Cardiac Transcriptome Analysis Reveals Nr4a1 Mediated Glucose Metabolism Dysregulation in Response to High-Fat Diet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Biochemical Analyses
2.3. Heart Histopathology
2.4. Total RNA Extraction and Validation
2.5. Transcriptome Data Mapping and Differential Expression Analysis
2.6. H9c2 Culture and Treatment
2.7. Quantitative Real-Time PCR (qPCR) Validation
2.8. Western Blot Analysis
2.9. Statistical Analysis
3. Results
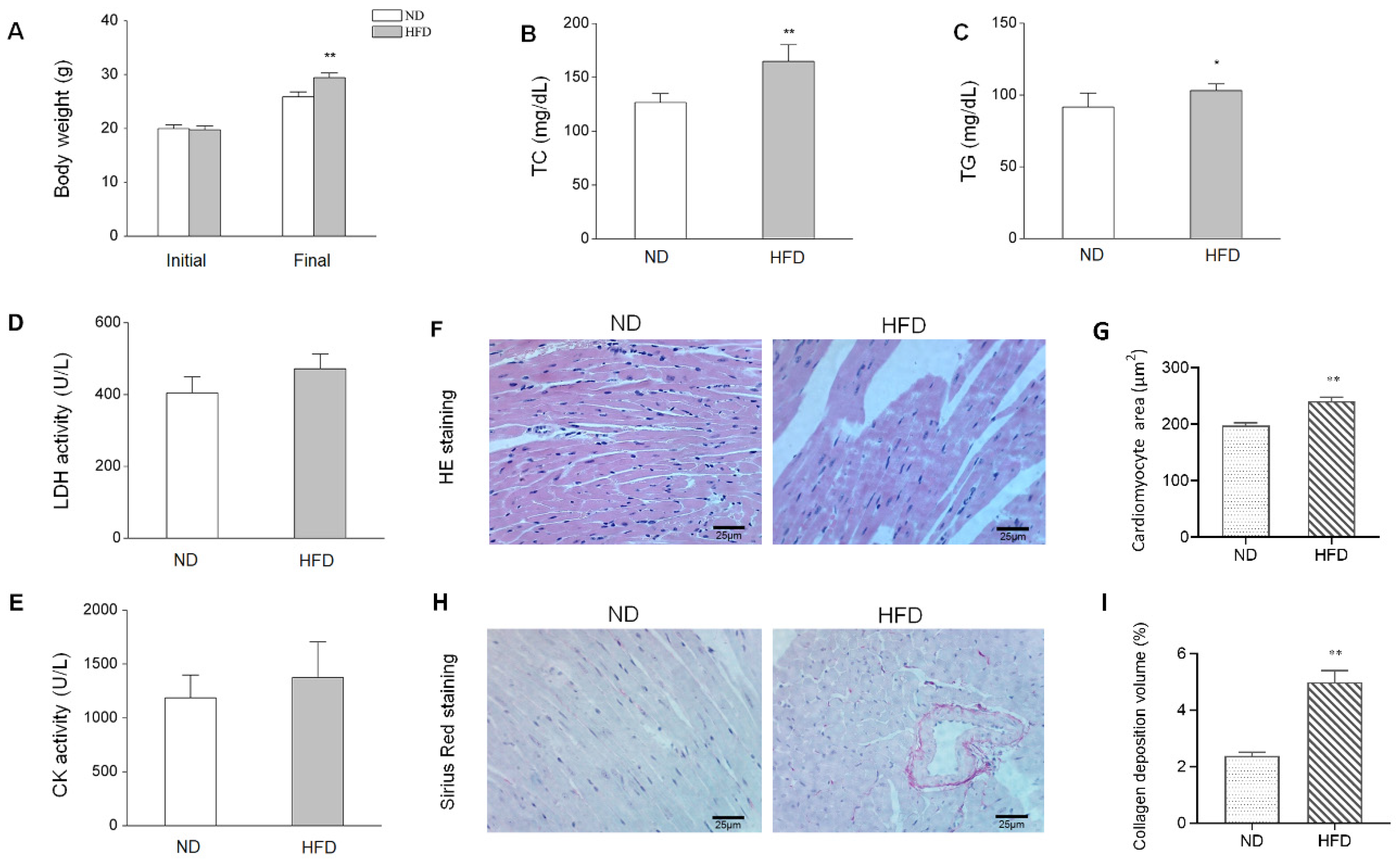
3.1. HFD-Induced Animal Characteristics, Alterations, and cardiac Injury
3.2. HFD-Induced Alteration of Myocardial Gene Expression
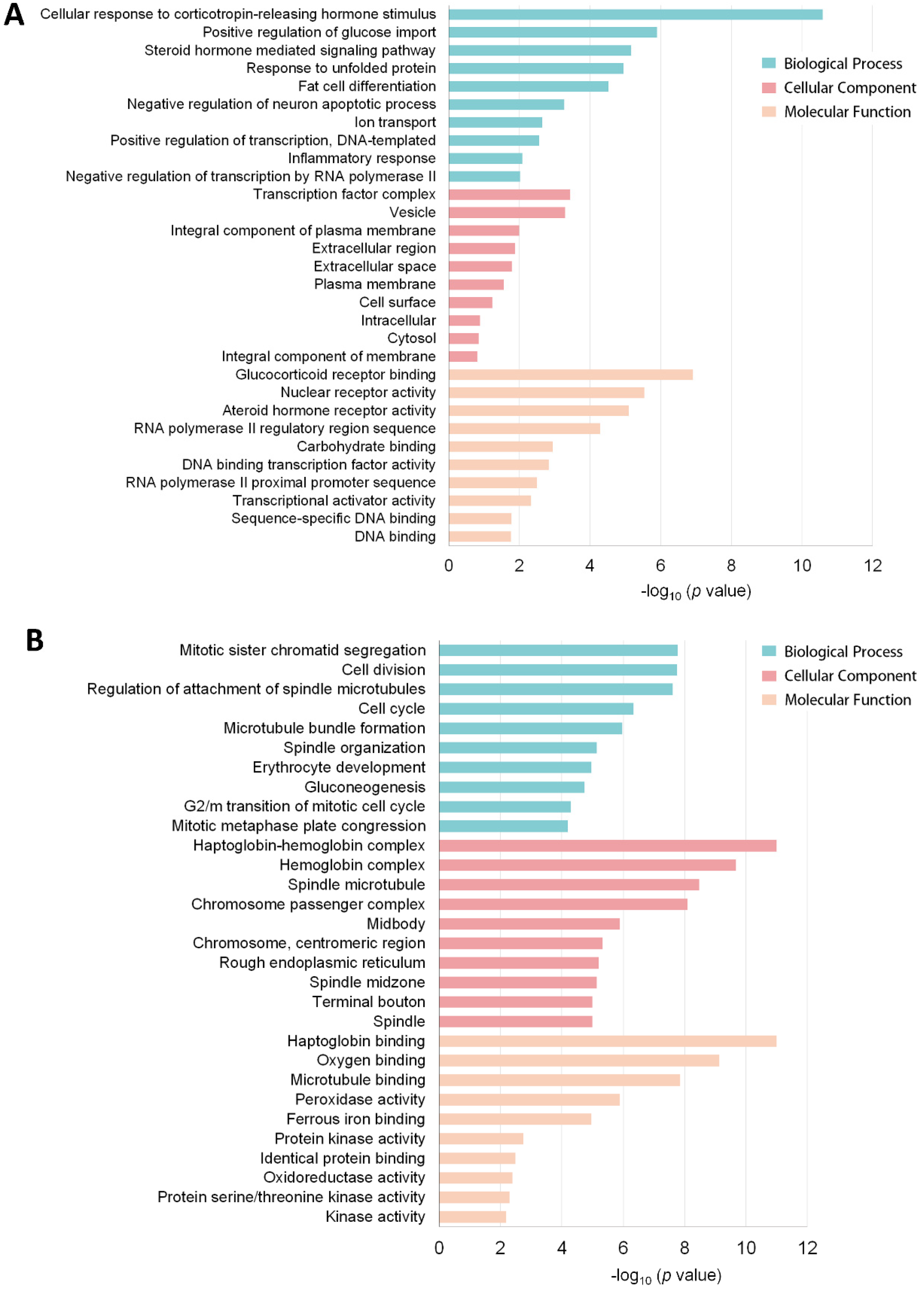
3.3. Functional Annotation of the DEG Profile
3.4. KEGG Pathway Enrichment and Network Analysis
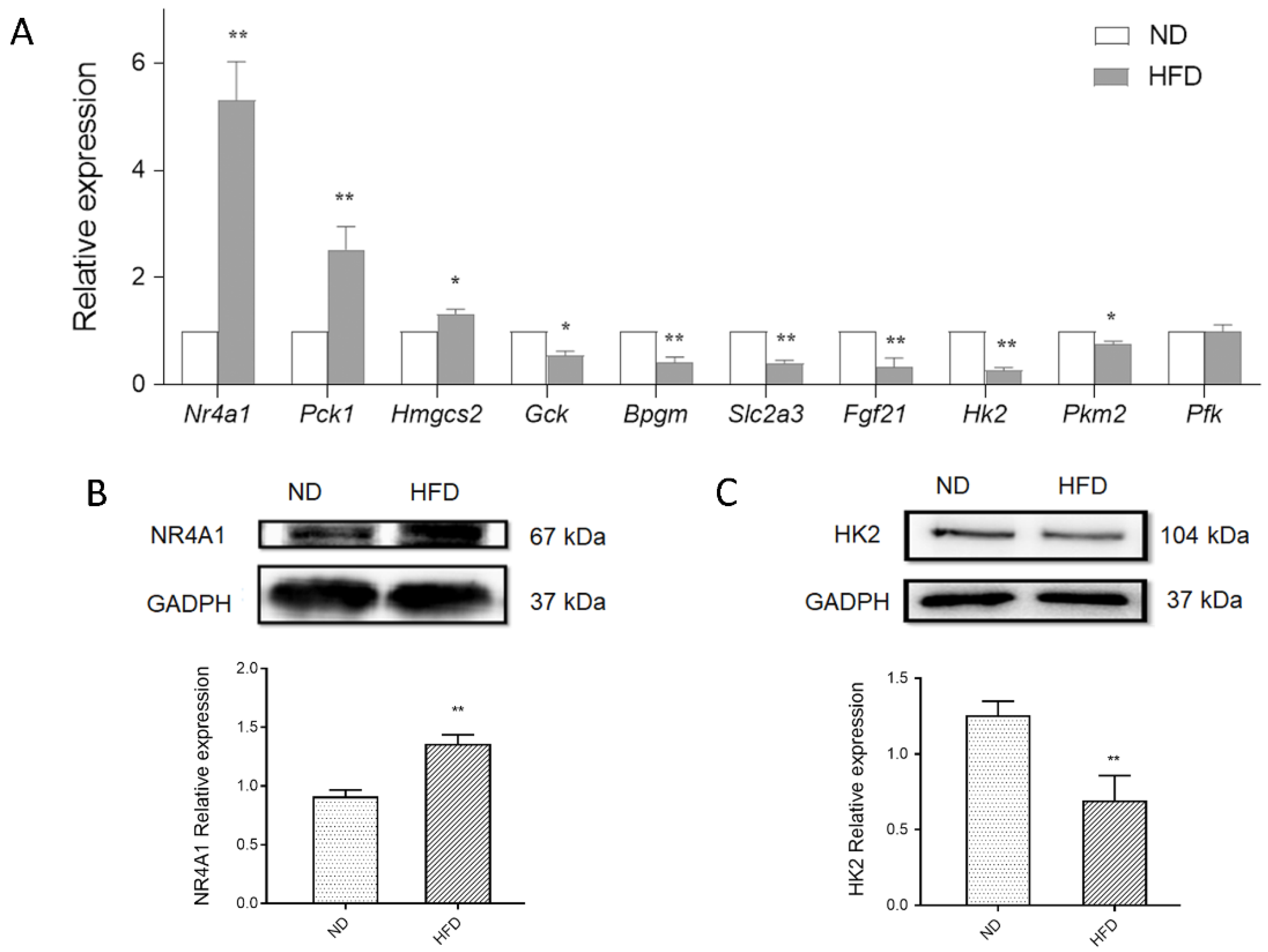
3.5. Validation of Glucose Metabolism-Related DEGs Using qPCR
3.6. Nr4a1 Regulates the Expression of DEGs in Glucose Homeostasis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- An, R.; Shen, J.; Bullard, T.; Han, Y.; Qiu, D.; Wang, S. A scoping review on economic globalization in relation to the obesity epidemic. Obes. Rev. 2019, 21, e12969. [Google Scholar] [CrossRef] [PubMed]
- Collaboration, N.C.D.R.F. Trends in adult body-mass index in 200 countries from 1975 to 2014: A pooled analysis of 1698 population-based measurement studies with 19.2 million participants. Lancet 2016, 387, 1377–1396. [Google Scholar]
- Koliaki, C.; Liatis, S.; Kokkinos, A. Obesity and cardiovascular disease: Revisiting an old relationship. Metabolism 2019, 92, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Krittanawong, C.; Tunhasiriwet, A.; Wang, Z.; Zhang, H.; Prokop, L.J.; Chirapongsathorn, S.; Sun, T.; Kitai, T.; Tang, W.H.W. Meta-Analysis Comparing Frequency of Overweight Versus Normal Weight in Patients with New-Onset Heart Failure. Am. J. Cardiol. 