Transcriptome Analysis of Genes Involved in Cold Hardiness of Peach Tree (Prunus persica) Shoots during Cold Acclimation and Deacclimation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Cold Hardiness Determination
2.3. Total RNA Extraction
2.4. cDNA Library Construction, Sequencing, and Assembly
2.5. Identification and Functional Annotation of the DEGs
2.6. Reverse Transcription Quantitative Polymerase Chain Reaction (qPCR) Analysis
2.7. Statistical Analysis
3. Results and Discussion
3.1. Changes in Cold Hardiness
3.2. Transcriptome Sequencing Data Statistics
3.3. DEG Analysis
3.4. Functional Annotation and GO Term Enrichment of the DEGs during CA, from Early to Late CA
3.5. Functional Annotation and GO Term Enrichment of the DEGs during DA, from Late CA to Late DA
3.6. KEGG Category Enrichment of the DEGs
3.7. Validation of In Silico DEG Data Using qPCR
3.8. Selection of the DEGs Involved in the Cold Hardiness of Peach Tree Shoots
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [Green Version]
- Kozlowski, T.T.; Pallardy, S.G. Acclimation and adaptive responses of woody plants to environmental stresses. Bot. Rev. 2002, 68, 270–334. [Google Scholar] [CrossRef]
- Kalberer, S.R.; Wisniewski, M.; Arora, R. Deacclimation and reacclimation of cold-hardy plants: Current understanding and emerging concepts. Plant Sci. 2006, 171, 3–16. [Google Scholar] [CrossRef]
- Welling, A.; Palva, E.T. Molecular control of cold acclimation in trees. Physiol. Plant. 2008, 127, 167–181. [Google Scholar] [CrossRef]
- Kim, S.A.; Ahn, S.Y.; Yun, H.K. Transcriptome analysis of grapevine shoots exposed to chilling temperature for four weeks. Hortic. Environ. Biotechnol. 2016, 57, 161–172. [Google Scholar] [CrossRef]
- Wang, Q.; Cao, K.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, L. Comparative transcriptome analysis of genes involved in the response of resistant and susceptible peach cultivars to water stress. Sci. Hortic. 2019, 245, 29–38. [Google Scholar] [CrossRef]
