Evolution of the T-Cell Receptor (TR) Loci in the Adaptive Immune Response: The Tale of the TRG Locus in Mammals
Abstract
1. Introduction
2. The TR Chains are Encoded by Separate Multigene Families
3. The Homo sapiens TRG Locus
4. The “Gene Cluster” Organization of the TRG Locus Is Predominant in the Outgroups
| TRGV Genes | TRGJ Genes | TRGC Genes | Chromosomal Localization | Miniature Locus | References | ||
|---|---|---|---|---|---|---|---|
| BIRDS | Chicken | TRGV1 (6) TRGV2 (7) TRGV3 (2) TRGV4 (5) TRGV5 (2) TRGV6 (4) TRGV7 (4) TRGV8 (2) TRGV9 (3) TRGV10 (1) TRGV11 (1) | TRGJ1 (1) TRGJ2 (1) TRGJ3 (1) | TRGC | one locus Chrom. 2 |  Gallus gallus | Liu et al., [69]; |
| Duck | TRGV1 (5) TRGV2 (2) TRGV3 (3) TRGV4 (1) TRGV5 (1) TRGV6 (1) | TRGJ1 (1) TRGJ2 (1) TRGJ3 (1) TRGJ4 (1) TRGJ5 (1) | TRGC | one locus no assigned |  Anas platyrynchos | Yang et al., [70]; | |
| FISHES | Shark | TRGV1 (1) TRGV2 (1) TRGV3 (1) TRGV4 (1) TRGV5 (1) | TRGJ1 (1) TRGJ2 (1) TRGJ3 (1) | TRGC | one locus no assigned |  Carcharhinus plumbeus | Chen et al., [71]; |
| Atlantic salmon | TRGV1 (4) TRGV2 (3) | TRGJ1 (1) TRGJ2 (1) TRGJ3 (1) TRGJ4 (1) TRGJ5 (1) | TRGC1 (1) TRGC2 (1) TRGC3 (1) TRGC4 (1) TRGC5 (1) | two loci no assigned |  Salmo salar | Yazawa et al., [73]; | |
| REPTILES | Alligator | TRGV1 (1) TRGV2 (1) TRGV3 (1) TRGV4 (1) TRGV5 (6) TRGV6 (3) TRGV7 (1) TRGV8 (2) TRGV9 (1) TRGV10 (1) | TRGJ1 (1) TRGJ2 (1) TRGJ3 (1) TRGJ4 (1) TRGJ5 (1) TRGJ6 (1) TRGJ7 (1) TRGJ8 (1) TRGJ9 (1) | TRGC | one locus no assigned |  Alligator sinensis | Wang et al., [74]; |
| MARSUPIALS | Opossum | TRGV1 (5) TRGV2 (1) TRGV3 (2) TRGV4 (1) | TRGJ1 (1) TRGJ2 (1) TRGJ3 (1) TRGJ4 (1) TRGJ5 (1) TRGJ6 (1) TRGJ7 (1) | TRGC | one locus Chrom. 6q |  Monodelphis domestica | Parra et al., [75]; |
5. The TRG Locus in Cetartiodactyla and Carnivora: The “Gene Cassette” Model
5.1. Genomic Organization of Ovine, Bovine, Camel, and Dolphin TRG Loci
5.2. Genomic Organization of Canine and Feline TRG Loci
| TRGV Genes | TRGJ Genes | TRGC Genes | Chromosomal Localization | Miniature Locus | References | ||
|---|---|---|---|---|---|---|---|
| CETARTIODACTYLA | Ovine | TRGV1 (1) TRGV2 (1) TRGV3 (2) TRGV4 (1) TRGV5 (2) TRGV6 (1) TRGV7 (1) TRGV8 (1) TRGV9 (1) TRGV10 (1P) TRGV11 (1P) | TRGJ1 (2) TRGJ2 (2) TRGJ3 (2) * TRGJ4 (2) * TRGJ5 (3) ° TRGJ6 (2) | TRGC1 (1) TRGC2 (1) TRGC3 (1) TRGC4 (1) TRGC5 (1) TRGC6 (1) | Two loci Chrom. 4q31 Chrom. 4q22 |  Ovis aries | Massari et al., [78]; Miccoli et al., [85]; Vaccarelli et al., [84,89]; |
| Bovine | TRGV1 (1) TRGV2 (1) TRGV3 (2) TRGV4 (1) TRGV5(1+1O) TRGV6 (2) TRGV7 (1) TRGV8 (4) TRGV9 (2) TRGV10 (1) | TRGJ1 (2) TRGJ2 (2) TRGJ3 (1) TRGJ4 (2) TRGJ5 (1) TRGJ6 (1) | TRGC1 (1) TRGC2 (1) TRGC3 (1) TRGC4 (1) TRGC5 (1) TRGC6 (1) TRGC7 (1P) | Two loci Chrom. 4q31 Chrom. 4q1.5-2.2 |  Bos taurus | Antonacci et al., [86]; Conrad et al., [92]; | |
| Camel | TRGV1 (1) TRGV2 (1) TRGV3 (1) TRGV4 (1O) TRGV7 (1) TRGV10 (1) TRGV11 (1) | TRGJ1 (2) * TRGJ2 (2) * TRGJ5 (3) ° | TRGC1 (1) TRGC2 (1) TRGC5 (1) | One locus Chrom. 7 q11.12 |  Camelus dromedarius | Vaccarelli et al., [97]; Antonacci et al., [96]; | |
| Dolphin | TRGV1 (1) TRGV2 (1) | TRGJ1 (1) TRGJ2 (1) TRGJ3 (1) | TRGC | One locus no assigned |  Tursiops truncatus | Linguiti et al., [38]; | |
| CARNIVORA | Canine | TRGV1(1P) TRGV2(4) TRGV3 (3P) TRGV4 (1O) TRGV5(3) § TRGV6(1P) TRGV7(3) # | TRGJ1 (2) * TRGJ2 (2) * TRGJ3 (2) * TRGJ4 (2) * TRGJ5 (2) TRGJ6 (2) TRGJ7 (2) * TRGJ8 (2) * | TRGC1 (1O) TRGC2 (1) TRGC3 (1) TRGC4 (1) TRGC5 (1) TRGC6 (1P) TRGC7 (1) TRGC8 (1) | One locus Chrom. 18 |  Canis lupus familiaris | Massari et al., [104]; Martin et al., [39]; |
| Feline | TRGV2(4) TRGV5(4) $ TRGVA(1P) TRGVB(1P) TRGV6(1P) TRGV7(1) | TRGJ1 (2) * TRGJ2 (3) ^ TRGJ3 (2) * TRGJ4 (1P) TRGJ5 (2) * TRGJ6 (2) | TRGC1 (1) TRGC2 (1) TRGC3 (1) TRGC4 (1) TRGC5 (1P) TRGC6 (1P) | One locus Chrom. A2 |  Felis catus | Radtanakatikanon et al., [27]; |
| TRG1 locus | Ovis aries 4q3.1 | Bos taurus 4q3.1 | ||||
| Cassettes names | IMGT gene names | Nb of alleles | CDR-IMGT lengths | IMGT gene names | Nb of alleles | CDR-IMGT lengths |
| TRGC5 | TRGV11-1 | 1 P | [8.6.4] | |||
| TRGV3-1 | 2 F | [8.7.4] | TRGV3-1 | 1 F, 1 (F) | [8.7.4] | |
| TRGV3-2 | 1 F | [8.7.5] | TRGV3-2 | 1 F, 1 (F) | [8.7.4] | |
| TRGV7 | 1 F | [8.6.6] | TRGV7-1 | 1 F, 1 (F) | [8.6.6] | |
| TRGV10-1 | 1P | [9.7.4] | TRGV10-1 | 1 F | [9.7.5] | |
| TRGV4 | 2 F | [8.4.5] | TRGV4-1 | 1 (F) | [8.4.x] | |