2018, 121, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Alpert, M.A.; Karthikeyan, K.; Abdullah, O.; Ghadban, R. Obesity and Cardiac Remodeling in Adults: Mechanisms and Clinical Implications. Prog. Cardiovasc. Dis. 2018, 61, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Kolwicz, S.C., Jr.; Purohit, S.; Fau-Tian, R.; Tian, R. Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ. Res. 2013, 113, 603–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalera, M.; Wang, J.; Frangogiannis, N.G. Obesity, metabolic dysfunction, and cardiac fibrosis: Pathophysiological pathways, molecular mechanisms, and therapeutic opportunities. Transl. Res. 2014, 164, 323–335. [Google Scholar] [CrossRef] [Green Version]
- Neely, J.R.; Morgan, H.E. Relationship between carbohydrate and lipid metabolism and the energy balance of heart muscle. Annu. Rev. Physiol. 1974, 36, 413–459. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, F.M.; Diczku, V.; Sherry, A.D.; Malloy, C.R. Substrate selection in the isolated working rat heart: Effects of reperfusion, afterload, and concentration. Basic. Res. Cardiol. 1995, 90, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Rider, O.J.; Cox, P.; Tyler, D.; Clarke, K.; Neubauer, S. Myocardial substrate metabolism in obesity. Int. J. Obes. 2013, 37, 972–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taegtmeyer, H.; Young, M.E.; Lopaschuk, G.D.; Abel, E.D.; Brunengraber, H.; Darley-Usmar, V.; Des Rosiers, C.; Gerszten, R.; Glatz, J.F.; Griffin, J.L.; et al. Assessing Cardiac Metabolism: A Scientific Statement From the American Heart Association. Circ. Res. 2016, 118, 1659–1701. [Google Scholar] [CrossRef] [PubMed]
- Gibb, A.A.; Hill, B.G. Metabolic Coordination of Physiological and Pathological Cardiac Remodeling. Circ. Res. 2018, 123, 107–128. [Google Scholar] [CrossRef] [PubMed]