- Artlip, T.S.; Wisniewski, M.E.; Bassett, C.L.; Norelli, J.L. CBF gene expression in peach leaf and bark tissues is gated by a circadian clock. Tree Physiol. 2013, 33, 866–877. [Google Scholar] [CrossRef] [Green Version]
- Bassett, C.L.; Fisher, K.M.; Farrell, R.E. The complete peach dehydrin family: Characterization of three recently recognized genes. Tree Genet. Genom. 2015, 11, 126. [Google Scholar] [CrossRef]
- Jiao, Y.; Shen, Z.; Yan, J. Transcriptome analysis of peach [Prunus persica (L.) Batsch] stigma in response to low-temperature stress with digital gene expression profiling. J. Plant Biochem. Biotechnol. 2017, 26, 141–148. [Google Scholar] [CrossRef]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F.; et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication, and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef] [Green Version]
- Pagter, M.; Jensen, C.R.; Peterson, K.K.; Liu, F.; Arora, R. Changes in carbohydrates, ABA, and bark proteins during seasonal cold acclimation and deacclimation in Hydrangea species differing in cold hardiness. Physiol. Plant. 2008, 134, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yu, D.J.; Kim, S.J.; Choi, D.; Lee, H.J. Intraspecies differences in cold hardiness, carbohydrate content, and β-amylase gene expression of Vaccinium corymbosum during cold acclimation and deacclimation. Tree Physiol. 2012, 32, 1533–1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, R.; Wisniewski, M.E.; Scorza, R. Cold acclimation in genetically related (sibling) deciduous and evergreen peach (Prunus persica [L.] Batsch). 1. Seasonal changes in cold hardiness and polypeptides of bark and xylem tissues. Plant Physiol. 