| TRGJ5-1 | 1 F | - | TRGJ5-1 | 1 F | - | |
| TRGJ5-2 | 1 P | - | ||||
| TRGJ5-3 | 1 F | - | ||||
| TRGC5 (EX2A) | 1 (F) | - | TRGC5 (EX2A) | 1 F, 2 (F) | - | |
| (TRGC7) | TRGV8-1 | 1 F, 1 F | [3.8.4] | TRGV8-3 | 1 F, (1 F) | [3.8.5] |
| TRGV8-4 | 1 F | [3.8.5] | ||||
| TGRV8-1 | 1 F | [3.8.5] | ||||
| TRGV8-2 | 1 F, (1 F) | [3.8.5] | ||||
| TRGV2-1 | 2 F, 1 (F) | [6.8.5] del81,82,83 | TRGV2-1 | 1 F | [6.8.5] del81,82,83 | |
| TRGC7 (EX2A,2C) | 1 P | - | ||||
| TRGC3 | TRGV9-1 | 1 F, 1 (F) | [6.8.4] | TRGV9-1 | 1 F | [6.8.5] |
| TRGV9-2 | 1 F | [6.8.5] | ||||
| TRGJ3-1 | 1 F | - | TRGJ3-1 | 1 F, 1 (F) | - | |
| TRGJ3-2 | 1 P | - | ||||
| TRGC3 (EX2A,2C) | 1 F, 1 (F) | - | TRGC3 (EX2A,2C) | 1 F, 1 (F) | - | |
| TRGC4 | TRGV1(L41) | 2 F | [5.8.5] | TRGV1-1(L41) | 1 F, 2 (F) | [5.8.5] |
| TRGJ4-1 | 1 P | - | TRGJ4-1 | 1 F | - | |
| TRGJ4-2 | 1 F | - | TRGJ4-2 | 1 F | - | |
| TRGC4 (EX2A,2B,2C) | 1 (F) | - | TRGC4 (EX2A) | 1 F, 1 (F) | - | |
| TRG2 locus | Ovis aries 4q2.1 | Bos taurus 4q1.5-2.2 | ||||
| Cassettes | IMGT gene names | Nb of alleles | CDR-IMGT | Bos taurus | Nb of alleles | CDR-IMGT |
| TRGC1 | TRGV5-1(L41) | 2 F | [5.8.5] del5-8 | TRGV5-1(L41) | 1 F, 4 (F) | [5.8.5] del5-8 |
| TRGJ1-1 | 1 F | - | TRGJ1-1 | 1 F | - | |
| TRGJ1-2 | 1 F | - | TRGJ1-2 | 1 F, 2 (F) | - | |
| TRGC1 (EX2A) | 1 F | - | TRGC1 (EX2A,2B,2C) | 1 F, 3 (F) | - | |
| TRGC2 | TRGV6-1 | 1 F, 1 (F) | [5.8.5] del16 | |||
| TRGV5-2 | 2 F | [5.8.5] del5-8 | TRGV5-2* | 10 (F) | [5.8.5] del5-8 | |
| TRGJ2-1 | 1 F | - | TRGJ2-1* | 1 F, 2 (F) | - | |
| TRGJ2-2 | 1 F | - | TRGJ2-2 | 1 F, 2 (F) | - | |
| TRGC2 (EX2A,2B,2C) | 1 F, 1 (F) | - | TRGC2 (EX2A,2B,2C) | 1 F, 4 (F) | - | |
| TRGC6 | TRGV6-1 | 1 F, 1 (F) | [5.8.5] del16 | TRGV6-2* | 1 F | [5.8.5] del16 |
| TRGJ6-1 | 1 F | - | TRGJ6-1* | 1 F | - | |
| TRGJ6-2 | 1 F | - | - | |||
| TRGC6 (EX2A,2B,2C) | - | TRGC6 (EX2A,2B,2C) | 1 F, 1 (F) | - | ||
6. The TRG Locus in Rodentia, Lagomorpha, and Primata
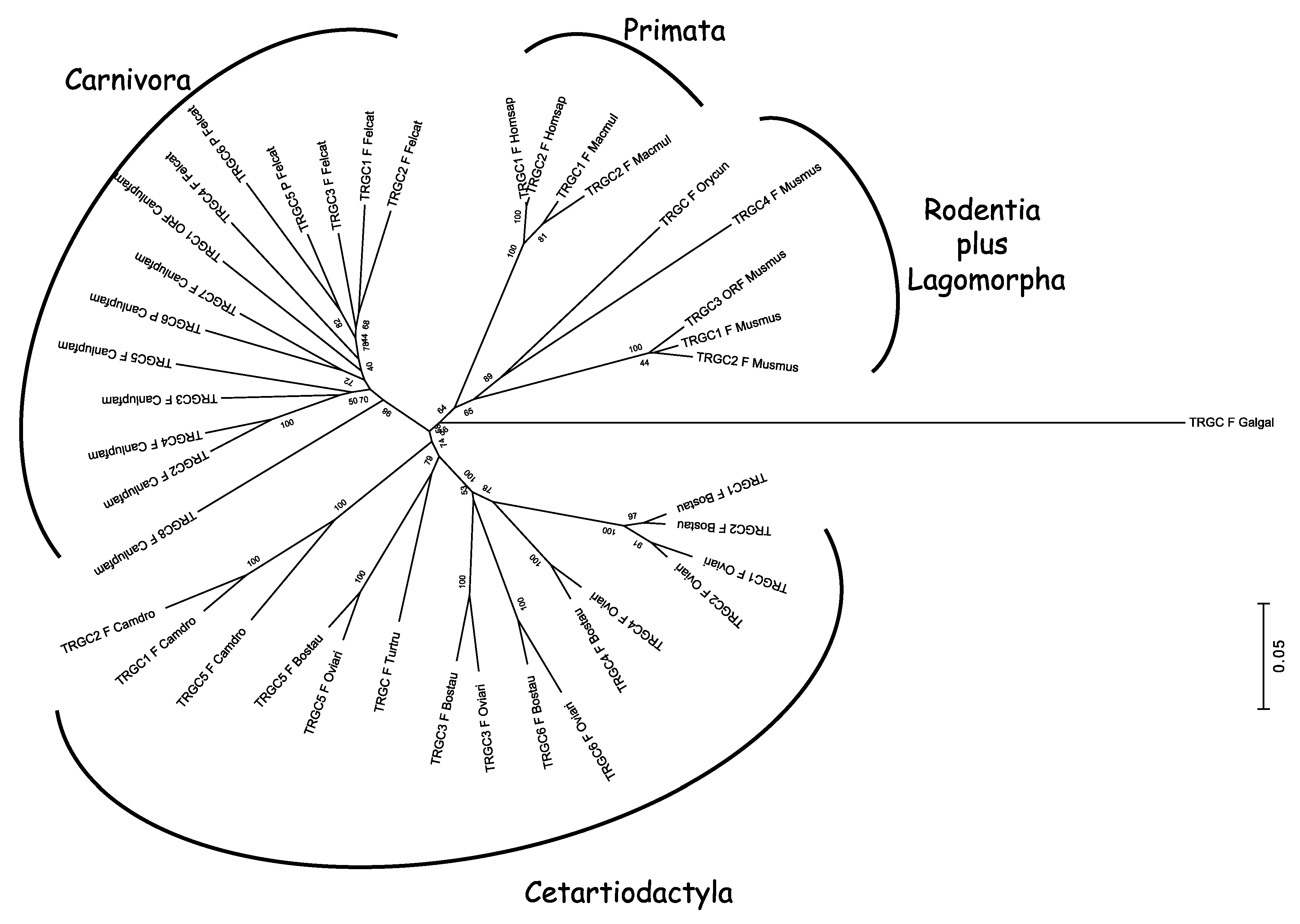
7. Phylogenetic Analysis Highlights a Diverse Mode of Evolution of the V, J, and C Subfamily Genes
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lefranc, M.-P.; Lefranc, G. The T Cell Receptor FactsBook; Academic Press. Harcourt Science and Technology Company: San Diego, CA, USA, 2001. [Google Scholar]
- Lefranc, M.-P. Immunoglobulin and T Cell Receptor Genes: IMGT® and the Birth and Rise of Immunoinformatics. Front. Immunol. 2014, 5, 22. [Google Scholar] [CrossRef]
- Janeway, C.A.; Jones, B.; Hayday, A. Specificity and function of T cells bearing γδ receptors. Immunol. Today 1988, 9, 73–76. [Google Scholar] [CrossRef]
- Sowder, J.T.; Yazawa, C.; Ager, L.L.; Chan, M.M.; Cooper, M. A large subpopulation of avian T cells express a homologue of the mammalian T gamma/delta receptor. J. Exp. Med. 1988, 167, 315–322. [Google Scholar] [CrossRef]
- Hein, W.R.; MacKay, C.R. Prominence of T-cell in the ruminant immune system. Immunol. Today 1991, 12, 30–34. [Google Scholar] [CrossRef]
- Cooper, M.D.; Chen, C.-L.H.; Bucy, R.P.; Thompson, C.B. Avian T cell ontogeny. Adv. Immunol. 1991, 50, 87–117. [Google Scholar] [PubMed]
- Binns, R.M.; Duncan, I.A.; Powis, S.J.; Hutchings, A.; Butcher, G.W. Subset of null and γδ T-cell receptor: T lymphocytes in the blood of young pigs identified by specific monoclonal antibodies. Immunology 1992, 77, 219–227. [Google Scholar] [PubMed]
- Ciccarese, S.; Lanave, C.; Saccone, C. Evolution of T-cell receptor gamma and delta constant region and other T-cell-related proteins in the human-rodentartiodactyl triplet. Genetics 1997, 145, 409–419. [Google Scholar] [PubMed]
- Lefranc, M.-P.; Lefranc, G. The Immunoglobulin FactsBook; Academic Press. Harcourt Science and Technology Company: San Diego, CA, USA, 2001. [Google Scholar]
- Lefranc, M.-P.; Duprat, E.; Kaas, Q.; Tranne, M.; Thiriot, A.; Lefranc, G. IMGT unique numbering for MHC groove G-Domain and MHC superfamily (MhcSF) G-Like-Domain. Dev. Comp. Immunol. 2005, 29, 917–938. [Google Scholar] [CrossRef]
- Wang, F.; Herzig, C.; Ozer, D.; Baldwin, C.L.; Telfer, J.C. Tyrosine phosphorylation of scavenger receptor cysteine-rich WC1 is required for the WC1-mediated potentiation of TCR-induced T-cell proliferation. Eur. J. Immunol. 2009, 39, 254–266. [Google Scholar] [CrossRef]
- Chen, C.; Hsu, H.; Hudgens, E.; Telfer, J.C.; Baldwin, C.L. Signal transduction by different forms of the gamma delta T cell-specific pattern recognition receptor WC1. J. Immunol. 2014, 193, 379–390. [Google Scholar] [CrossRef]
