Computational Analysis of Transcriptomic and Proteomic Data for Deciphering Molecular Heterogeneity and Drug Responsiveness in Model Human Hepatocellular Carcinoma Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Pre-Processing
2.2. Exploratory Analysis of Transcriptomic and Proteomic Data
2.3. Between-Group Differential Gene and Protein Expression Analysis
2.4. Functional Enrichment Analysis of Differentially Expressed Genes
2.5. Drug-Specific Sensitivity in Association with Differentiation Status of HCC lines
2.6. HCC Tumor Clustering Based on SU_LIVER Gene-Set Expression Data From TCGA
3. Results
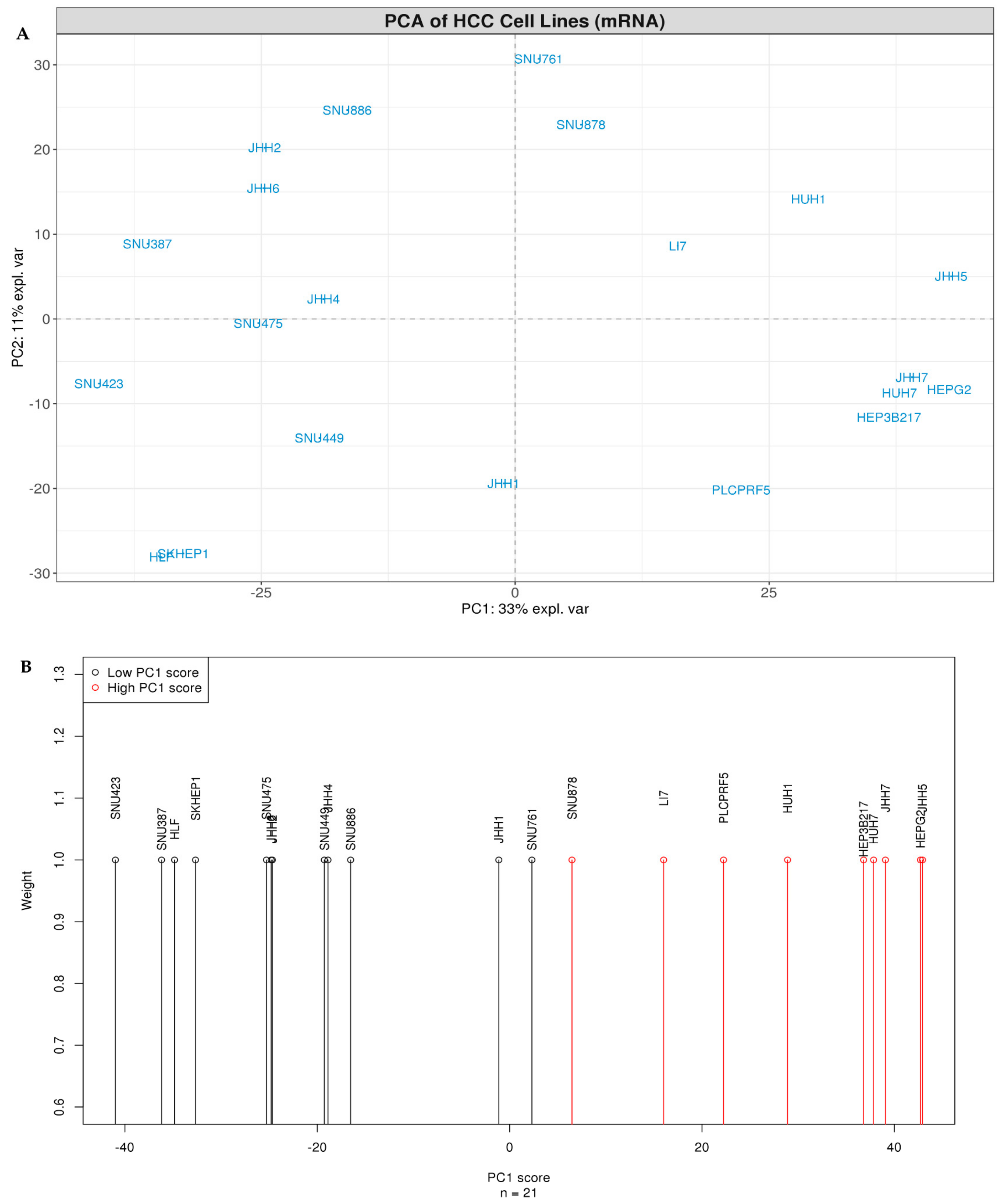
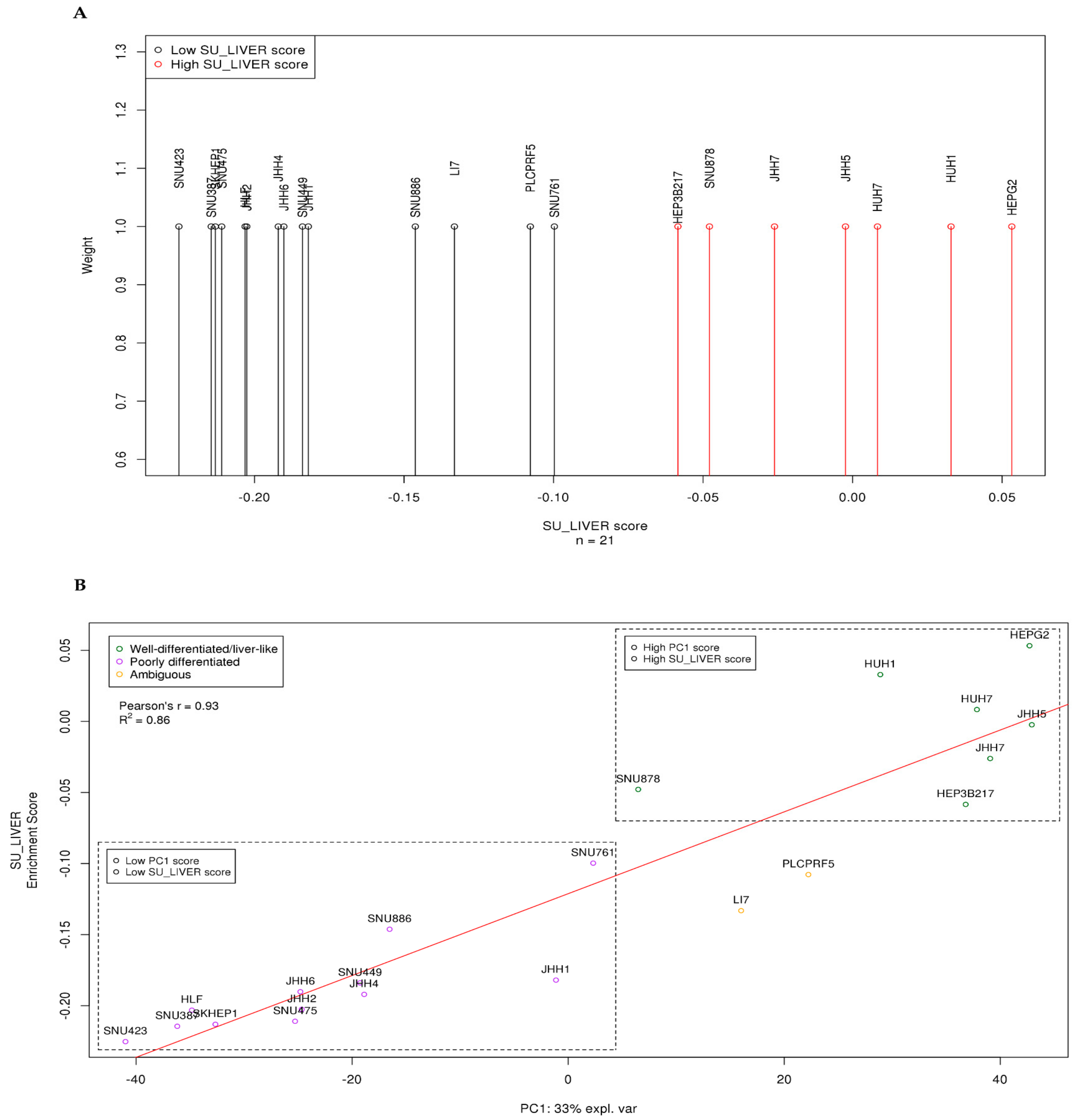
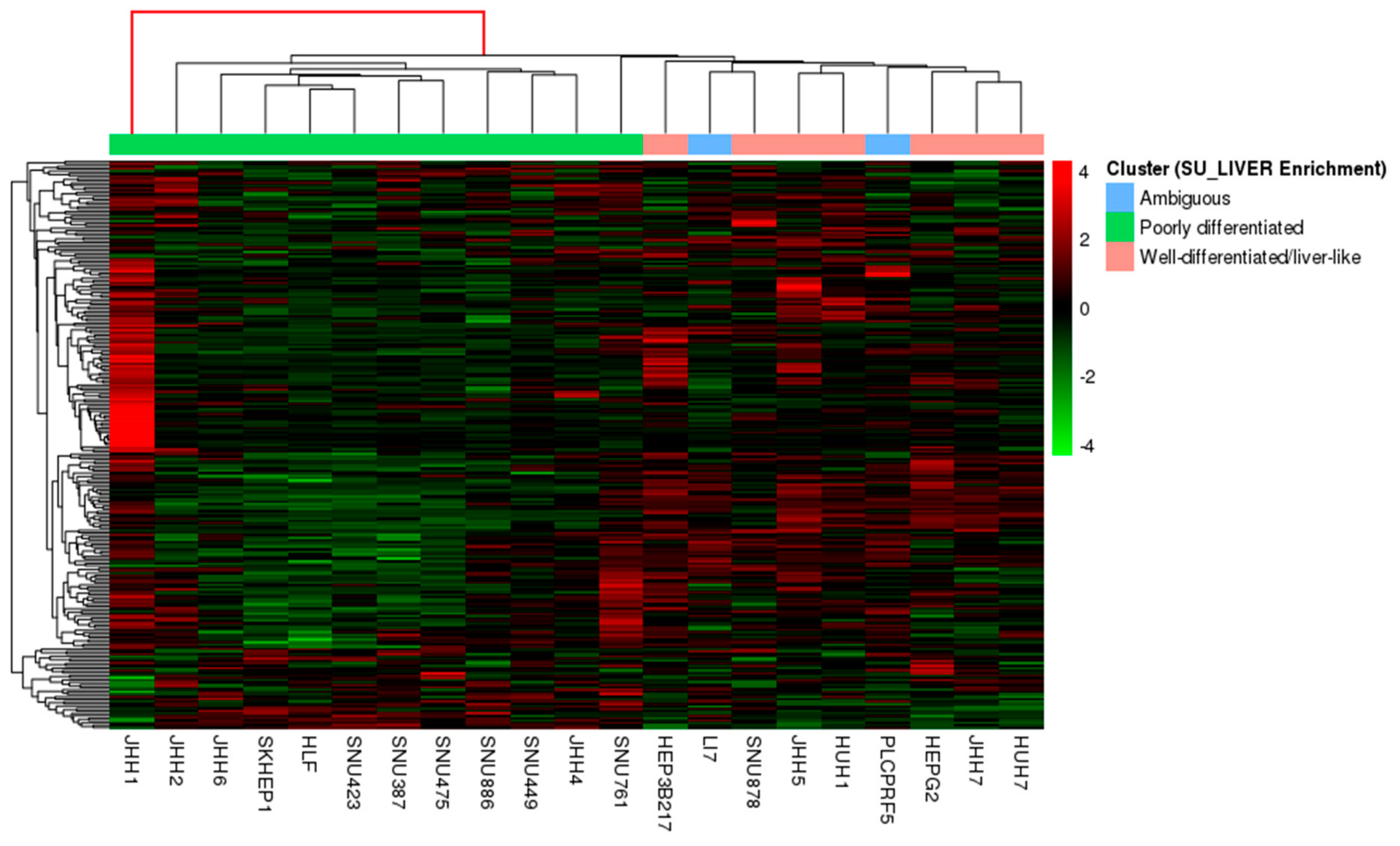
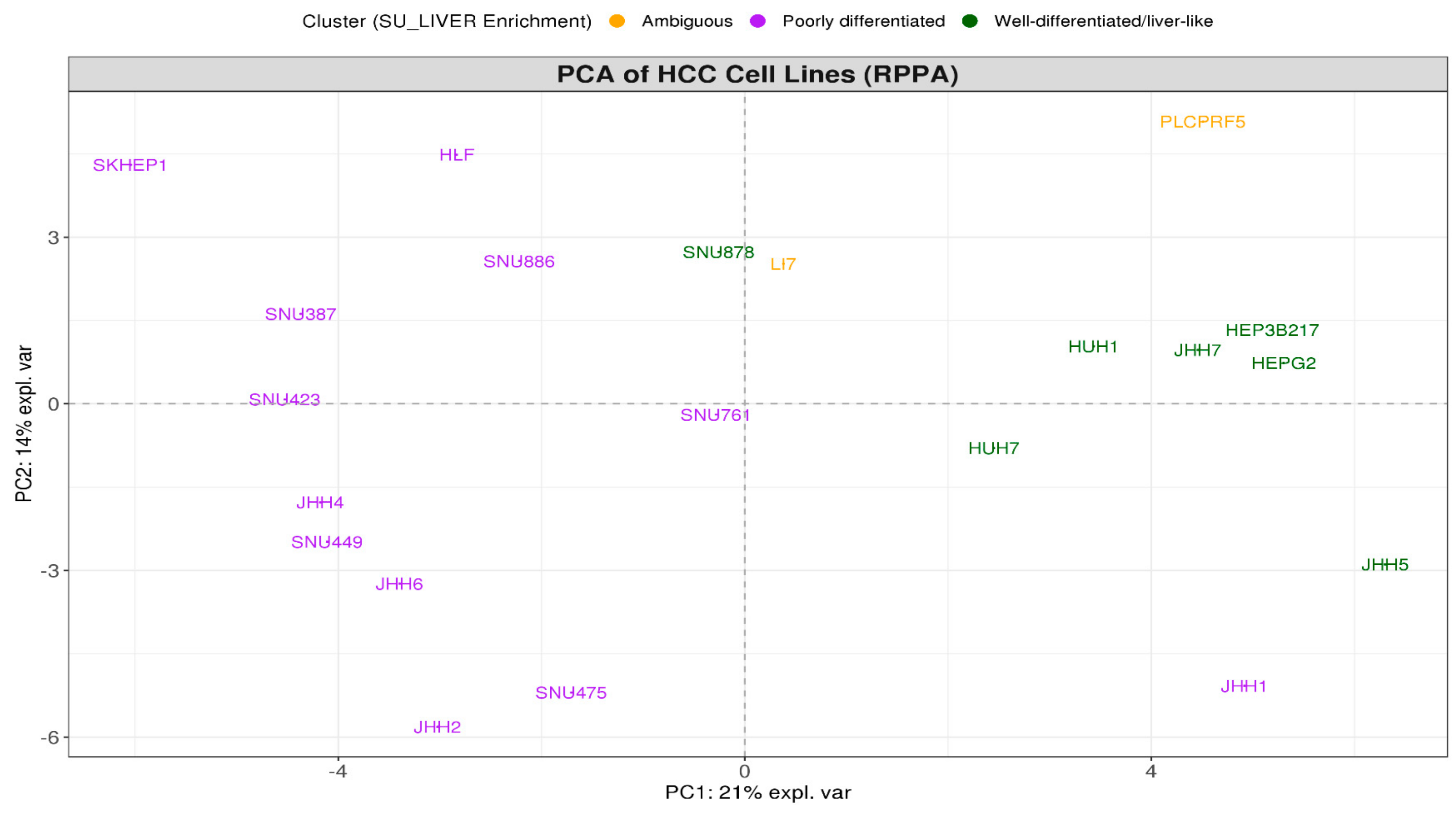