1992, 99, 1562–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, D.J.; Hwang, J.Y.; Chung, S.W.; Oh, H.D.; Yun, S.K.; Lee, H.J. Changes in cold hardiness and carbohydrate content in peach (Prunus persica) trunk bark and wood tissues during cold acclimation and deacclimation. Sci. Hortic. 2017, 219, 45–52. [Google Scholar] [CrossRef]
- Arora, R.; Rowland, L.J.; Ogden, E.L.; Dhanaraj, A.L.; Marian, C.O.; Ehlenfeldt, M.K.; Vinyard, B. Dehardening kinetics, bud development, and dehydrin metabolism in blueberry cultivars during deacclimation at constant, warm temperatures. J. Amer. Soc. Hortic. Sci. 2004, 129, 667–674. [Google Scholar] [CrossRef] [Green Version]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef]
- Zifkin, M.; Jin, A.; Ozga, J.A.; Zaharia, L.I.; Schernthaner, J.P.; Gesell, A.; Abrams, S.R.; Kennedy, J.A.; Constabel, C.P. Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 2012, 158, 200–224. [Google Scholar] [CrossRef] [Green Version]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Cur. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef] [Green Version]
- Gall, H.L.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [Green Version]
- Dutta, A.; Bahar, I. Metal-binding sites are designed to achieve optimal mechanical and signaling properties. Structure 2010, 18, 1140–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, P.K.; Gupta, K.; Lopato, S.; Agarwal, P. Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 2017, 68, 2135–2148. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, T.; Xia, H.; Liu, J.; Ma, F. The gene family of dehydration responsive element-binding transcription factors in grape (Vitis vinifera): Genome-wide identification and analysis, expression profiles, and involvement in abiotic stress resistance. Mol. Biol. Rep. 2014, 41, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhu, J.K. The broad roles of CBF genes: From development to abiotic stress. Plant Signal. Behav. 2016, 11, e1215794. [Google Scholar] [CrossRef] [Green Version]