- Havran, W.L.; Jameson, J.M. Epidermal T cells and wound healing. J. Immunol. 2010, 184, 5423–5428. [Google Scholar] [CrossRef] [PubMed]
- Gober, H.J.; Kistowska, M.; Angman, L.; Jen€o, P.; Mori, L.; De Libero, G. Human T cell receptor gamma/delta cells recognize endogenous mevalonate metabolites in tumor cells. J. Exp. Med. 2003, 197, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Willcox, C.R.; Pitard, V.; Netzer, S.; Couzi, L.; Salim, M.; Silberzahn, T.; Moreau, J.-C.; Hayday, A.C.; Willcox, B.E.; Déchanet-Merville, J. Cytomegalovirus and tumor stress surveillance by binding of a human γ δ T cell antigen receptor to endothelial protein C receptor. Nat. Immunol. 2012, 13, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Uldrich, A.P.; Le Nours, J.; Pellicci, D.G.; Gherardin, N.A.; McPherson, K.G.; Lim, R.T.; Patel, O.; Beddoe, T.; Gras, S.; Rossjohn, J.; et al. CD1d-lipid antigen recognition by the γ δ TCR. Nat. Immunol. 2013, 14, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Vantourout, P.; Hayday, A. Six-of-the-best: Unique contributions of gamma/delta T cells to immunology. Nat. Rev. Immunol. 2013, 13, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Holderness, J.; Hedges, J.F.; Ramstead, A.; Jutila, M.A. Comparative biology of γ δ T cell function in humans, mice, and domestic animals. Annu. Rev. Anim. Biosci. 2013, 1, 99–124. [Google Scholar] [CrossRef]
- Telfer, J.C.; Baldwin, C.L. Bovine gamma delta T cells and the function of gamma/delta T cell specific WC1 co-receptors. Cell. Immunol. 2015, 296, 76–86. [Google Scholar] [CrossRef]
- Tunnacliffe, A.; Kefford, R.; Milstein, C.; Forster, A.; Rabbitts, T.H. Sequence and evolution of the human T-cell antigen receptor beta-chain genes. Proc. Natl. Acad. Sci. USA 1985, 82, 5068–5072. [Google Scholar] [CrossRef] [PubMed]
- Toyonaga, B.; Yoshikai, Y.; Vadasz, V.; Chin, B.; Mak, T.W. Organization and sequences of the diversity, joining, and constant region genes of the human T-cell receptor beta chain. Proc. Natl. Acad. Sci. USA 1985, 82, 8624–8628. [Google Scholar] [CrossRef]
- Gascoigne, N.R.; Chien, Y.; Becker, D.M.; Kavaler, J.; Davis, M.M. Genomic organization and sequence of T-cell receptor beta-chain constant- and joining-region genes. Nature 1984, 310, 387–391. [Google Scholar] [CrossRef]
- Jaeger, E.E.; Bontrop, R.E.; Lanchbury, J.S. Nucleotide sequences, polymorphism and gene deletion of T cell receptor beta-chain constant regions of Pan troglodytes and Macaca mulatta. J. Immunol. 1993, 151, 5301–5309. [Google Scholar] [PubMed]
- Mineccia, M.; Massari, S.; Linguiti, G.; Ceci, L.; Ciccarese, S.; Antonacci, R. New insight into the genomic structure of dog T cell receptor beta (TRB) locus inferred from expression analysis. Dev. Comp. Immunol. 2012, 37, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Antonacci, R.; Giannico, F.; Ciccarese, S.; Massari, S. Genomic characteristics of the T cell receptor (TRB) locus in the rabbit (Oryctolagus cuniculus) revealed by comparative and phylogenetic analyses. Immunogenetics 2014, 66, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Gerritsen, B.; Pandit, A.; Zaaraoui-Boutahar, F.; van den Hout, M.C.; van IJcken, W.F.; de Boer, R.J.; Andeweg, A.C. Characterization of the ferret TRB locus guided by V, D, J, and C gene expression analysis. Immunogenetics 2020, 72, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Radtanakatikanon, A.; Keller, S.M.; Darzentas, N.; Moore, P.F.; Folch, G.; Ngoune, V.N.; Lefranc, M.-P.; Vernau, W. Topology and expressed repertoire of the Felis catus T cell receptor loci. BMC Genom. 2020, 21, 20. [Google Scholar] [CrossRef]
- Antonacci, R.; Di Tommaso, S.; Lanave, C.; Cribiu, E.P.; Ciccarese, S.; Massari, S. Organization, structure and evolution of 41 kb of genomic DNA spanning the D-J-C region of the sheep TRB locus. Mol. Immunol. 2008, 45, 493–509. [Google Scholar] [CrossRef]
- Connelley, T.; Aerts, J.; Law, A.; Morrison, W.I. Genomic analysis reveals extensive gene duplication within the bovine TRB locus. BMC Genom. 2009, 10. [Google Scholar] [CrossRef]
- Eguchi-Ogawa, T.; Toki, D.; Uenishi, H. Genomic structure of the whole D-J-C clusters and the upstream region coding V segments of the TRB locus in pig. Dev. Comp. Immunol. 2009, 33, 1111–1119. [Google Scholar] [CrossRef]