3.1. HCC Lines Clustered into Two Distinct Differentiation Subtypes
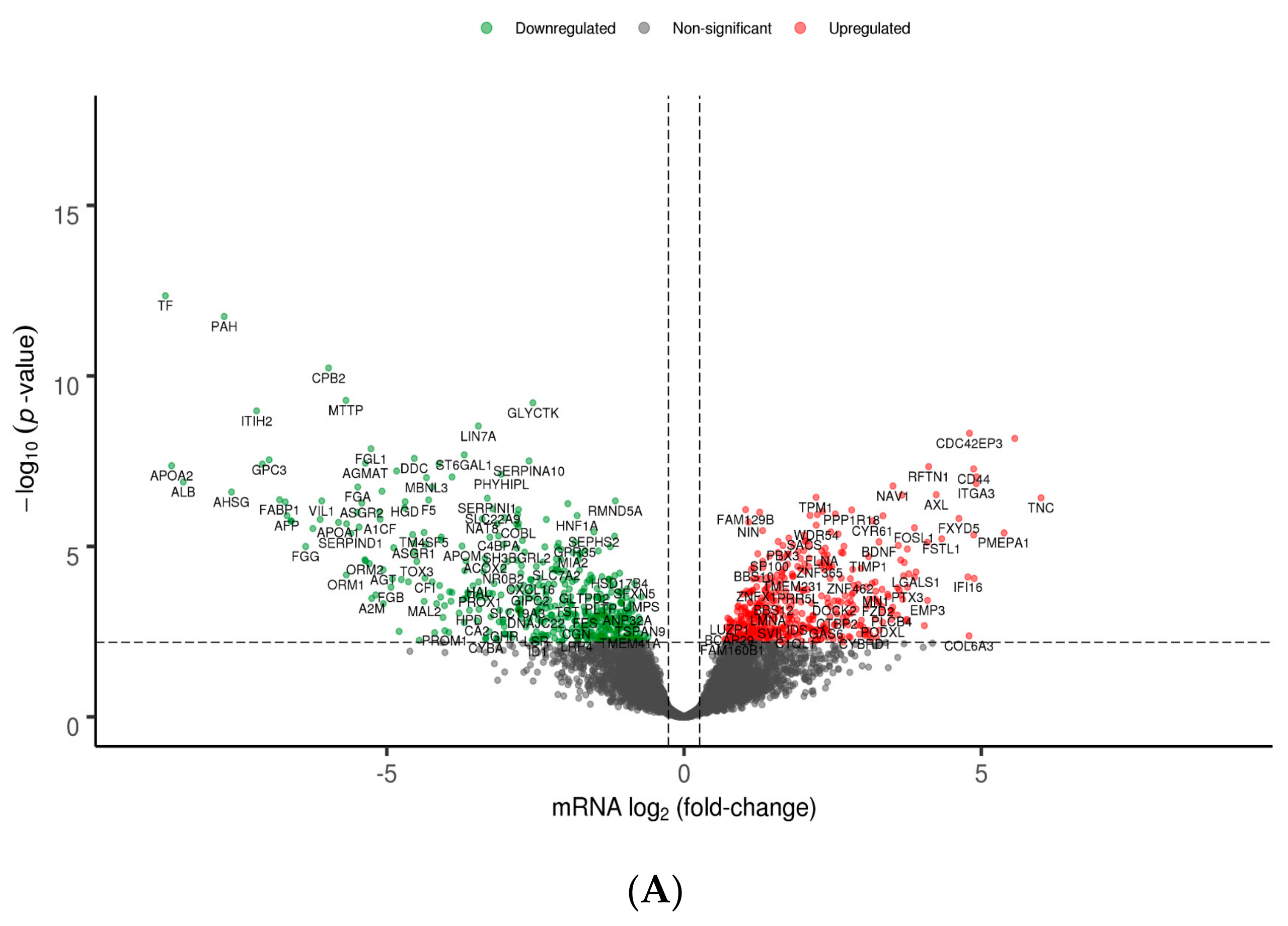
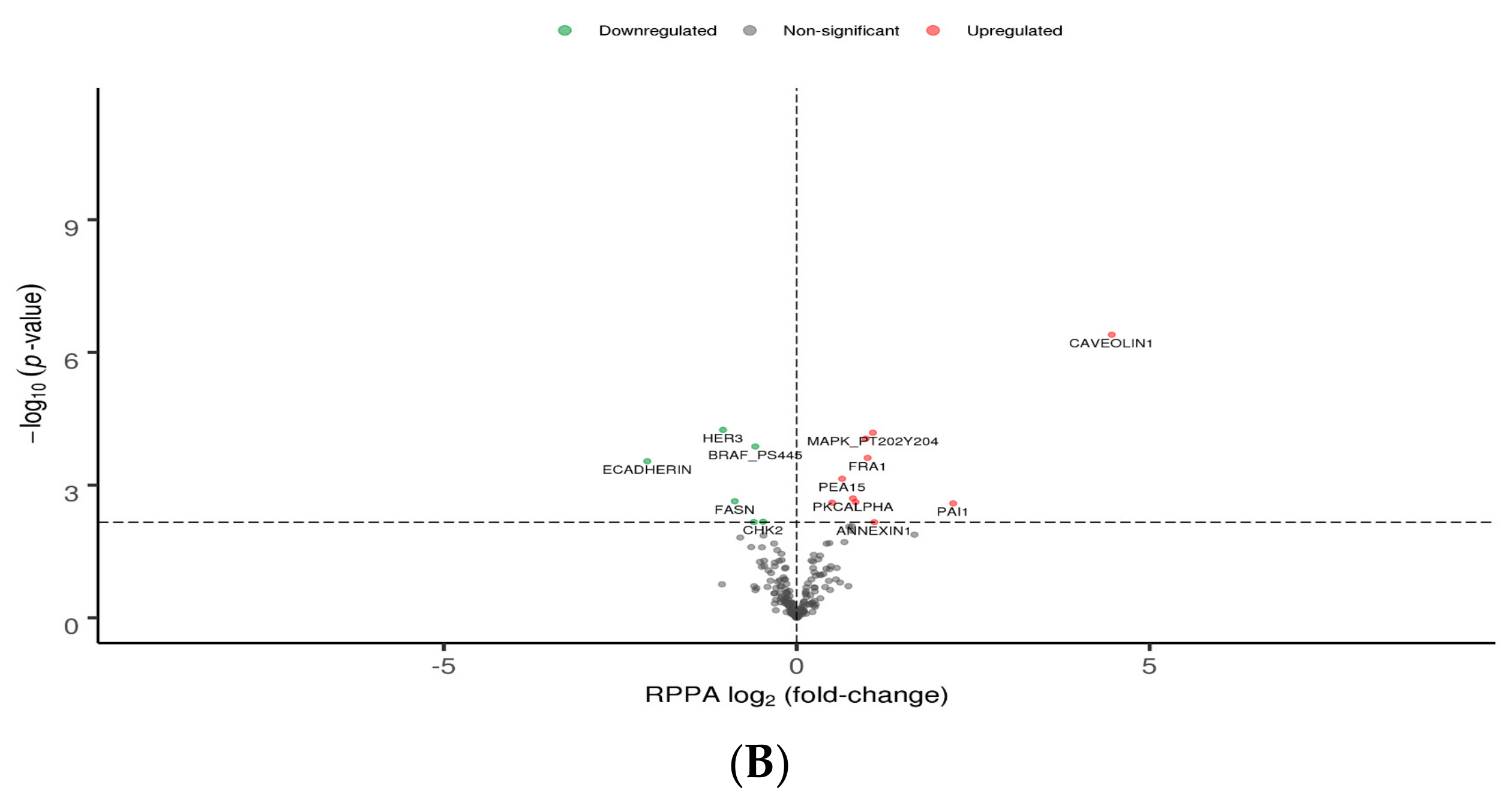
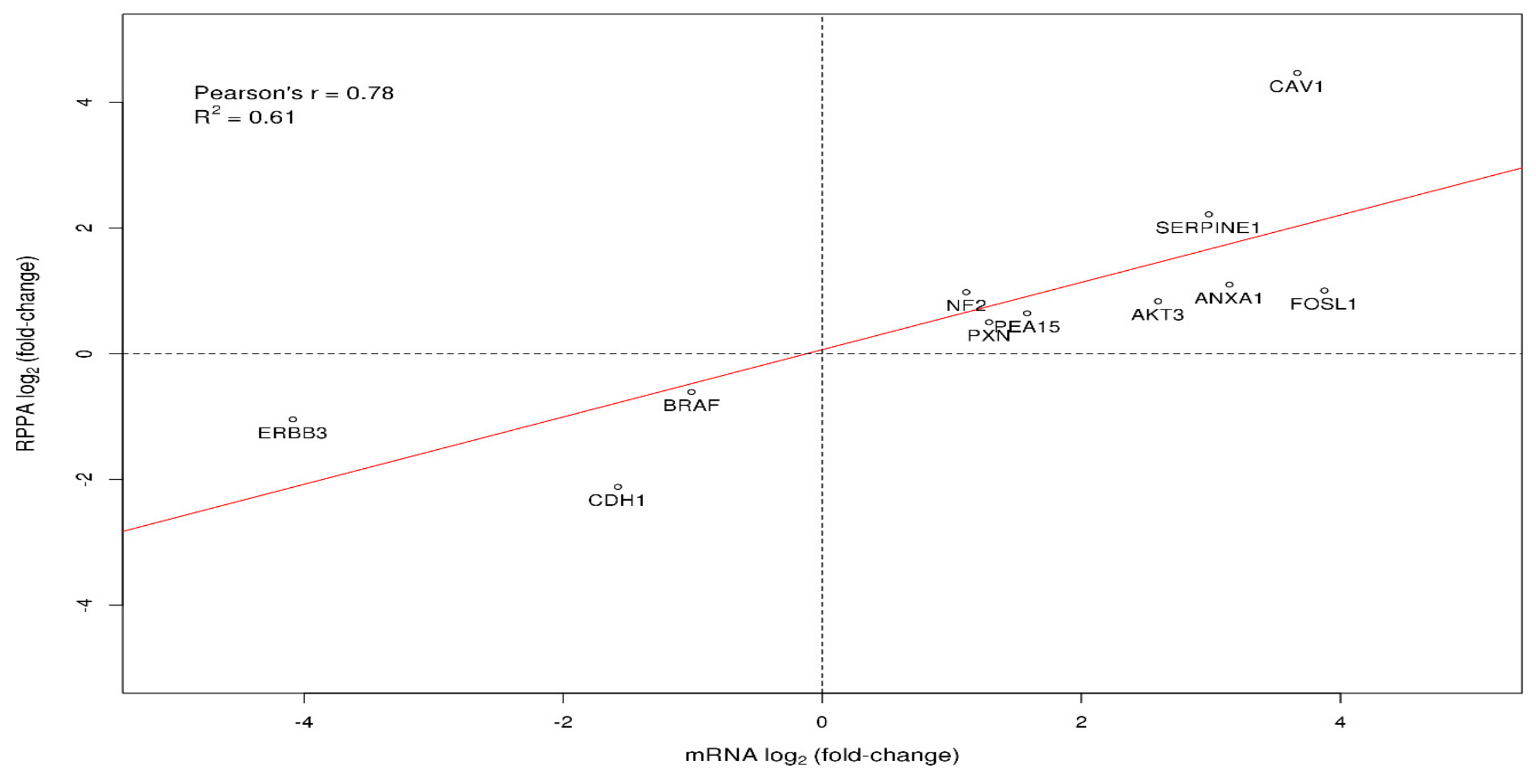
3.2. Differential Gene and Protein Expression between Poorly and Well-Differentiated HCC Lines
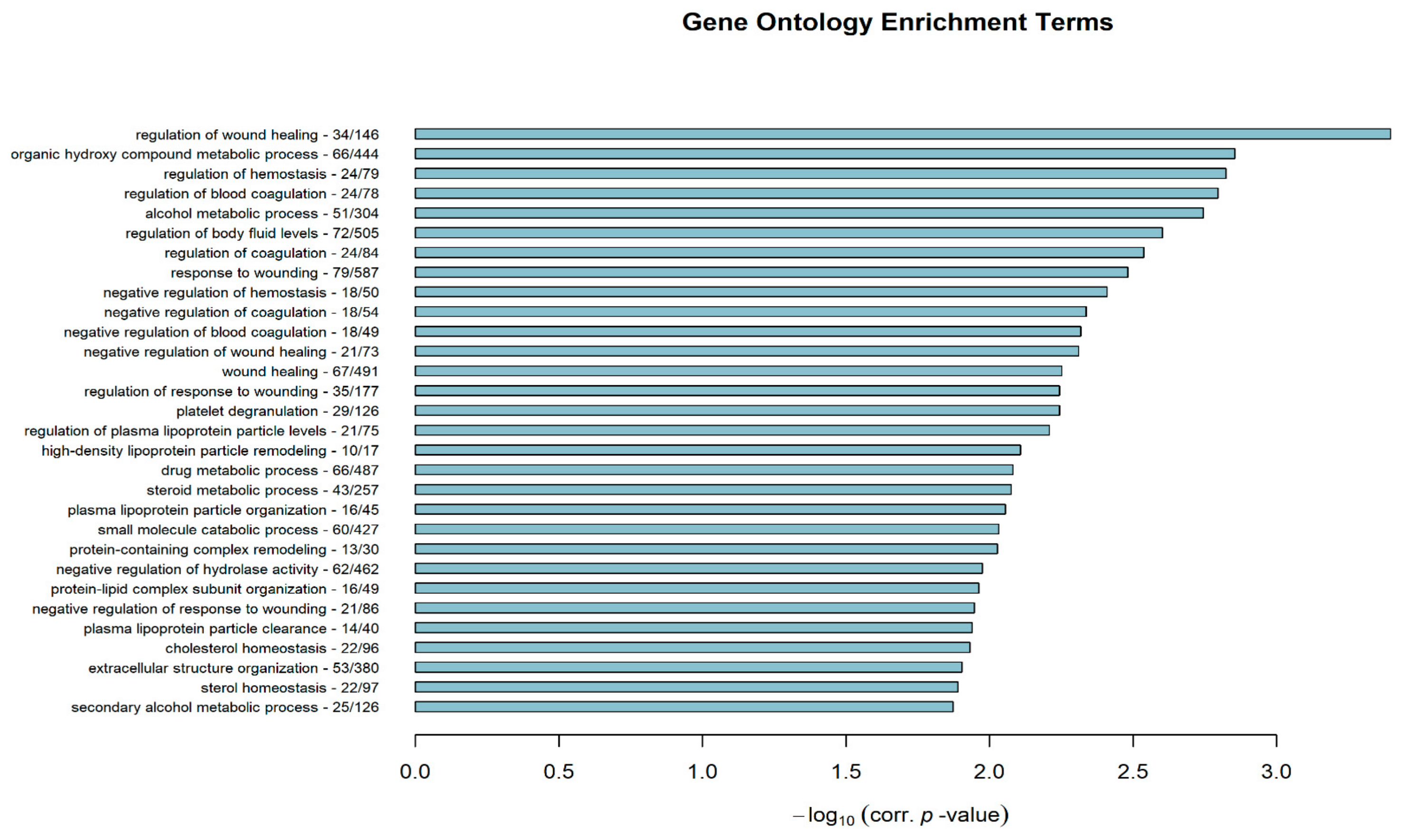
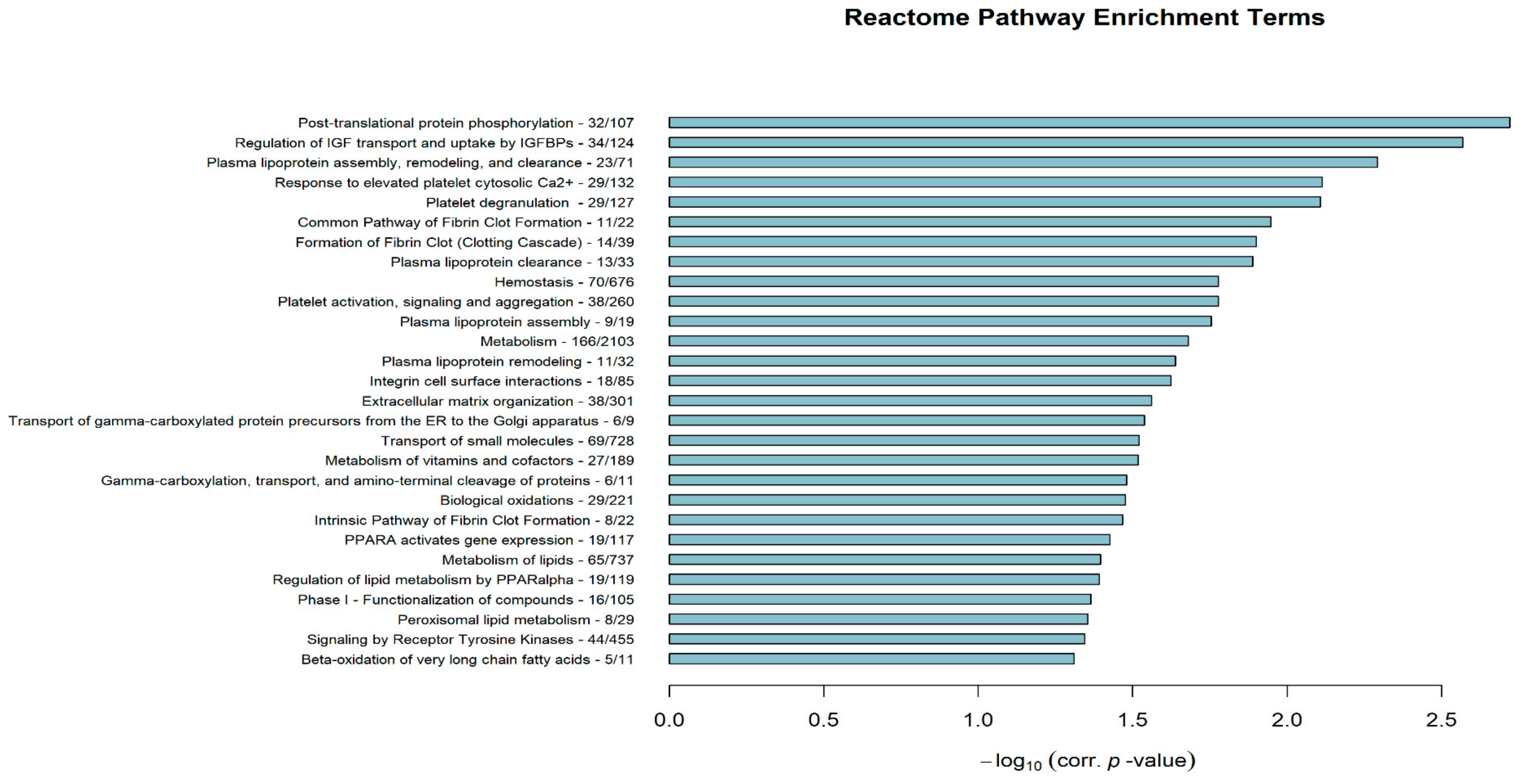
3.3. Differentially Enriched Biological Processes/Pathways and Hub Driver-Gene Signatures between Poorly and Well-Differentiated HCC Lines
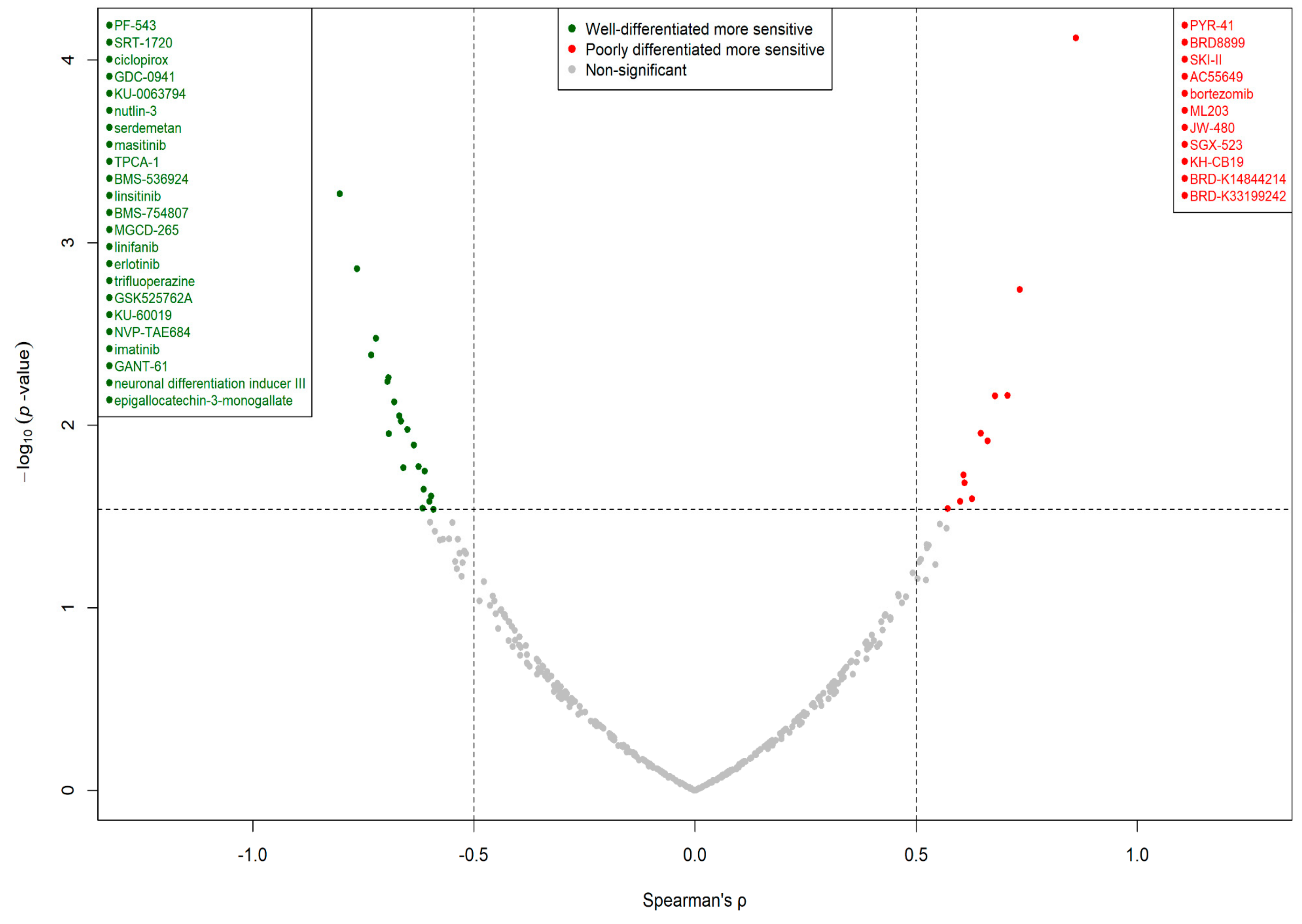
3.4. Cell Line Differentiation Status Correlated with Drug-Specific Sensitivity
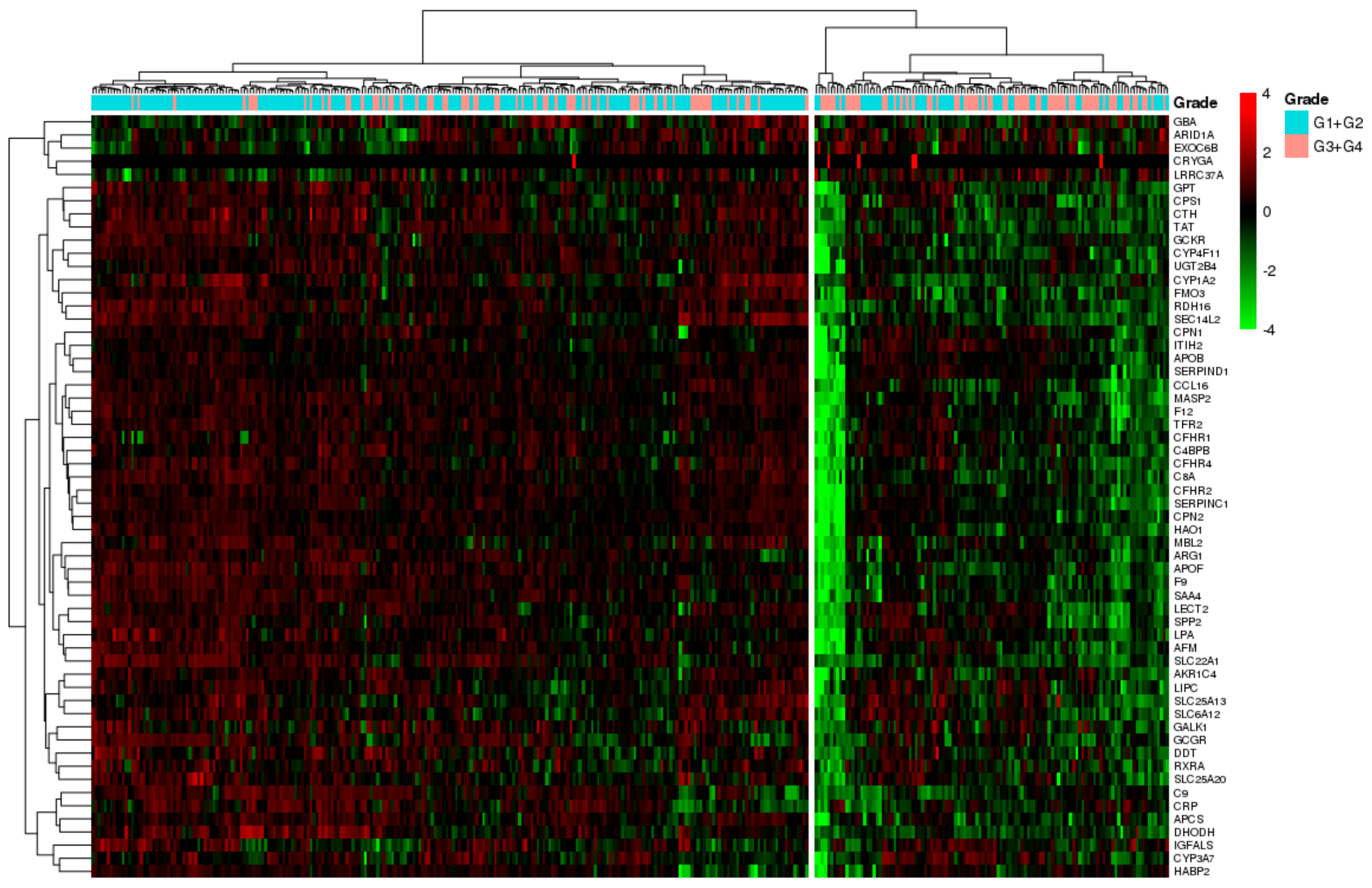
3.5. SU_LIVER-Based Clustering of HCC Patients Associated with the Assigned Tumor Grade