- Men, L.; Pi, Z.; Zhou, Y.; Wei, M.; Liu, Y.; Song, F.; Liu, Z. Urine metabolomics of high-fat diet induced obesity using UHPLC-Q-TOF-MS. J. Pharm. Biomed. Anal. 2017, 132, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Airaksinen, K.; Jokkala, J.; Ahonen, I.; Auriola, S.; Kolehmainen, M.; Hanhineva, K.; Tiihonen, K. High-Fat Diet, Betaine, and Polydextrose Induce Changes in Adipose Tissue Inflammation and Metabolism in C57BL/6J Mice. Mol. Nutr. Food Res. 2018, 62, e1800455. [Google Scholar] [CrossRef] [PubMed]
- Barallobre-Barreiro, J.; Lynch, M.; Yin, X.; Mayr, M. Systems biology-opportunities and challenges: The application of proteomics to study the cardiovascular extracellular matrix. Cardiovasc. Res. 2016, 112, 626–636. [Google Scholar] [CrossRef] [Green Version]
- Badimon, L.; Vilahur, G.; Padro, T. Systems biology approaches to understand the effects of nutrition and promote health. Br. J. Clin. Pharmacol. 2017, 83, 38–45. [Google Scholar] [CrossRef] [Green Version]
- Herring, J.A.; Elison, W.S.; Tessem, J.S. Function of Nr4a Orphan Nuclear Receptors in Proliferation, Apoptosis and Fuel Utilization across Tissues. Cells 2019, 8, 1373. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Calvo, R.; Tajes, M.; Vazquez-Carrera, M. The NR4A subfamily of nuclear receptors: Potential new therapeutic targets for the treatment of inflammatory diseases. Expert. Opin. Ther. Tar. 2017, 21, 291–304. [Google Scholar] [CrossRef]
- You, X.; Guo, Z.F.; Cheng, F.; Yi, B.; Yang, F.; Liu, X.; Zhu, N.; Zhao, X.; Yan, G.; Ma, X.L.; et al. Transcriptional up-regulation of relaxin-3 by Nur77 attenuates beta-adrenergic agonist-induced apoptosis in cardiomyocytes. J. Biol. Chem. 2018, 293, 14001–14011. [Google Scholar] [CrossRef] [Green Version]
- Xu, A.; Liu, J.; Liu, P.; Jia, M.; Wang, H.; Tao, L. Mitochondrial translocation of Nur77 induced by ROS contributed to cardiomyocyte apoptosis in metabolic syndrome. Biochem. Biophys. Res. Commun. 2014, 446, 1184–1189. [Google Scholar] [CrossRef]