- Antonacci, R.; Bellini, M.; Pala, A.; Mineccia, M.; Hassanane, M.S.; Ciccarese, S.; Massari, S. The occurrence of three D-J-C clusters within the dromedary TRB locus highlights a shared evolution in Tylopoda, Ruminantia and Suina. Dev. Comp. Immunol. 2017, 76, 105–119. [Google Scholar] [CrossRef]
- Antonacci, R.; Bellini, M.; Castelli, V.; Ciccarese, S.; Massari, S. Data charactering the genomic structure of the T cell receptor (TRB) locus in Camelus dromedarius. Data Brief 2017, 14, 507–514. [Google Scholar] [CrossRef]
- Massari, S.; Bellini, M.; Ciccarese, S.; Antonacci, R. Overview of the germline and expressed repertoires of the TRB genes in Sus scrofa. Front. Immunol. 2018, 9, 2526. [Google Scholar] [CrossRef]
- Antonacci, R.; Bellini, M.; Ciccarese, S.; Massari, S. Comparative analysis of the TRB locus in the Camelus genus. Front. Genet. 2019, 10, 482. [Google Scholar] [CrossRef]
- Ciccarese, S.; Burger, P.A.; Ciani, E.; Castelli, V.; Linguiti, G.; Plasil, M.; Massari, S.; Horin, P.; Antonacci, R. The Camel Adaptive Immune Receptors Repertoire as a Singular Example of Structural and Functional Genomics. Front. Genet. 2019, 10, 997. [Google Scholar] [CrossRef] [PubMed]
- Herzig, C.T.; Lefranc, M.-P.; Baldwin, C.L. Annotation and classification of the bovine T cell receptor delta genes. BMC Genom. 2010, 11, 100. [Google Scholar] [CrossRef] [PubMed]
- Piccinni, B.; Massari, S.; Caputi Jambrenghi, A.; Giannico, F.; Lefranc, M.-P.; Ciccarese, S.; Antonacci, R. Sheep (Ovis aries) T cell receptor alpha (TRA) and delta (TRD) genes and genomic organization of the TRA/TRD locus. BMC Genom. 2015, 16, 709. [Google Scholar] [CrossRef]
- Linguiti, G.; Antonacci, R.; Tasco, G.; Grande, F.; Casadio, R.; Massari, S.; Castelli, V.; Consiglio, A.; Lefranc, M.P.; Ciccarese, S. Genomic and expression analyses of Tursiops truncatus T cell receptor gamma (TRG) and alpha/delta (TRA/TRD) loci reveal a similar basic public γδ repertoire in dolphin and human. BMC Genom. 2016, 17, 634. [Google Scholar]
- Martin, J.; Ponstingl, H.; Lefranc, M.-P.; Archer, J.; Sargan, D.; Bradley, A. Comprehensive annotation and evolutionary insights into the canine (Canis lupus familiaris) antigen receptor loci. Immunogenetics 2018, 70, 223–236. [Google Scholar] [CrossRef]
- Mondot, S.; Lantz, O.; Lefranc, M.-P.; Boudinot, P. The T cell receptor (TRA) locus in the rabbit (Oryctolagus cuniculus): Genomic features and consequences for invariant T cells. Eur. J. Immunol. 2019, 49, 2146–2158. [Google Scholar] [CrossRef]
- Lefranc, M.-P.; Chuchana, P.; Dariavach, P.; Nguyen, C.; Huck, S.; Brockly, F.; Jordan, B.; Lefranc, G. Molecular mapping of the human T cell receptor gamma (TRG) genes and linkage of the variable and constant regions. Eur. J. Immunol. 1989, 19, 989–994. [Google Scholar] [CrossRef]
- Lefranc, M.-P.; Rabbitts, T.H. Two tandemly organized human genes encoding the T-cell gamma constant-region sequences show multiple rearrangement in different T.-cell types. Nature 1985, 316, 464–466. [Google Scholar] [CrossRef]
- Lefranc, M.-P.; Forster, A.; Rabbitts, T.H. Rearrangement of two distinct T-cell gamma-chain variable-region genes in human DNA. Nature 1986, 319, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, M.-P.; Forster, A.; Baer, R.; Stinson, M.A.; Rabbitts, T.H. Diversity and rearrangement of the human T cell rearranging gamma genes: Nine germ-line variable genes belonging to two subgroups. Cell 1986, 45, 237–246. [Google Scholar] [CrossRef]
- Lefranc, M.-P.; Forster, A.; Rabbitts, T.H. Genetic polymorphism and exon changes of the constant regions of the human T-cell rearranging gene gamma. Proc. Natl. Acad. Sci. USA 1986, 83, 9596–9600. [Google Scholar] [CrossRef] [PubMed]
- Forster, A.; Huck, S.; Ghanem, N.; Lefranc, M.-P.; Rabbitts, T.H. New subgroups in the human T cell rearranging Vgamma gene locus. EMBO J. 1987, 6, 1945–1950. [Google Scholar] [CrossRef] [PubMed]
- Huck, S.; Lefranc, M.-P. Rearrangements to the JP1, JP and JP2 segments in the human T-cell rearranging gamma gene (TRGgamma) locus. FEBS Lett. 1987, 224, 291–296. [Google Scholar] [CrossRef]
- Huck, S.; Dariavach, P.; Lefranc, M.-P. Variable region genes in the human T-cell rearranging gamma (TRG) locus: V-J junction and homology with the mouse genes. EMBO J. 1988, 7, 719–726. [Google Scholar] [CrossRef]
- Lefranc, M.P.; Rabbitts, T.H. The human T-cell receptor gamma (TRG) genes. TIBS 1989, 14, 214–218. [Google Scholar]