- Wentzinger, L.F.; Bach, T.J.; Hartmann, M.A. Inhibition of squalene synthase and squalene epoxidase in tobacco cells triggers an up-regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Physiol. 2002, 130, 334–346. [Google Scholar] [CrossRef]
- Dubois, F.; Tercé-Laforgue, T.; Gonzalez-Moro, M.B.; Estavillo, J.M.; Sangwan, R.; Gallais, A.; Hirel, B. Glutamate dehydrogenase in plants: Is there a new story for an old enzyme? Plant Physiol. Biochem. 2003, 42, 565–576. [Google Scholar] [CrossRef]
- Rylott, E.L.; Gilday, A.D.; Graham, I.A. The gluconeogenic enzyme phosphoenolpyruvate carboxykinase in Arabidopsis is essential for seedling establishment. Plant Physiol. 2003, 131, 1834–1842. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Witten, D.M.; Johnstone, I.M.; Tibshirani, R. Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics 2012, 13, 523–538. [Google Scholar] [CrossRef]
- Nordin, K.; Vahala, T.; Palva, E.T. Differential expression of two related, low-temperature genes in Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 1993, 21, 641–653. [Google Scholar] [CrossRef]
- Visconti, S.; D’Ambrosio, C.; Fiorillo, A.; Arena, S.; Muzi, C.; Zottini, M.; Aducci, P.; Marra, M.; Scaloni, A.; Camoni, L. Overexpression of 14-3-3 proteins enhances cold tolerance and increases level of stress-responsive proteins of Arabidopsis plants. Plant Sci. 2019, 289, 110215. [Google Scholar] [CrossRef] [PubMed]
- Olvera-Carrillo, Y.; Reyes, J.L.; Covarrubias, A.A. Late embryogenesis abundant proteins: Versatile players in the plant adaptation to water limiting environments. Plant Signal. Behav. 2011, 6, 586–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoundi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.C.S.; Edsgärd, D.; Hussain, S.S.; Alquezar, D.; Rasmussen, M.; Gilbert, T.; Nielsen, B.H.; Bartels, D.; Mundy, J. Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 2010, 63, 212–228. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.W.; Yu, D.J.; Oh, H.D.; Ahn, J.H.; Huh, J.H.; Lee, H.J. Transcriptional regulation of abscisic acid biosynthesis and signal transduction, and anthocyanin biosynthesis in ‘Bluecrop’ highbush blueberry fruit during ripening. PLoS ONE 2019, 14, e0220015. [Google Scholar] [CrossRef] [Green Version]
- Grün, S.; Lindermayr, C.; Sell, S.; Durner, J. Nitric oxide and gene regulation in plants. J. Exp. Bot. 2006, 57, 507–516. [Google Scholar] [CrossRef] [Green Version]
- Ouellet, F.; Charron, J.B. Cold Acclimation and Freezing Tolerance in Plants; eLS. John Wiley & Sons, Ltd.: Chichester, UK, 2013. [Google Scholar]
- Thomashow, M.F. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 1998, 118, 1–7. [Google Scholar] [CrossRef] [Green Version]
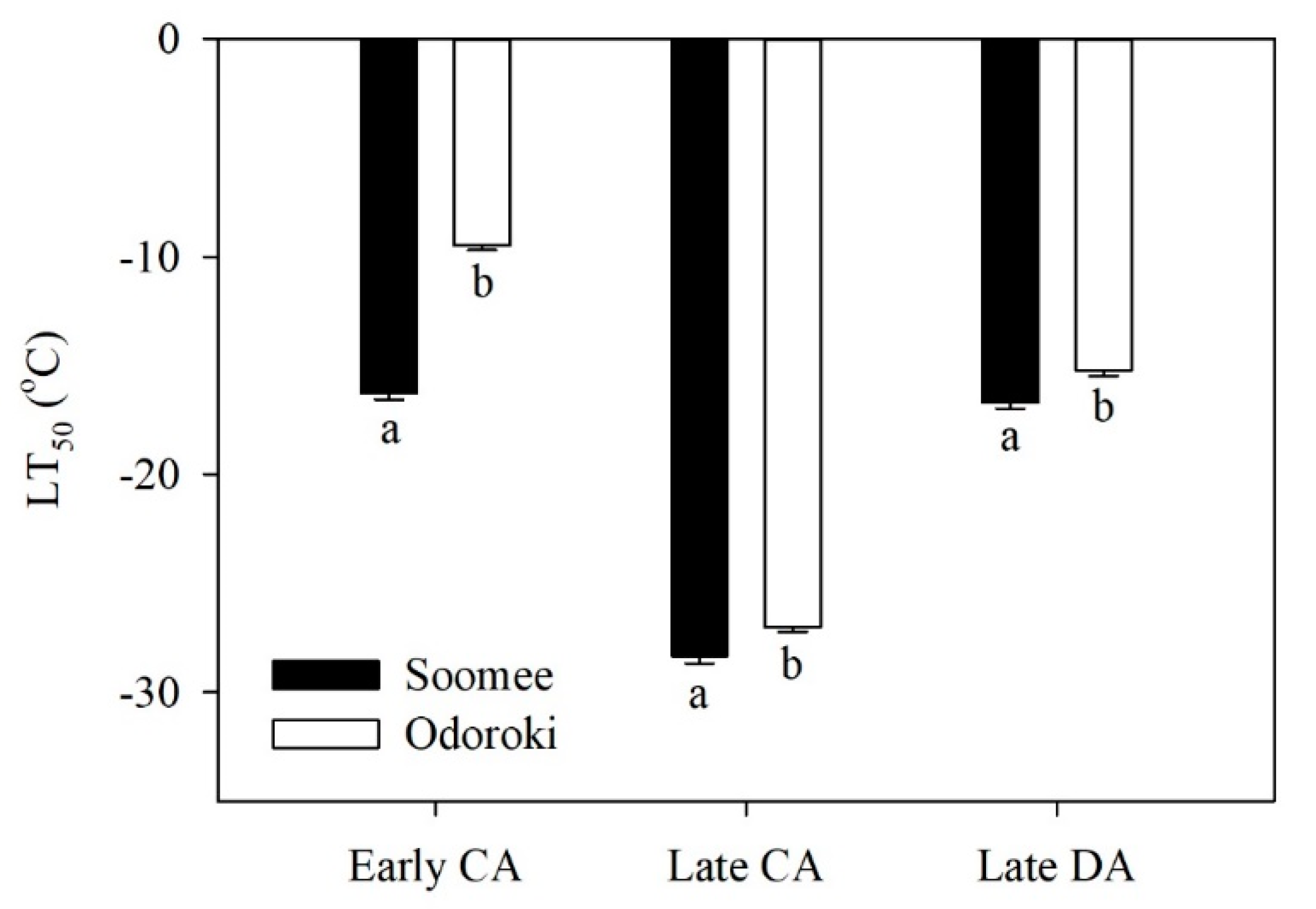
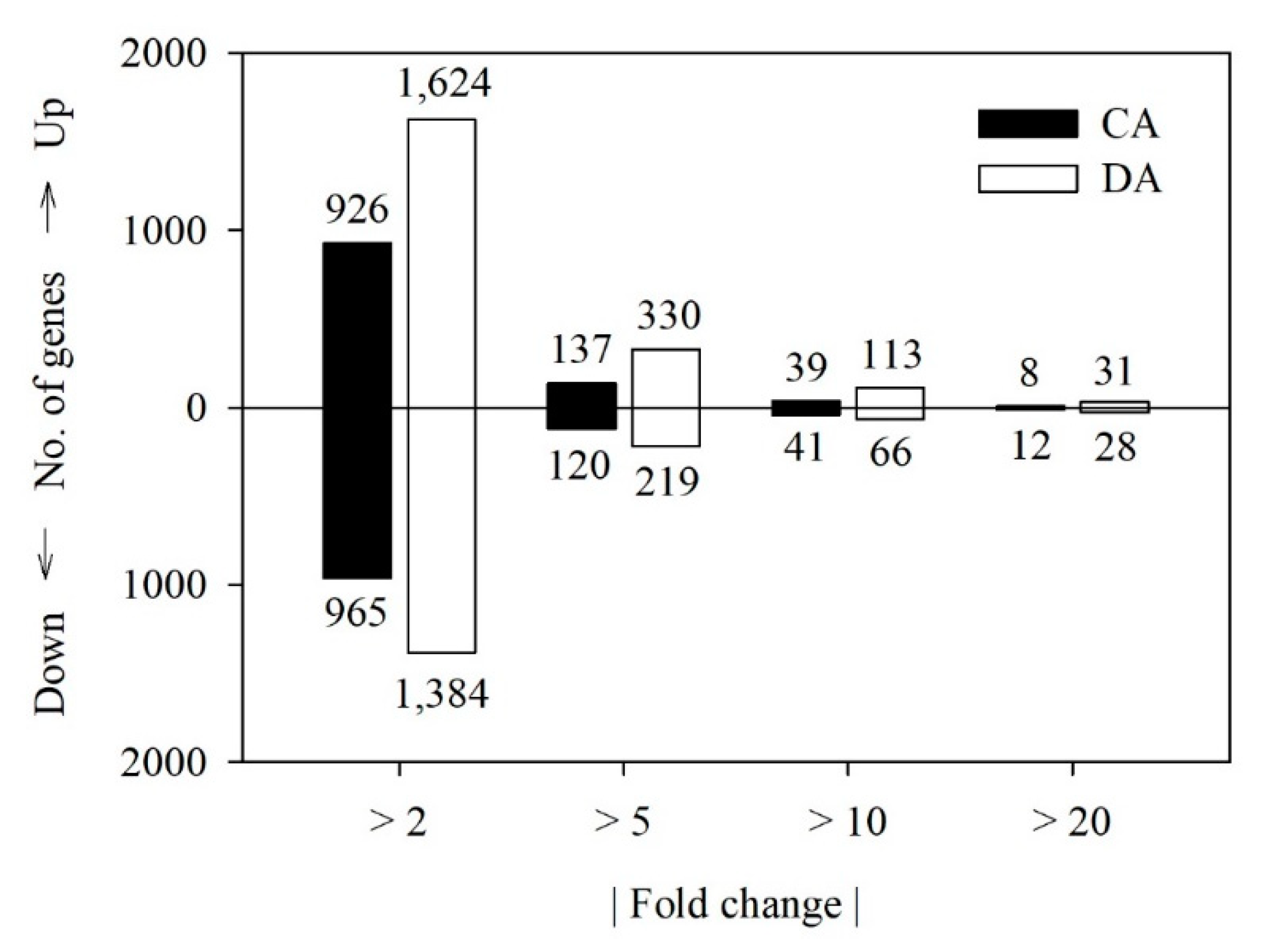

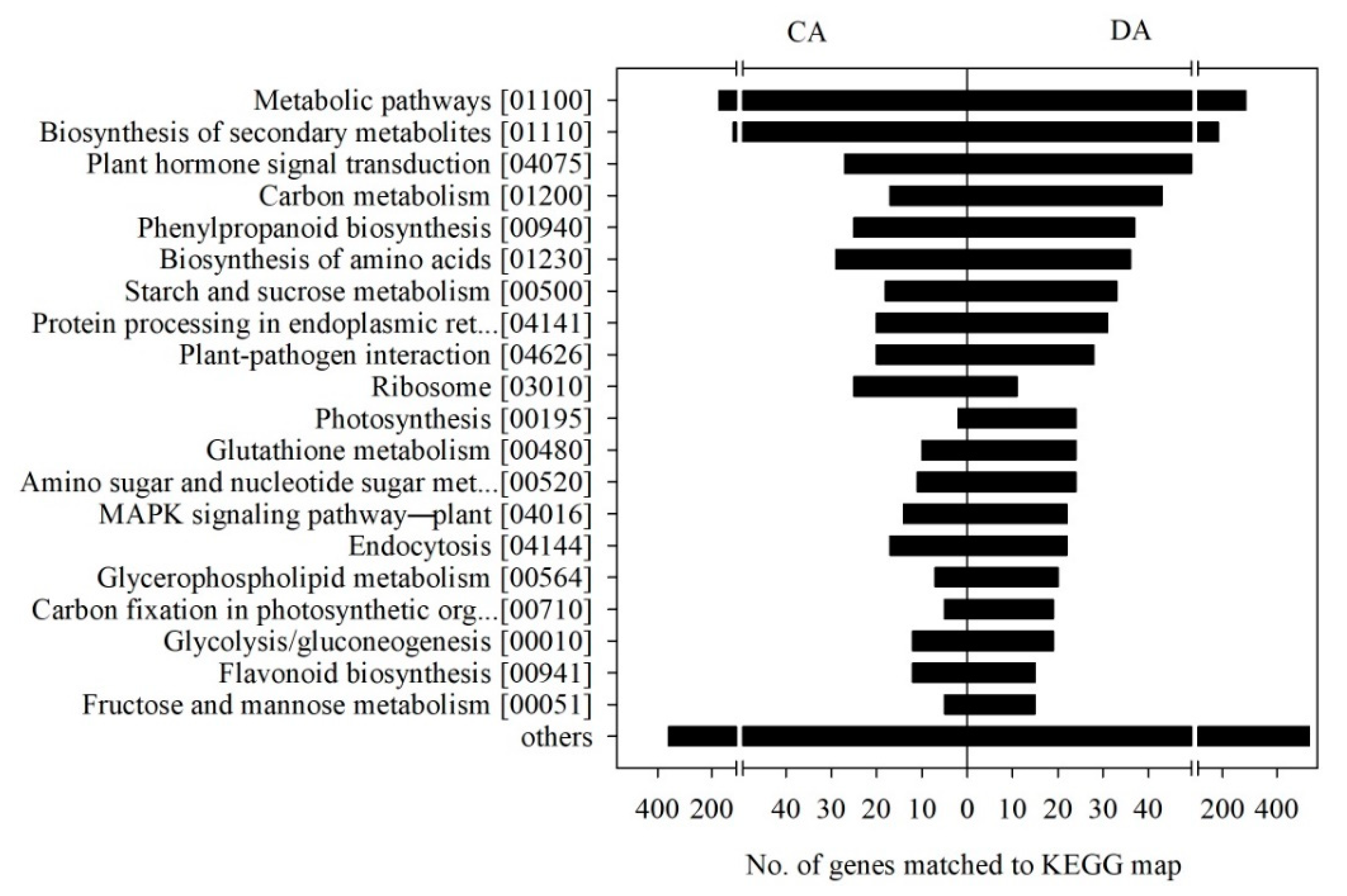

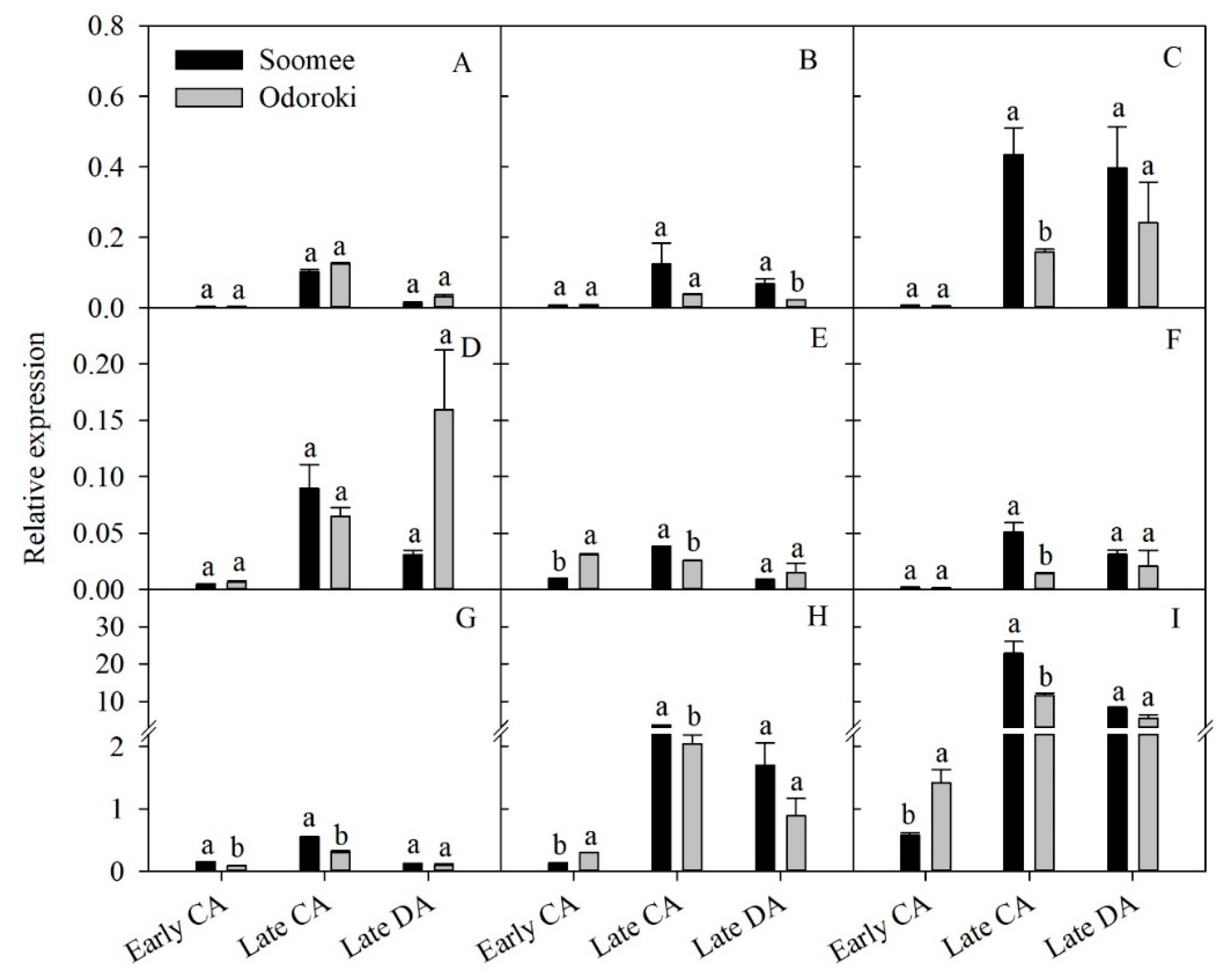
| Physiological Stage | Total Read | Total Base | GC Content (%) | Q30 (%) | Mapping Ratio (%) |
|---|---|---|---|---|---|
| Early CA | 66,451,463 | 6,683,605,100 | 46.3 | 95.4 | 96.9 |
| Late CA | 54,420,355 | 5,463,673,888 | 45.8 | 93.9 | 96.7 |
| Late DA | 50,845,192 | 5,122,738,506 | 45.6 | 96.3 | 97.2 |
| Gene Symbol (LOC No.) | Fold Change | Correlation Coefficient (r) | |||
|---|---|---|---|---|---|
| CA | DA | LT50 and log2(FPKM + 1) | log2(FPKM + 1) and RE | RE and LT50 | |
| 18766910 | 61.9 * | −18.7 * | −0.81 ns | 0.84 ns | −0.99 * |
| 18768153 | 12.4 * | −7.1 ** | −0.62 ns | 0.86 ns | −0.92 ns |
| 18769194 | 47.3 ** | −2.4 ns | −0.98 ns | 0.97 ns | −0.99 * |
| 18780397 | 20.9 * | −6.7 ** | −0.99 * | 0.99 ns | −0.99 * |