- Bux, A.S.; Lindsey, M.L.; Vasquez, H.G.; Taegtmeyer, H.; Harmancey, R. Glucose regulates the intrinsic inflammatory response of the heart to surgically induced hypothermic ischemic arrest and reperfusion. Physiol. Genom. 2017, 49, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, L.H.; Jebessa, Z.H.; Kreusser, M.M.; Horsch, A.; He, T.; Kronlage, M.; Dewenter, M.; Sramek, V.; Oehl, U.; Krebs-Haupenthal, J.; et al. A proteolytic fragment of histone deacetylase 4 protects the heart from failure by regulating the hexosamine biosynthetic pathway. Nat. Med. 2018, 24, 62–72. [Google Scholar] [CrossRef]
- Maxwell, M.A.; Cleasby, M.E.; Harding, A.; Stark, A.; Cooney, G.J.; Muscat, G.E. Nur77 regulates lipolysis in skeletal muscle cells. Evidence for cross-talk between the beta-adrenergic and an orphan nuclear hormone receptor pathway. J. Biol. Chem. 2005, 280, 12573–12584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Gene Ontology, C. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic. Acids Res. 2019, 47, D330–D338. [Google Scholar]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.X.; Dong, F.; Thomas, D.P.; Ma, H.; He, L.; Ren, J. Hypertrophic cardiomyopathy in high-fat diet-induced obesity: Role of suppression of forkhead transcription factor and atrophy gene transcription. Am. J. Physiol. Heart Circ. Physiol. 2008, 295, H1206–H1215. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.P.; Tsai, C.W.; Shen, C.Y.; Day, C.H.; Yeh, Y.L.; Chen, R.J.; Ho, T.J.; Padma, V.V.; Kuo, W.W.; Huang, C.Y. Palmitic acid interferes with energy metabolism balance by adversely switching the SIRT1-CD36-fatty acid pathway to the PKC zeta-GLUT4-glucose pathway in cardiomyoblasts. J. Nutr. Biochem. 2016, 31, 137–149. [Google Scholar] [CrossRef]
- Miranda, J.J.; Barrientos-Gutierrez, T.; Corvalan, C.; Hyder, A.A.; Lazo-Porras, M.; Oni, T.; Wells, J.C.K. Understanding the rise of cardiometabolic diseases in low- and middle-income countries. Nat. Med. 2019, 25, 1667–1679. [Google Scholar] [CrossRef]