- Lefranc, M.-P.; Rabbitts, T.H. A nomenclature to fit the organization of the human T cell receptor gamma and delta genes. Res. Immunol. 1990, 141, 615–618. [Google Scholar] [CrossRef]
- Lefranc, M.-P.; Rabbitts, T.H. Genetic organization of the human T-cell receptor gamma and delta loci. Res. Immunol. 1990, 141, 565–577. [Google Scholar] [CrossRef]
- Zhang, X.M.; Tonnelle, C.; Lefranc, M.-P.; Huck, S. T cell receptor gamma cDNA in human fetal liver and thymus: Variable regions of gamma chains are restricted to VgammaI or V9, due to the absence of splicing of the V10 and V11 leader intron. Eur. J. Immunol. 1994, 24, 571–578. [Google Scholar] [CrossRef]
- Zhang, X.M.; Cathala, G.; Soua, Z.; Lefranc, M.-P.; Huck, S. The human T-cell receptor gamma variable pseudogene V10 is a distinctive marker of human speciation. Immunogenetics 1996, 43, 196–203. [Google Scholar] [CrossRef]
- Lefranc, M.-P. Locus maps and genomic repertoire of the human T cell receptor genes. Immunologist 2000, 8, 72–79. [Google Scholar]
- Lefranc, M.-P. Nomenclature of the human T cell receptor genes. In Current Protocols in Immunology; John Wiley and Sons: Hoboken, NJ, USA, 2000; pp. A.1O.1–A.1O.23. [Google Scholar]
- Lefranc, M.-P. Unique database numbering system for immunogenetic analysis. Immunol. Today 1997, 18, 509. [Google Scholar] [CrossRef]
- Lefranc, M.-P. The IMGT unique numbering for Immunoglobulins, T cell receptors and Ig-like domains. Immunologist 1999, 7, 132–136. [Google Scholar]
- Lefranc, M.-P.; Pommié, C.; Ruiz, M.; Giudicelli, V.; Foulquier, E.; Truong, L.; Thouvenin-Contet, V.; Lefranc, G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003, 27, 55–77. [Google Scholar] [CrossRef]
- Lefranc, M.-P. IMGT Collier de Perles for the Variable (V), Constant (C), and Groove (G) Domains of IG, TR, MH, IgSF, and MhSF. Cold Spring Harb. Protoc. 2011, 6, 643–651. [Google Scholar] [CrossRef]
- Lefranc, M.-P.; Pommié, C.; Kaas, Q.; Duprat, E.; Bosc, N.; Guiraudou, D.; Jean, C.; Ruiz, M.; Da Piedade, I.; Rouard, M.; et al. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev. Comp. Immunol. 2005, 29, 185–203. [Google Scholar] [CrossRef]
- Ghanem, N.; Buresi, C.; Moisan, J.-P.; Bensmana, M.; Chuchana, P.; Huck, S.; Lefranc, G.; Lefranc, M.-P. Deletion, insertion, and restriction site polymorphism of the T-cell receptor gamma variable locus in French, Lebanese, Tunisian and Black African populations. Immunogenetics 1989, 30, 350–360. [Google Scholar] [CrossRef]
- Ghanem, N.; Soua, Z.; Zhang, X.G.; Zijun, M.; Zhiwei, Y.; Lefranc, G.; Lefranc, M.-P. Polymorphism of the T-cell receptor gamma variable and constant region genes in a Chinese population. Hum. Genet. 1991, 86, 450–456. [Google Scholar] [CrossRef]
- Buresi, C.; Ghanem, N.; Huck, S.; Lefranc, G.; Lefranc, M.-P. Exon duplication and triplication in the human T-cell receptor gamma constant region genes and RFLP in French, Lebanese, Tunisian, and Black African populations. Immunogenetics 1989, 29, 161–172. [Google Scholar] [CrossRef]
- Lefranc, M.-P.; Alexandre, D. Gamma/delta lineage-specific transcription of human T cell receptor gamma genes by a combination of a non-lineage-specific enhancer and silencers. Eur. J. Immunol. 1995, 25, 617–622. [Google Scholar] [CrossRef]
- Six, A.; Rast, J.P.; McCormack, W.T.; Dunon, D.; Courtois, D.; Li, Y.; Chen, C.-L.H.; Cooper, M.D. Characterization of avian T-cell receptor γ genes. Proc. Natl. Acad. Sci. USA 1996, 96, 15329–15334. [Google Scholar] [CrossRef]
- Hayday, A.C.; Saito, H.; Gillies, S.D.; Kranz, D.M.; Tanigawa, G.; Eisen, H.N.; Tonegawa, S. Structure, organization, and somatic rearrangement of T cell gamma genes. Cell 1985, 40, 259–269. [Google Scholar] [CrossRef]
- Raulet, D.H. The structure, function, and molecular genetics of the gamma/delta T cell receptor. Ann. Rev. Immunol. 1989, 7, 175–207. [Google Scholar] [CrossRef]
- Vernooij, B.T.; Lenstra, J.A.; Wang, K.; Hood, L. Organization of the murine T-cell receptor gamma locus. Genomics 1993, 17, 566–574. [Google Scholar] [CrossRef]
- Liu, F.; Li, J.; Lin, I.; Yang, X.; Ma, J.; Chen, Y.; Lv, N.; Shi, Y.; Gao, G.F.; Zhu, B. The Genome Resequencing of TCR Loci in Gallus gallus Revealed Their Distinct Evolutionary Features in Avians. ImmunoHorizons 2020, 4, 33–46. [Google Scholar] [CrossRef]
- Yang, Z.; Sun, Y.; Ma, Y.; Li, Z.; Zhao, Y.; Ren, L.; Han, H.; Jiang, Y.; Zhao, Y. A comprehensive analysis of the germline and expressed TCR repertoire in White Peking duck. Sci. Rep. 2017, 7, 41426. [Google Scholar] [CrossRef]