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Fujiwara, N.; Friedman, S.L.; Goossens, N.; Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018, 68, 526–549. [Google Scholar] [CrossRef]
- Aravalli, R.N.; Cressman, E.N.K.; Steer, C.J. Cellular and molecular mechanisms of hepatocellular carcinoma: An update. Arch. Toxicol. 2013, 87, 227–247. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In vitro tumor models: Advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Berg, K.C.G.; Eide, P.W.; Eilertsen, I.A.; Johannessen, B.; Bruun, J.; Danielsen, S.A.; Bjørnslett, M.; Meza-Zepeda, L.A.; Eknæs, M.; Lind, G.E.; et al. Multi-omics of 34 colorectal cancer cell lines—A resource for biomedical studies. Mol. Cancer 2017, 16, 1–16. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Gregory, V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modeling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R.; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Park, J.; Lee, J.; Kang, M.; Park, K.; Jeon, Y.; Lee, H.; Kwon, H.; Park, H.; Yeo, K.; Lee, K.; et al. Characterization of cell lines established from human hepatocellular carcinoma. Int. J. Cancer 1995, 62, 276–282. [Google Scholar] [CrossRef]
- Heffelfinger, S.C.; Hawkins, H.H.; Barrish, J.; Taylor, L.; Darlington, G.J. SK HEP-1: A human cell line of endothelial origin. Cell. Dev. Biol. Anim. 1992, 28, 136–142. [Google Scholar] [CrossRef]
- Rebouissou, S.; Zucman-Rossi, J.; Moreau, R.; Qiu, Z.; Hui, L. Note of caution: Contaminations of hepatocellular cell lines. J. Hepatol. 2017, 67, 896–897. [Google Scholar] [CrossRef] [PubMed]