| 18784088 | 6.0 ** | −53.4 ** | −0.90 ns | 0.93 ns | −0.99 ns |
| 18793247 | 20.6 * | −14.8 ** | −0.96 ns | 0.94 ns | −0.99 * |
| 18768928 | 3.1 * | −93.7 ** | −0.69 ns | 0.90 ns | −0.93 ns |
| 18785901 | 13.5 * | −58.7 * | −0.96 ns | 0.96 ns | −0.99 * |
| 109947416 | 16.4 ** | −7.5 ns | −0.94 ns | 0.95 ns | −0.99 * |
| 18770886 | −10.3 * | 5.7 ** | 0.99 ns | 0.87 ns | 0.79 ns |
| 18775166 | −6.2 ** | 15.6 ** | 0.90 ns | 0.96 ns | 0.77 ns |
| 18780494 | −6.2 ** | 15.2 ** | 0.89 ns | 0.88 ns | 0.58 ns |
| 18780769 | −9.5 * | 32.5 ** | 0.91 ns | 0.83 ns | 0.53 ns |
| 18789909 | −16.7 * | 63.4 ** | 0.91 ns | 0.99 ** | 0.91 ns |
| 18790018 | −6.8 * | 12.7 ** | 0.94 ns | 0.87 ns | 0.67 ns |
| 18793298 | −5.2 * | 27.0 ** | 0.80 ns | 0.91 ns | 0.50 ns |
| 18793783 | −52.2 ** | 16.3 ** | 0.98 ns | 0.97 ns | 0.91 ns |
| 109948145 | −7.5 ** | 9.7 ** | 0.97 ns | 0.87 ns | 0.75 ns |
| Gene Symbol (LOC No.) | Gene Description | Fold Change (Expression Volume) | Correlation Coefficient (r) | |
|---|---|---|---|---|
| CA | DA | |||
| 18792263 | Early light-induced protein 1, chloroplastic | 15.7 (10.3) * | −53.0 (9.4) * | −0.92 ns |
| 18785901 | 14 kDa proline-rich protein DC2.15 | 13.5 (8.3) * | −58.7 (6.9) * | −0.90 ns |
| 18784998 | 21 kDa protein | 9.0 (8.6) ** | −5.6 (9.0) ** | −0.99 ns |
| 18788919 | F-box/Kelch-repeat protein At1g67480 | 6.9 (7.3) ** | −7.0 (7.3) ** | −0.99 ns |
| 18789866 | Chaperone protein dnaJ 11, chloroplastic | 8.9 (6.0) ** | −16.2 (5.4) ** | −0.97 ns |
| 18767775 | Indole-3-acetic acid-induced protein ARG2 | 3.9 (7.8) * | −12.8 (6.8) * | −0.83 ns |
| 18778639 | Stem-specific protein TSJT1 | 11.7 (5.3) ** | −20.0 (4.8) * | −0.98 ns |
| 18785018 | Glutamate dehydrogenase 2 | 11.9 (5.3) ** | −9.4 (5.6) ** | −0.99 * |
| 18766910 | Non-symbiotic hemoglobin | 61.9 (2.9) * | −18.7 (4.6) * | −0.96 ns |