- Saxton, S.N.; Clark, B.J.; Withers, S.B.; Eringa, E.C.; Heagerty, A.M. Mechanistic Links Between Obesity, Diabetes, and Blood Pressure: Role of Perivascular Adipose Tissue. Physiol. Rev. 2019, 99, 1701–1763. [Google Scholar] [CrossRef]
- Neubauer, S. Mechanisms of disease-The failing heart-An engine out of fuel. New Engl. J. Med. 2007, 356, 1140–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, M.A.; Murray, A.J.; Cochlin, L.E.; Heather, L.C.; McAleese, S.; Knight, N.S.; Sutton, E.; Abd Jamil, A.; Parassol, N.; Clarke, K. A high fat diet increases mitochondrial fatty acid oxidation and uncoupling to decrease efficiency in rat heart. Basic Res. Cardiol. 2011, 106, 447–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sternisha, S.M.; Miller, B.G. Molecular and cellular regulation of human glucokinase. Arch. Biochem. Biophys. 2019, 663, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Calmettes, G.; Ribalet, B.; John, S.; Korge, P.; Ping, P.; Weiss, J.N. Hexokinases and cardioprotection. J. Mol. Cell. Cardiol. 2015, 78, 107–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulquiney, P.J.; Bubb, W.A.; Kuchel, P.W. Model of 2,3-bisphosphoglycerate metabolism in the human erythrocyte based on detailed enzyme kinetic equations: In vivo kinetic characterization of 2,3-bisphosphoglycerate synthase/phosphatase using 13C and 31P NMR. Biochem. J. 1999, 342, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Oslund, R.C.; Su, X.; Haugbro, M.; Kee, J.M.; Esposito, M.; David, Y.; Wang, B.; Ge, E.; Perlman, D.H.; Kang, Y.; et al. Bisphosphoglycerate mutase controls serine pathway flux via 3-phosphoglycerate. Nat. Chem. Biol. 2017, 13, 1081–1087. [Google Scholar] [CrossRef]
- Wei, S.N.; Zhao, W.J.; Zeng, X.J.; Kang, Y.M.; Du, J.; Li, H.H. Microarray and Co-expression Network Analysis of Genes Associated with Acute Doxorubicin Cardiomyopathy in Mice. Cardiovasc. Toxicol. 2015, 15, 377–393. [Google Scholar] [CrossRef] [PubMed]