- Seashore-Ludlow, B.; Rees, M.G.; Cheah, J.H.; Coko, M.; Price, E.V.; Coletti, M.E.; Jones, V.; Bodycombe, N.E.; Soule, C.K.; Gould, J.; et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015, 5, 1210–1223. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Bodycombe, N.E.; Cheah, J.H.; Price, E.V.; Liu, K.; Schaefer, G.I.; Ebright, R.Y.; Stewart, M.L.; Ito, D.; Wang, S.; et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 2013, 154, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Ally, A.; Balasundaram, M.; Carlsen, R.; Chuah, E.; Clarke, A.; Dhalla, N.; Holt, R.A.; Jones, S.J.M.; Lee, D.; Ma, Y.; et al. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017, 169, 1327–1341.e23. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, 1–19. [Google Scholar] [CrossRef]
- Wang, H.; Song, M. Ckmeans.1d.dp: Optimal k-means clustering in one dimension by dynamic programming. R Journal 2011, 3, 29–33. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Koplev, S.; Lin, K.; Dohlman, A.B.; Ma’ayan, A. Integration of pan-cancer transcriptomics with RPPA proteomics reveals mechanisms of epithelial-mesenchymal transition. PLoS Comput. Biol. 2018, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Mccarthy, D.J.; Smyth, G.K. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 2009, 25, 765–771. [Google Scholar] [CrossRef]
- Blighe, K. Publication-ready volcano plots with enhanced colouring and labeling. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 5 June 2020). (R-Package Version 1.0.1).