- Chen, H.; Kshirsagar, S.; Jensen, I.; Lau, K.; Covarrubias, R.; Schluter, S.F.; Marchalonis, J.J. Characterization of arrangement and expression of the T cell receptor gamma locus in the sandbar shark. Proc. Natl. Acad. Sci. USA 2009, 106, 8591–8596. [Google Scholar] [CrossRef]
- Chen, H.; Bernstein, H.; Ranganathan, P.; Schluter, S.F. Somatic hypermutation of TCR γ V genes in the sandbar shark. Dev. Comp. Immunol. 2012, 37, 176–183. [Google Scholar] [CrossRef]
- Yazawa, R.; Cooper, G.A.; Beetz-Sargent, M.; Robb, A.; McKinnel, L.; Davidson, W.S.; Koop, B.F. Functional adaptive diversity of the Atlantic salmon T-cell receptor gamma locus. Mol. Immunol. 2008, 45, 2150–2157. [Google Scholar] [CrossRef]
- Wang, X.; Wang, P.; Wang, R.; Wang, C.; Bai, J.; Ke, C.; Yu, D.; Li, K.; Ma, Y.; Han, H.; et al. Analysis of TCRβ and TCRγ genes in Chinese alligator provides insights into the evolution of TCR genes in jawed vertebrates. Dev. Comp. Immunol. 2018, 85, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Parra, Z.E.; Baker, M.L.; Hathaway, J.; Lopez, A.M.; Trujillo, J.; Sharp, A.; Miller, R.D. Comparative genomic analysis and evolution of the T cell receptor loci in the opossum Monodelphis domestica. BMC Genom. 2008, 9, 111. [Google Scholar] [CrossRef]
- Deakin, J.E.; Parra, Z.E.; Graves, J.A.M.; Miller, R.D. Physical Mapping of T cell receptor loci (TRA, TRB, TRD and TRG) in the opossum (Monodelphis domestica). Cytogenet. Genome Res. 2006, 112, 342K. [Google Scholar] [CrossRef] [PubMed]
- Parra, Z.E.; Arnold, T.; Nowak, M.A.; Hellman, L.; Miller, R.D. TCR gamma chain diversity in the spleen of the duckbill platypus (Ornithorhynchus anatinus). Dev. Comp. Immunol. 2006, 30, 699–710. [Google Scholar] [CrossRef]
- Massari, S.; Lipsi, M.R.; Vonghia, G.; Antonacci, R.; Ciccarese, S. T-cell receptor TCRG1 and TCRG2 clusters map separately in two different regions of sheep chromosome 4. Chromosome Res. 1998, 6, 419–420. [Google Scholar] [CrossRef]
- Bensmana, M.; Mattei, M.G.; Lefranc, M.-P. Localization of the human T-cell receptor gamma locus (TCRG) to 7p14-p15 by in situ hybridization. Cytogenet. Cell Genet. 1991, 56, 31–32. [Google Scholar] [CrossRef]
- Robinson, M.H.; Mitchell, M.P.; Wei, S.; Day, C.E.; Zhao, T.M.; Concannon, P. Organization of human T-cell receptor beta chain genes: Clusters of V beta genes are present on chromosomes 7 and 9. Proc. Natl. Acad. Sci. USA 1993, 90, 2433–2437. [Google Scholar] [CrossRef]
- Rowen, L.; Koop, B.F.; Hood, L. The complete 685-kilobase DNA sequence of the human beta T-cell receptor locus. Science 1996, 272, 1755–1762. [Google Scholar] [CrossRef]
- Hein, W.R.; Dudler, L. Divergent evolution of T-cell repertoire: Extensive diversity and developmentally regulated expression of the sheep γδ T-cell receptor. EMBO J. 1993, 12, 715–724. [Google Scholar] [CrossRef]
- Hein, W.R.; Dudler, L. TCR γ δ cells are prominent in normal bovine skin and express a diverse repertoire of antigen receptors. Immunology 1997, 91, 58–64. [Google Scholar] [CrossRef]
- Vaccarelli, G.; Miccoli, M.C.; Lanave, C.; Massari, S.; Cribiu, E.P.; Ciccarese, S. Genomic organization of the sheep TRG1 locus and comparative analyses of Bovidae and human variable genes. Gene 2005, 357, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Miccoli, M.C.; Antonacci, R.; Vaccarelli, G.; Lanave, C.; Massari, S.; Cribiu, E.; Ciccarese, S. Evolution of TRG clusters in cattle and sheep genomes as drawn from the structural analysis of the ovine TRG2@ locus. J. Mol. Evol. 2003, 57, 52–62. [Google Scholar] [CrossRef]
- Antonacci, R.; Vaccarelli, G.; Di Meo, G.P.; Piccinni, B.; Miccoli, M.C.; Cribiu, E.P.; Perucatti, A.; Iannuzzi, L.; Ciccarese, S. Molecular in situ hybridization analysis of sheep and goat BAC clones identifies the transcriptional orientation of T cell receptor gamma genes on chromosome 4 in bovids. Vet. Res. Commun. 2007, 31, 977–983. [Google Scholar] [CrossRef]
- Lee, H.C.; Ye, S.K.; Honjo, T.; Ikuta, K. Induction of germline transcription in the human TCR gamma locus by STAT5. J. Immunol. 2001, 167, 320–326. [Google Scholar] [CrossRef]
- Miccoli, M.C.; Vaccarelli, G.; Lanave, C.; Cribiu, E.P.; Ciccarese, S. Comparative analyses of sheep and human TRG joining regions: Evolution of J genes in Bovidae is driven by sequence conservation in their promoters for germline transcription. Gene 2005, 355, 67–78. [Google Scholar] [CrossRef]
