosal glands coated with the MUC5AC mucin. Biochem. Biophys. Res. Commun. 2017, 492, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Schogler, A.; Blank, F.; Brugger, M.; Beyeler, S.; Tschanz, S.A.; Regamey, N.; Casaulta, C.; Geiser, T.; Alves, M.P. Characterization of pediatric cystic fibrosis airway epithelial cell cultures at the air-liquid interface obtained by non-invasive nasal cytology brush sampling. Respir. Res. 2017, 18, 215. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.P.; Bear, C.E.; Chin, S.; Pasceri, P.; Thompson, T.O.; Huan, L.J.; Ratjen, F.; Ellis, J.; Rossant, J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012, 30, 876–882. [Google Scholar] [CrossRef]
- de Winter-de Groot, K.M.; Janssens, H.M.; van Uum, R.T.; Dekkers, J.F.; Berkers, G.; Vonk, A.; Kruisselbrink, E.; Oppelaar, H.; Vries, R.; Clevers, H.; et al. Stratifying infants with cystic fibrosis for disease severity using intestinal organoid swelling as a biomarker of CFTR function. Eur. Respir. J. 2018, 52. [Google Scholar] [CrossRef] [PubMed]
- Schwank, G.; Koo, B.K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Berkers, G.; Kruisselbrink, E.; Vonk, A.; de Jonge, H.R.; Janssens, H.M.; Bronsveld, I.; van de Graaf, E.A.; Nieuwenhuis, E.E.; Houwen, R.H.; et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 2016, 8, 344ra84. [Google Scholar] [CrossRef]
- Muller, L.; Brighton, L.E.; Carson, J.L.; Fischer, W.A., 2nd; Jaspers, I. Culturing of human nasal epithelial cells at the air liquid interface. J. Vis. Exp. 2013. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Anderson, J.D.; Deng, L.; Mackay, S.; Bailey, J.; Kersh, L.; Rowe, S.M.; Guimbellot, J.S. Human Nasal Epithelial Organoids for Therapeutic Development in Cystic Fibrosis. Genes 2020, 11, 603. https://doi.org/10.3390/genes11060603
Liu Z, Anderson JD, Deng L, Mackay S, Bailey J, Kersh L, Rowe SM, Guimbellot JS. Human Nasal Epithelial Organoids for Therapeutic Development in Cystic Fibrosis. Genes. 2020; 11(6):603. https://doi.org/10.3390/genes11060603
Chicago/Turabian StyleLiu, Zhongyu, Justin D. Anderson, Lily Deng, Stephen Mackay, Johnathan Bailey, Latona Kersh, Steven M. Rowe, and Jennifer S. Guimbellot. 2020. "Human Nasal Epithelial Organoids for Therapeutic Development in Cystic Fibrosis" Genes 11, no. 6: 603. https://doi.org/10.3390/genes11060603
APA StyleLiu, Z., Anderson, J. D., Deng, L., Mackay, S., Bailey, J., Kersh, L., Rowe, S. M., & Guimbellot, J. S. (2020). Human Nasal Epithelial Organoids for Therapeutic Development in Cystic Fibrosis. Genes, 11(6), 603. https://doi.org/10.3390/genes11060603