- Koutsandreas, T.; Binenbaum, I.; Pilalis, E.; Valavanis, I.; Papadodima, O.; Chatziioannou, A. Analyzing and visualizing genomic complexity for the derivation of the emergent molecular networks. Int. J. Monit. Surveill. Technol. Res. 2016, 4, 30–49. [Google Scholar] [CrossRef][Green Version]
- Lhomond, S.; Avril, T.; Dejeans, N.; Voutetakis, K.; Doultsinos, D.; McMahon, M.; Pineau, R.; Obacz, J.; Papadodima, O.; Jouan, F.; et al. Dual IRE1 RNase functions dictate glioblastoma development. EMBO Mol. Med. 2018, 10, 1–19. [Google Scholar] [CrossRef]
- Wu, D.; Smyth, G.K. Camera: A competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012, 40, 1–12. [Google Scholar] [CrossRef]
- Wang, H.; Unternaehrer, J.J. Epithelial-mesenchymal transition and cancer stem cells: At the crossroads of differentiation and dedifferentiation. Dev. Dyn. 2019, 248, 10–20. [Google Scholar] [CrossRef]
- Su, A.I.; Cooke, M.P.; Ching, K.A.; Hakak, Y.; Walker, J.R.; Wiltshire, T.; Orth, A.P.; Vega, R.G.; Sapinoso, L.M.; Moqrich, A.; et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA 2002, 99, 4465–4470. [Google Scholar] [CrossRef]
- Hsiao, L.L.; Dangond, F.; Yoshida, T.; Hong, R.; Jensen, R.V.; Misra, J.; Dillon, W.; Lee, K.F.; Clark, K.E.; Haverty, P.; et al. A compendium of gene expression in normal human tissues. Physiol. Genom. 2002, 2002, 97–104. [Google Scholar] [CrossRef]
- Anastassiou, D.; Rumjantseva, V.; Cheng, W.; Huang, J.; Canoll, P.D.; Yamashiro, D.J.; Kandel, J.J. Human cancer cells express Slug-based epithelial-mesenchymal transition gene expression signature obtained in vivo. BMC Cancer 2011, 11, 529. [Google Scholar] [CrossRef] [PubMed]
- Elvidge, G.P.; Glenny, L.; Appelhoff, R.J.; Ratcliffe, P.J.; Ragoussis, J.; Gleadle, J.M. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: The role of HIF-1α, HIF-2α, and other pathways. J. Biol. Chem. 2006, 281, 15215–15226. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Siadaty, M.S.; Berens, M.E.; Hampton, G.M.; Theodorescu, D. Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene 2008, 27, 6679–6689. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Nijman, S.M.B.; Kobayashi, M.; Chan, J.A.; Brunet, J.P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M.; et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009, 69, 7385–7392. [Google Scholar] [CrossRef] [PubMed]
- Caruso, S.; Calatayud, A.L.; Pilet, J.; La Bella, T.; Rekik, S.; Imbeaud, S.; Letouzé, E.; Meunier, L.; Bayard, Q.; Rohr-Udilova, N.; et al. Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response. Gastroenterology 2019, 157, 760–776. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, W.; Akbani, R.; Liu, W.; Ju, Z.; Ling, S.; Vellano, C.P.; Roebuck, P.; Yu, Q.; Eterovic, A.K.; et al. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell 2017, 31, 225–239. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Martins-Filho, S.N.; Paiva, C.; Azevedo, R.S.; Alves, V.A.F. Histological grading of hepatocellular carcinoma-a systematic review of literature. Front. Med. 2017, 4, 1–9. [Google Scholar] [CrossRef]
- Williams, E.D.; Gao, D.; Redfern, A.; Thompson, E.W. Controversies around epithelial–mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer 2019. [Google Scholar] [CrossRef]
- Giannelli, G.; Koudelkova, P.; Dituri, F.; Mikulits, W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J. Hepatol. 2016, 65, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Schmalhofer, O.; Brabletz, S.; Brabletz, T. E-cadherin, β-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009, 28, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Ting Tse, E.Y.; Fat Ko, F.C.; Kwan Tung, E.K.; Chan, L.K.; Wah Lee, T.K.; Wai Ngan, E.S.; Man, K.; Tsai Wong, A.S.; Ng, I.O.L.; Ping Yam, J.W. Caveolin-1 overexpression is associated with hepatocellular carcinoma tumourigenesis and metastasis. J. Pathol. 2012, 226, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wei, X.; He, J.; Tian, X.; Yuan, S.; Sun, L. Plasminogen activator inhibitor-1 in cancer research. Biomed. Pharmacother. 2018, 105, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, H.F. Tumors: Wounds that do not heal-redux. Cancer Immunol. Res. 2015, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Foster, D.S.; Jones, R.E.; Ransom, R.C.; Longaker, M.T.; Norton, J.A. The evolving relationship of wound healing and tumor stroma. JCI Insight 2018, 3, 1–17. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Nwosu, Z.C.; Battello, N.; Rothley, M.; Piorońska, W.; Sitek, B.; Ebert, M.P.; Hofmann, U.; Sleeman, J.; Wölfl, S.; Meyer, C.; et al. Liver cancer cell lines distinctly mimic the metabolic gene expression pattern of the corresponding human tumours. J. Exp. Clin. Cancer Res. 2018, 37, 211. [Google Scholar] [CrossRef]
- Lee, H.J.; Jeng, Y.M.; Chen, Y.L.; Chung, L.; Yuan, R.H. Gas6/Axl pathway promotes tumor invasion through the transcriptional activation of slug in hepatocellular carcinoma. Carcinogenesis 2014, 35, 769–775. [Google Scholar] [CrossRef]
- Antony, J.; Huang, R.Y.J. AXL-driven EMT state as a targetable conduit in cancer. Cancer Res. 2017, 77, 3725–3732. [Google Scholar] [CrossRef]
- Reichl, P.; Dengler, M.; van Zijl, F.; Huber, H.; Führlinger, G.; Reichel, C.; Sieghart, W.; Peck-Radosavljevic, M.; Grubinger, M.; Mikulits, W. Axl activates autocrine transforming growth factor-β signaling in hepatocellular carcinoma. Hepatology 2015, 61, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Ikushima, H.; Miyazono, K. TGFΒ 2 signalling: A complex web in cancer progression. Nat. Rev. Cancer 2010, 10, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Kwaan, H.C.; Lindholm, P.F. Fibrin and fibrinolysis in cancer. Semin. Thromb. Hemost. 2019, 45, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Kurz, S.; Thieme, R.; Amberg, R.; Groth, M.; Jahnke, H.G.; Pieroh, P.; Horn, L.C.; Kolb, M.; Huse, K.; Platzer, M.; et al. The anti-tumorigenic activity of A2M—A lesson from the naked mole-rat. PLoS ONE 2017, 12, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Akl, M.R.; Nagpal, P.; Ayoub, N.M.; Tai, B.; Prabhu, S.A.; Capac, C.M.; Gliksman, M.; Goy, A.; Suh, K.S. Molecular and clinical significance of fibroblast growth factor 2 (FGF2/bFGF) in malignancies of solid and hematological cancers for personalized therapies. Oncotarget 2016, 7, 44735–44762. [Google Scholar] [CrossRef]
- Ning, B.F.; Ding, J.; Yin, C.; Zhong, W.; Wu, K.; Zeng, X.; Yang, W.; Chen, Y.X.; Zhang, J.P.; Zhang, X.; et al. Hepatocyte nuclear factor 4α suppresses the development of hepatocellular carcinoma. Cancer Res. 2010, 70, 7640–7651. [Google Scholar] [CrossRef]