- Vaccarelli, G.; Miccoli, M.C.; Antonacci, R.; Pesole, G.; Ciccarese, S. Genomic organization and recombinational unit duplication-driven evolution of ovine and bovine T cell receptor gamma loci. BMC Genom. 2008, 9, 81. [Google Scholar] [CrossRef]
- Miccoli, M.C.; Lipsi, M.R.; Massari, S.; Lanave, C.; Ciccarese, S. Exon-intron organization of TRGC genes in sheep. Immunogenetics 2001, 53, 416–422. [Google Scholar] [CrossRef]
- Herzig, C.; Blumerman, S.; Lefranc, M.P.; Baldwin, C. Bovine T cell receptor gamma variable and constant genes: Combinatorial usage by circulating gammadelta T cells. Immunogenetics 2006, 58, 138–151. [Google Scholar] [CrossRef]
- Conrad, M.L.; Mawer, M.A.; Lefranc, M.P.; McKinnell, L.; Whitehead, J.; Davis, S.K.; Pettman, R.; Koop, B.F. The genomic sequence of the bovine T cell receptor gamma TRG loci and localization of the TRGC5 cassette. Vet. Immunol. Immunopathol. 2007, 115, 346–356. [Google Scholar] [CrossRef]
- Lefranc, M.-P. WHO-IUIS Nomenclature Subcommittee for Immunoglobulins and T cell receptors report. Immunogenetics 2007, 59, 899–902. [Google Scholar] [CrossRef]
- Blumerman, S.L.; Herzig, C.T.; Rogers, A.N.; Telfer, J.C.; Baldwin, C.L. Differential TCR gene usage between WC1- and WC1+ ruminant gammadelta T cell subpopulations including those responding to bacterial antigen. Immunogenetics 2006, 58, 680–692. [Google Scholar] [CrossRef]
- Damani-Yokota, P.; Gillespie, A.; Pasman, Y.; Merico, D.; Connelley, T.K.; Kaushik, A.; Baldwin, C.L. Bovine T cell receptors and γ δ WC1 co-receptor transcriptome analysis during the first month of life. Dev. Comp. Immunol. 2018, 88, 190–199. [Google Scholar] [CrossRef]
- Antonacci, R.; Linguiti, G.; Burger, P.A.; Castelli, V.; Pala, A.; Fitak, R.; Massari, S.; Ciccarese, S. Comprehensive genomic analysis of the dromedary T cell receptor gamma (TRG) locus and identification of a functional TRGC5 cassette. Dev. Comp. Immunol. 2020, 106, 103614. [Google Scholar] [CrossRef]
- Vaccarelli, G.; Antonacci, R.; Tasco, G.; Yang, F.; El Ashmaoui, H.M.; Hassanane, M.S.; Massari, S.; Casadio, R.; Ciccarese, S. Generation of diversity by somatic mutation in the Camelus dromedarius T-cell receptor gamma (TCRG) variable domains. Eur. J. Immunol. 2012, 42, 1–13. [Google Scholar] [CrossRef]
- Ciccarese, S.; Vaccarelli, G.; Lefranc, M.-P.; Tasco, G.; Consiglio, A.; Casadio, R.; Linguiti, G.; Antonacci, R. Characteristics of the somatic hypermutation in the Camelus dromedarius T cell receptor gamma (TRG) and delta (TRD) variable domains. Dev. Comp. Immunol. 2014, 46, 300–313. [Google Scholar] [CrossRef]
- Kotani, A.; Okazaki, I.M.; Muramatsu, M.; Kinoshita, K.; Begum, N.A.; Nakajima, T.; Saito, H.; Honjo, T. A target selection of somatic hypermutations is regulated similarly between T and B cells upon activation induced cytidine deaminase expression. Proc. Natl. Acad. Sci. USA 2005, 102, 4506–4511. [Google Scholar] [CrossRef]
- Antonacci, R.; Mineccia, M.; Lefranc, M.-P.; Ashmaoui, H.M.; Lanave, C.; Piccinni, B.; Pesole, G.; Hassanane, M.S.; Massari, S.; Ciccarese, S. Expression and genomic analyses of Camelus dromedarius T cell receptor delta (TRD) genes reveal a variable domain repertoire enlargement due to CDR3 diversification and somatic mutation. Mol. Immunol. 2011, 48, 1384–1396. [Google Scholar] [CrossRef]
- Venturi, V.; Kedzierska, K.; Tanaka, M.M.; Turner, S.J.; Doherty, P.C.; Davenport, M.P. Method for assessing the similarity between subsets of the T cell receptor repertoire. J. Immunol. Methods 2008, 329, 67–80. [Google Scholar] [CrossRef]
- Dariavach, P.; Lefranc, M.-P. The promoter regions of the T-cell receptor V9 gamma (TRGV9) and V2 delta (TRDV2) genes display short direct repeats but no TATA box. FEBS Lett. 1989, 256, 185–191. [Google Scholar] [CrossRef]
- Pauza, C.D.; Cairo, C. Evolution and function of the TCR Vgamma9 chain repertoire: It’s good to be public. Cell. Immunol. 2015, 296, 22–30. [Google Scholar] [CrossRef]
- Massari, S.; Bellahcene, F.; Vaccarelli, G.; Carelli, G.; Mineccia, M.; Lefranc, M.-P.; Antonacci, R.; Ciccarese, S. The deduced structure of the T cell receptor gamma locus in Canis lupus familiaris. Mol. Immunol. 2009, 46, 2728–2736. [Google Scholar] [CrossRef]
- Garman, R.D.; Doherty, P.J.; Raulet, D.H. Diversity, rearrangement, and expression of murine T cell gamma genes. Cell 1986, 45, 733–742. [Google Scholar] [CrossRef]