- Yanez, A.J.; Nualart, F.; Droppelmann, C.; Bertinat, R.; Brito, M.; Concha, I.I.; Slebe, J.C. Broad expression of fructose-1,6-bisphosphatase and phosphoenolpyruvate carboxykinase provide evidence for gluconeogenesis in human tissues other than liver and kidney. J. Cell. Physiol. 2003, 197, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Anousis, N.; Carvalho, R.A.; Zhao, P.Y.; Malloy, C.R.; Sherry, A.D. Compartmentation of glycolysis and glycogenolysis in the perfused rat heart. NMR Biomed. 2004, 17, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Simpson, I.A.; Dwyer, D.; Malide, D.; Moley, K.H.; Travis, A.; Vannucci, S.J. The facilitative glucose transporter GLUT3: 20 years of distinction. Am. J. Physiol.-Endoc. M. 2008, 295, 242–253. [Google Scholar] [CrossRef] [Green Version]
- Abel, E.D. Glucose transport in the heart. Front. Biosci. 2004, 9, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Markan, K.R.; Naber, M.C.; Ameka, M.K.; Anderegg, M.D.; Mangelsdorf, D.J.; Kliewer, S.A.; Mohammadi, M.; Potthoff, M.J. Circulating FGF21 is liver derived and enhances glucose uptake during refeeding and overfeeding. Diabetes 2014, 63, 4057–4063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruperez, C.; Lerin, C.; Ferrer-Curriu, G.; Cairo, M.; Mas-Stachurska, A.; Sitges, M.; Villarroya, J.; Giralt, M.; Villarroya, F.; Planavila, A. Autophagic control of cardiac steatosis through FGF21 in obesity-associated cardiomyopathy. Int. J. Cardiol. 2018, 260, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Iroz, A.; Montagner, A.; Benhamed, F.; Levavasseur, F.; Polizzi, A.; Anthony, E.; Regnier, M.; Fouche, E.; Lukowicz, C.; Cauzac, M.; et al. A Specific ChREBP and PPARalpha Cross-Talk Is Required for the Glucose-Mediated FGF21 Response. Cell Rep. 2017, 21, 403–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iizuka, K.; Takeda, J.; Horikawa, Y. Glucose