- Wang, M.; Han, J.; Xing, H.; Zhang, H.; Li, Z.; Liang, L.; Li, C.; Dai, S.; Wu, M.; Shen, F.; et al. Dysregulated fatty acid metabolism in hepatocellular carcinoma. Hepatic Oncol. 2016, 3, 241–251. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, L.; Liu, L.; Niu, X. EMT-Mediated Acquired EGFR-TKI Resistance in NSCLC: Mechanisms and Strategies. Front. Oncol. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Terai, H.; Soejima, K.; Yasuda, H.; Nakayama, S.; Hamamoto, J.; Arai, D. Activation of the FGF2-FGFR1 autocrine pathway: A novel mechanism of acquired resistance to gefitinib in NSCLC. Mol. Cancer Res. 2013, 1, 759–768. [Google Scholar] [CrossRef]
- Michailidou, M.; Melas, I.; Messinis, D.; Klamt, S.; Alexopoulos, L.; Kolisis, F.; Loutrari, H. Network-based analysis of nutraceuticals in human hepatocellular carcinomas reveals mechanisms of chemopreventive action. CPT Pharmacomet. Syst. Pharmacol. 2015, 4, 350–361. [Google Scholar] [CrossRef]
- Agioutantis, P.C.; Kotsikoris, V.; Kolisis, F.N.; Loutrari, H. RNA-seq data analysis of stimulated hepatocellular carcinoma cells treated with epigallocatechin gallate and fisetin reveals target genes and action mechanisms. Comput. Struct. Biotechnol. J. 2020, 18, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, A.; Affatato, R.; Centritto, F.; Fratelli, M.; Kurosaki, M.; Barzago, M.M.; Bolis, M.; Terao, M.; Garattini, E.; Paroni, G. All-trans-retinoic acid modulates the plasticity and inhibits the motility of breast cancer cells role of notch1 and transforming growth factor (TGF β). J. Biol. Chem. 2015, 290, 17690–17709. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ma, Y.; He, H.W.; Zhao, W.L.; Shao, R.G. SPHK1 (sphingosine kinase 1) induces epithelial-mesenchymal transition by promoting the autophagy-linked lysosomal degradation of CDH1/E-cadherin in hepatoma cells. Autophagy 2017, 13, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.C.; Hung, W.C. Pyruvate kinase M2 fuels multiple aspects of cancer cells: From cellular metabolism, transcriptional regulation to extracellular signaling. Mol. Cancer 2018, 17, 1–9. [Google Scholar] [CrossRef]
- Tamada, M.; Nagano, O.; Tateyama, S.; Ohmura, M.; Yae, T.; Ishimoto, T.; Sugihara, E.; Onishi, N.; Yamamoto, T.; Yanagawa, H.; et al. Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer Res. 2012, 72, 1438–1448. [Google Scholar] [CrossRef] [PubMed]

| Cell Line Name | Cancer Type | Cell Line Name | Cancer Type | Cell Line Name | Cancer Type |
|---|---|---|---|---|---|
| HEP3B217 | HCC | JHH4 | HCC | SNU387 | HCC |
| HEPG2 | HCC | JHH5 | HCC | SNU423 | HCC |
| HLF | HCC | JHH6 | HCC | SNU449 | HCC |
| HUH1 | HCC | JHH7 | HCC | SNU475 | HCC |
| HUH7 | HCC | LI7 | HCC | SNU761 | HCC |
| JHH1 | HCC | PLCPRF5 | HCC | SNU878 | HCC |
| JHH2 | HCC | SKHEP1 | Adenocarcinoma | SNU886 | HCC |
| Gene Symbol | Gene Name | Systemic Processes | log2 (Fold-Change) |
|---|---|---|---|
| APOA1 | apolipoprotein A1 | 19 | –5.81 |
| APOE | apolipoprotein E | 14 | –4.11 |
| APOA2 | apolipoprotein A2 | 12 | –8.62 |
| CAV1 | caveolin 1 | 12 | 3.67 |
| SERPINF2 | serpin family F member 2 | 12 | –2.43 |
| TGFB2 | transforming growth factor beta 2 | 11 | 2.84 |
| AGTR1 | angiotensin II receptor type 1 | 11 | –2.95 |
| ANXA1 | annexin A1 | 11 | 3.14 |
| AGT | angiotensinogen | 10 | –5.06 |
| APOH | apolipoprotein H | 10 | –7.09 |
| FGF2 | fibroblast growth factor 2 | 10 | 2.84 |
| SCARB1 | scavenger receptor class B member 1 | 10 | –1.57 |
| APOC3 | apolipoprotein C3 | 10 | –4.89 |
| APOC1 | apolipoprotein C1 | 10 | –5.47 |
| THBS1 | thrombospondin 1 | 10 | 2.21 |
| APOB | apolipoprotein B | 9 | –6.71 |
| NRP1 | neuropilin 1 | 9 | 1.80 |
| FGG | fibrinogen gamma chain | 9 | –6.37 |
| FGA | fibrinogen alpha chain | 9 | –5.49 |
| FGB | fibrinogen beta chain | 9 | –4.93 |
| SERPINE1 | serpin family E member 1 | 9 | 2.98 |
| CEACAM1 | carcinoembryonic antigen related cell adhesion molecule 1 | 8 | –2.67 |
| DYSF | dysferlin | 8 | 1.76 |
| NR1H4 | nuclear receptor subfamily 1 group H member 4 | 8 | –4.08 |
| TSPO | translocator protein | 8 | 2.48 |
| CPB2 | carboxypeptidase B2 | 8 | –5.98 |
| HNF4A | hepatocyte nuclear factor 4 alpha | 8 | –1.24 |
| XBP1 | X-box binding protein 1 | 8 | –1.38 |
| ANGPTL3 | angiopoietin like 3 | 7 | –3.90 |
| NR1H3 | nuclear receptor subfamily 1 group H member 3 | 7 | –1.42 |
| FLNA | filamin A | 7 | 2.32 |
| F2 | coagulation factor II, thrombin | 7 | –5.12 |
| BAD | BCL2 associated agonist of cell death | 7 | 0.96 |
| LIPC | lipase C, hepatic type | 7 | –4.24 |
| GAS6 | growth arrest specific 6 | 7 | 2.39 |
| VTN | vitronectin | 7 | –5.08 |
| FGFR1 | fibroblast growth factor receptor 1 | 7 | 1.49 |
| ARG1 | arginase 1 | 7 | –2.79 |
| CYBA | cytochrome b-245 alpha chain | 7 | –3.33 |
| SULT1E1 | sulfotransferase family 1E member 1 | 4 | –2.07 |
| ACOX1 | acyl-CoA oxidase 1 | 4 | –1.10 |
| Gene Symbol | Gene Name | Systemic Processes | log2(Fold- Change) |
|---|---|---|---|
| APOA1 | apolipoprotein A1 | 5 | –5.81 |
| APOA2 | apolipoprotein A2 | 4 | –8.62 |
| APOB | apolipoprotein B | 4 | –6.71 |
| ALB | albumin | 4 | –8.42 |
| SERPINC1 | serpin family C member 1 | 3 | –2.66 |
| GNG11 | G protein subunit gamma 11 | 3 | 3.24 |
| GNG12 | G protein subunit gamma 12 | 3 | 2.41 |
| FGG | fibrinogen gamma chain | 3 | –6.37 |
| FGA | fibrinogen alpha chain | 3 | –5.49 |
| F2 | coagulation factor II, thrombin | 3 | –5.12 |
| KNG1 | kininogen 1 | 3 | –1.82 |
| APOE | apolipoprotein E | 3 | –4.11 |
| NR1H3 | nuclear receptor subfamily 1 group H member 3 | 3 | –1.42 |
| GAS6 | growth arrest specific 6 | 3 | 2.39 |
| PROC | protein C, inactivator of coagulation factors Va and VIIIa | 3 | –3.25 |
| A2M | alpha-2-macroglobulin | 3 | –5.26 |
| SERPIND1 | serpin family D member 1 | 3 | –5.60 |
| F5 | coagulation factor V | 3 | –4.30 |
| ACOX1 | acyl-CoA oxidase 1 | 3 | –1.10 |
| PRKACB | protein kinase cAMP-activated catalytic subunit beta | 3 | 1.11 |
| TF | transferrin | 3 | –8.73 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agioutantis, P.C.; Loutrari, H.; Kolisis, F.N. Computational Analysis of Transcriptomic and Proteomic Data for Deciphering Molecular Heterogeneity and Drug Responsiveness in Model Human Hepatocellular Carcinoma Cell Lines. Genes 2020, 11, 623. https://doi.org/10.3390/genes11060623
Agioutantis PC, Loutrari H, Kolisis FN. Computational Analysis of Transcriptomic and Proteomic Data for Deciphering Molecular Heterogeneity and Drug Responsiveness in Model Human Hepatocellular Carcinoma Cell Lines. Genes. 2020; 11(6):623. https://doi.org/10.3390/genes11060623
Chicago/Turabian StyleAgioutantis, Panagiotis C., Heleni Loutrari, and Fragiskos N. Kolisis. 2020. "Computational Analysis of Transcriptomic and Proteomic Data for Deciphering Molecular Heterogeneity and Drug Responsiveness in Model Human Hepatocellular Carcinoma Cell Lines" Genes 11, no. 6: 623. https://doi.org/10.3390/genes11060623
APA StyleAgioutantis, P. C., Loutrari, H., & Kolisis, F. N. (2020). Computational Analysis of Transcriptomic and Proteomic Data for Deciphering Molecular Heterogeneity and Drug Responsiveness in Model Human Hepatocellular Carcinoma Cell Lines. Genes, 11(6), 623. https://doi.org/10.3390/genes11060623