| 18768928 | Bidirectional sugar transporter SWEET1 | 3.1 (7.2) * | −93.7 (3.6) ** | −0.62 ns |
| 18780397 | Tonoplast dicarboxylate transporter | 20.9 (4.3) * | −6.7 (5.5) ** | −0.94 ns |
| 18793304 | GEM-like protein 5 | 2.6 (7.3) * | −16.1 (5.8) ** | −0.69 ns |
| 18768308 | Protein EXORDIUM-like 2 | 4.7 (7.2) * | −5.3 (7.1) ** | −0.99 ns |
| 18767434 | Glutaredoxin-C11 | 5.5 (5.7) * | −42.7 (3.5) ** | −0.82 ns |
| 18782474 | Triacylglycerol lipase 2 | 5.7 (6.3) ** | −5.7 (6.4) ** | −0.99 * |
| 18784501 | ABC transporter C family member 8 | 20.7 (4.1) * | −5.0 (5.6) * | −0.90 ns |
| 18792321 | Probable xyloglucan endotransglucosylase/hydrolase protein 23 | 2.0 (8.8) * | −7.1 (7.8) * | −0.70 ns |
| 18787602 | Sorbitol dehydrogenase | 4.6 (7.6) ** | −3.2 (7.9) ** | −0.98 ns |
| 18770266 | LOW QUALITY PROTEIN: protein PIN-LIKES 7 | 26.9 (3.6) ** | −5.4 (5.3) * | −0.89 ns |
| 18792372 | Zinc finger protein ZAT10 | 8.2 (5.1) ** | −7.0 (5.2) ** | −0.99 * |
| Gene Symbol (LOC No.) | Gene Description | Primer Sequence |
|---|---|---|
| DEGs up- and down-regulated during CA and DA, respectively | ||
| 18766910 | Non-symbiotic hemoglobin | F: AGAGCAGGAAACATTGGTGG R: CTTCTGAGCTGATGGTGCAA |
| 18768153 | Squalene monooxygenase | F: CCAAGCATGCTTCGACTACT R: TCAGTCTAGCTCCAACCCAA |
| 18769194 | Polygalacturonase inhibitor | F: ACCATCCTAAACCCAGCTCT R: AGTACCAGTCACAGCAGTCT |
| 18780397 | Tonoplast dicarboxylate transporter | F: ATGGATGATGTCATTGCGCT R: CTCCGCAGAATAGCATGGTT |
| 18784088 | Late embryogenesis abundant protein 2 | F: AAACAGAGGGCAGAAGAAGC R: CTTTTCACAGCATCAGCAGC |
| 18793247 | Extracellular ribonuclease LE | F: GAATGGGAAAAGCATGGCAC R: TGGTTGTATGCCTGCACTTT |
| 18768928 | Bidirectional sugar transporter SWEET1 | F: CCTTTTGTTGCTGTGCCAAA R: TTCATCAGCAGTAGCAGCAG |
| 18785901 | 14 kDa proline-rich protein DC2.15 | F: CACCAACACCACCAAAAACC R: TAGGGTCGAAGGAGGTTGTT |