induces FGF21 mRNA expression through ChREBP activation in rat hepatocytes. FEBS Lett. 2009, 583, 2882–2886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, X.L.; Chen, H.Z.; Yang, P.B.; Li, Y.P.; Zhang, F.N.; Zhang, J.Y.; Wang, W.J.; Zhao, W.X.; Zhang, S.; Chen, Q.T.; et al. Nur77 suppresses hepatocellular carcinoma via switching glucose metabolism toward gluconeogenesis through attenuating phosphoenolpyruvate carboxykinase sumoylation. Nat. Commun. 2017, 8, 14420. [Google Scholar] [CrossRef] [Green Version]
- Chao, L.C.; Zhang, Z.; Pei, L.; Saito, T.; Tontonoz, P.; Pilch, P.F. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol. Endocrinol. 2007, 21, 2152–2163. [Google Scholar] [CrossRef] [Green Version]
- Medzikovic, L.; Schumacher, C.A.; Verkerk, A.O.; van Deel, E.D.; Wolswinkel, R.; van der Made, I.; Bleeker, N.; Cakici, D.; van den Hoogenhof, M.M.; Meggouh, F.; et al. Orphan nuclear receptor Nur77 affects cardiomyocyte calcium homeostasis and adverse cardiac remodelling. Sci. Rep. 2015, 5, 15404. [Google Scholar] [CrossRef] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Men, L.; Hui, W.; Guan, X.; Song, T.; Wang, X.; Zhang, S.; Chen, X. Cardiac Transcriptome Analysis Reveals Nr4a1 Mediated Glucose Metabolism Dysregulation in Response to High-Fat Diet. Genes 2020, 11, 720. https://doi.org/10.3390/genes11070720
Men L, Hui W, Guan X, Song T, Wang X, Zhang S, Chen X. Cardiac Transcriptome Analysis Reveals Nr4a1 Mediated Glucose Metabolism Dysregulation in Response to High-Fat Diet. Genes. 2020; 11(7):720. https://doi.org/10.3390/genes11070720
Chicago/Turabian StyleMen, Lihui, Wenting Hui, Xin Guan, Tongtong Song, Xuan Wang, Siwei Zhang, and Xia Chen. 2020. "Cardiac Transcriptome Analysis Reveals Nr4a1 Mediated Glucose Metabolism Dysregulation in Response to High-Fat Diet" Genes 11, no. 7: 720. https://doi.org/10.3390/genes11070720