- Traunecker, A.; Oliveri, F.; Allen, N.; Karjalainen, K. Normal T cell development is possible without ‘functional’ gamma chain genes. EMBO J. 1986, 5, 1589–1593. [Google Scholar] [CrossRef]
- Pelkonen, J.; Traunecker, A.; Karjalainen, K. A new mouse TCR V gamma gene that shows remarkable evolutionary conservation. EMBO J. 1987, 6, 1941–1944. [Google Scholar] [CrossRef]
- Spencer, D.M.; Hsiang, Y.H.; Goldman, J.P.; Raulet, D.H. Identification of a T-cell-specific transcriptional enhancer located 3′ of C gamma 1 in the murine T-cell receptor gamma locus. Proc. Natl. Acad. Sci. USA 1991, 88, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Massari, S.; Ciccarese, S.; Antonacci, R. Structural and comparative analysis of the T cell receptor gamma (TRG) locus in Oryctolagus cuniculus. Immunogenetics 2012, 64, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Kazen, A.R.; Adams, E.J. Evolution of the V, D, and J gene segments used in the primate gammadelta T-cell receptor reveals a dichotomy of conservation and diversity. Proc. Natl. Acad. Sci. USA 2011, 108, E332–E340. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment with reduced time and space complexity. BMC Bioinf. 2004, 5, 113. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Ohno, S. Evolution by Gene Duplication; Springer: New York, NY USA, 1970. [Google Scholar]
- Thome, M.; Saalmuller, A.; Pfaff, E. Molecular cloning of porcine T cell receptor α, β, γ and δ chains using polymerase chain reaction fragments of the constant regions. Eur. J. Immunol. 1993, 23, 1005–1010. [Google Scholar] [CrossRef]
- Schrenzel, M.D.; Ferrick, D.A. Horse (Equus caballus) T-cell receptor alpha, gamma, and delta chain genes: Nucleotide sequences and tissue specific gene expression. Immunogenetics 1995, 42, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Bäckström, B.T.; Milia, E.; Peter, A.; Jaureguiberry, B.; Baldari, C.T.; Palmer, E. A motif within the T cell receptor alpha chain constant region connecting peptide domain controls antigen responsiveness. Immunity 1996, 5, 437–447. [Google Scholar] [CrossRef]
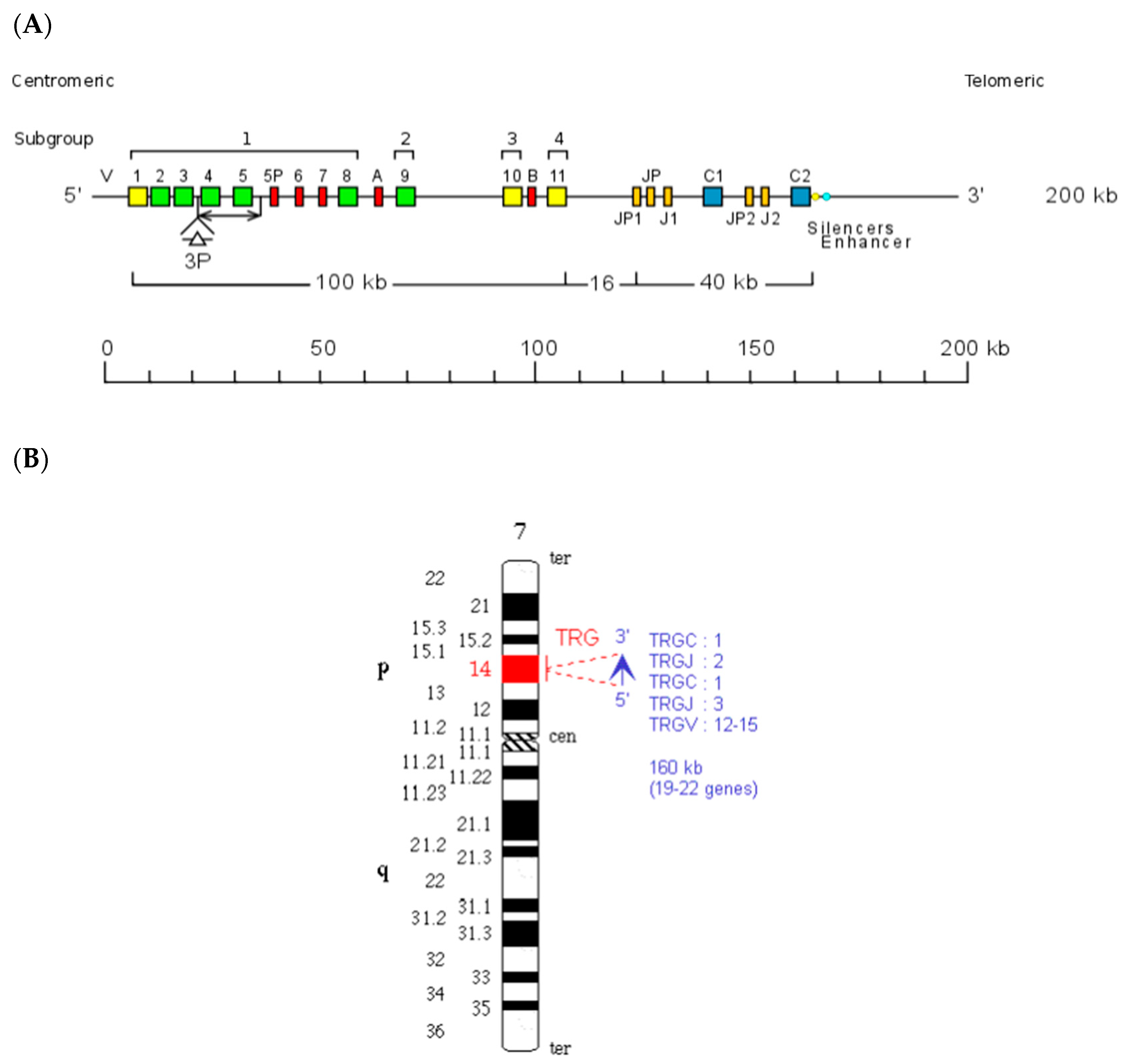



| TRGV Genes | TRGJ Genes | TRGC Genes | Chromosomal Localization | Miniature Locus | References | ||
|---|---|---|---|---|---|---|---|
| RODENTIA | Mouse | TRGV1 TRGV2 TRGV3 TRGV4 TRGV5 TRGV6 TRGV7 | TRGJ1 TRGJ2 TRGJ3 TRGJ4 | TRGC1 TRGC2 TRGC3 (P) TRGC4 | One locus Chrom. 13A2 |  Mus musculus | Hayday et al., [66]; Garman et al., [105]; Traunecker et al., [106]; Pelkonen et al., [107]; Vernooij et al., [68]; |
| LAGOMORPHA | Rabbit | TRGV1 (7+1O) TRGV2 (O) TRGV3 TRGV4 (O) | TRGJ1 TRGJ2 | TRGC | One locus Chrom. 10 |  Oryctolagus cuniculus | Massari et al., [109]; |
| PRIMATA | Rhesus monkey | TRGV1 (O) TRGV2 TRGV3 TRGV6 (P) TRGV8 TRBVA (P) TRGV9 TRGV10 (O) TRGVB (P) TRGV11 (P) TRGVC (P) TRGVD (P) | TRGJ1-1 TRGJ1-2 TRGJ2-1 TRGJ2-2 (P) TRGJ2-3 | TRGC1 TRGC2 | One locus Chrom. 3 |  Macaca mulatta | Lefranc M-P., [2]; |
| Human | TRGV1 (O) TRGV2 TRGV3 TRGV4 TRGV5 TRGV5P (P) TRGV6 (P) TRGV7 (P) TRGV8 TRGVA (P) TRGV9 TRGV10 (O) TRGVB (P) TRGV11 (O) | TRGJP1 TRGJP TRGJ1 TRGJP2 TRGJ2 | TRGC1 TRGC2 | One locus Chrom. 7p14 |  Homo sapiens | Lefranc, and Rabbitts, [42]; Lefranc, [43,44,45]; Lefranc and Rabbitts, [49]; |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonacci, R.; Massari, S.; Linguiti, G.; Caputi Jambrenghi, A.; Giannico, F.; Lefranc, M.-P.; Ciccarese, S. Evolution of the T-Cell Receptor (TR) Loci in the Adaptive Immune Response: The Tale of the TRG Locus in Mammals. Genes 2020, 11, 624. https://doi.org/10.3390/genes11060624
Antonacci R, Massari S, Linguiti G, Caputi Jambrenghi A, Giannico F, Lefranc M-P, Ciccarese S. Evolution of the T-Cell Receptor (TR) Loci in the Adaptive Immune Response: The Tale of the TRG Locus in Mammals. Genes. 2020; 11(6):624. https://doi.org/10.3390/genes11060624
Chicago/Turabian StyleAntonacci, Rachele, Serafina Massari, Giovanna Linguiti, Anna Caputi Jambrenghi, Francesco Giannico, Marie-Paule Lefranc, and Salvatrice Ciccarese. 2020. "Evolution of the T-Cell Receptor (TR) Loci in the Adaptive Immune Response: The Tale of the TRG Locus in Mammals" Genes 11, no. 6: 624. https://doi.org/10.3390/genes11060624
APA StyleAntonacci, R., Massari, S., Linguiti, G., Caputi Jambrenghi, A., Giannico, F., Lefranc, M.-P., & Ciccarese, S. (2020). Evolution of the T-Cell Receptor (TR) Loci in the Adaptive Immune Response: The Tale of the TRG Locus in Mammals. Genes, 11(6), 624. https://doi.org/10.3390/genes11060624