| 109947416 | pEARLI1-like lipid-transfer protein 3 | F: TGATGCTGCTGTGTGTCTTT R: GCATTGGAAGTCTGTTGGGA |
| DEGs down- and up-regulated during CA and DA, respectively | ||
| 18770886 | O-Acyltransferase WSD1 | F: TCTCAGGTTTCTGCAACACC R: AATGTATTTCCCGCATGCCT |
| 18775166 | Phosphoenolpyruvate carboxykinase (ATP) | F: ATTCTGGCTGCAATATGGGG R: CAGCATCTTATCAGCCAGCA |
| 18780494 | Protein SRG1 | F: AGACGTGGAAGGTTTTGGAC R: ACTGGACCAATTTCACCACC |
| 18780769 | Protein E6 | F: CACCACCAACAACAACAACC R: TGGCACGAACTCTTCTTCAC |
| 18789909 | Non-specific lipid-transfer protein 2 | F: TTCTTGTCTTTGGGGGAAGC R: AAATGGGGAGCCACAAGTG |
| 18790018 | Aspartyl protease AED3 | F: GCATCAATTCCATCTCGGGT R: AAGCTGTTCTCCAATGGCAA |
| 18793298 | Cyclin-D1-1 | F: ATGTCATCGGTTTGGAGCTC R: CCTCCAATCCAAAACACCCA |
| 18793783 | Heterogeneous nuclear ribonucleoprotein A2 homolog 1 | F: GCCGATGTAGTGACTTTTCCT R: GGTTCTTGTTCCCGTTCAGT |
| 109948145 | Vinorine synthase-like | F: ACTGCTTGCTATCTGACGTG R: CCATCCTGGTCCGTAAGTTG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, D.J.; Jun, S.H.; Park, J.; Kwon, J.H.; Lee, H.J. Transcriptome Analysis of Genes Involved in Cold Hardiness of Peach Tree (Prunus persica) Shoots during Cold Acclimation and Deacclimation. Genes 2020, 11, 611. https://doi.org/10.3390/genes11060611
Yu DJ, Jun SH, Park J, Kwon JH, Lee HJ. Transcriptome Analysis of Genes Involved in Cold Hardiness of Peach Tree (Prunus persica) Shoots during Cold Acclimation and Deacclimation. Genes. 2020; 11(6):611. https://doi.org/10.3390/genes11060611
Chicago/Turabian StyleYu, Duk Jun, Sung Hoon Jun, Junhyung Park, Jung Hyun Kwon, and Hee Jae Lee. 2020. "Transcriptome Analysis of Genes Involved in Cold Hardiness of Peach Tree (Prunus persica) Shoots during Cold Acclimation and Deacclimation" Genes 11, no. 6: 611. https://doi.org/10.3390/genes11060611




