Summary of BARD1 Mutations and Precise Estimation of Breast and Ovarian Cancer Risks Associated with the Mutations
Abstract
:1. Introduction
2. Methods
2.1. Literature Search and Study Selection
2.2. Data Extraction and Analysis
3. Results
3.1. Eligible Studies
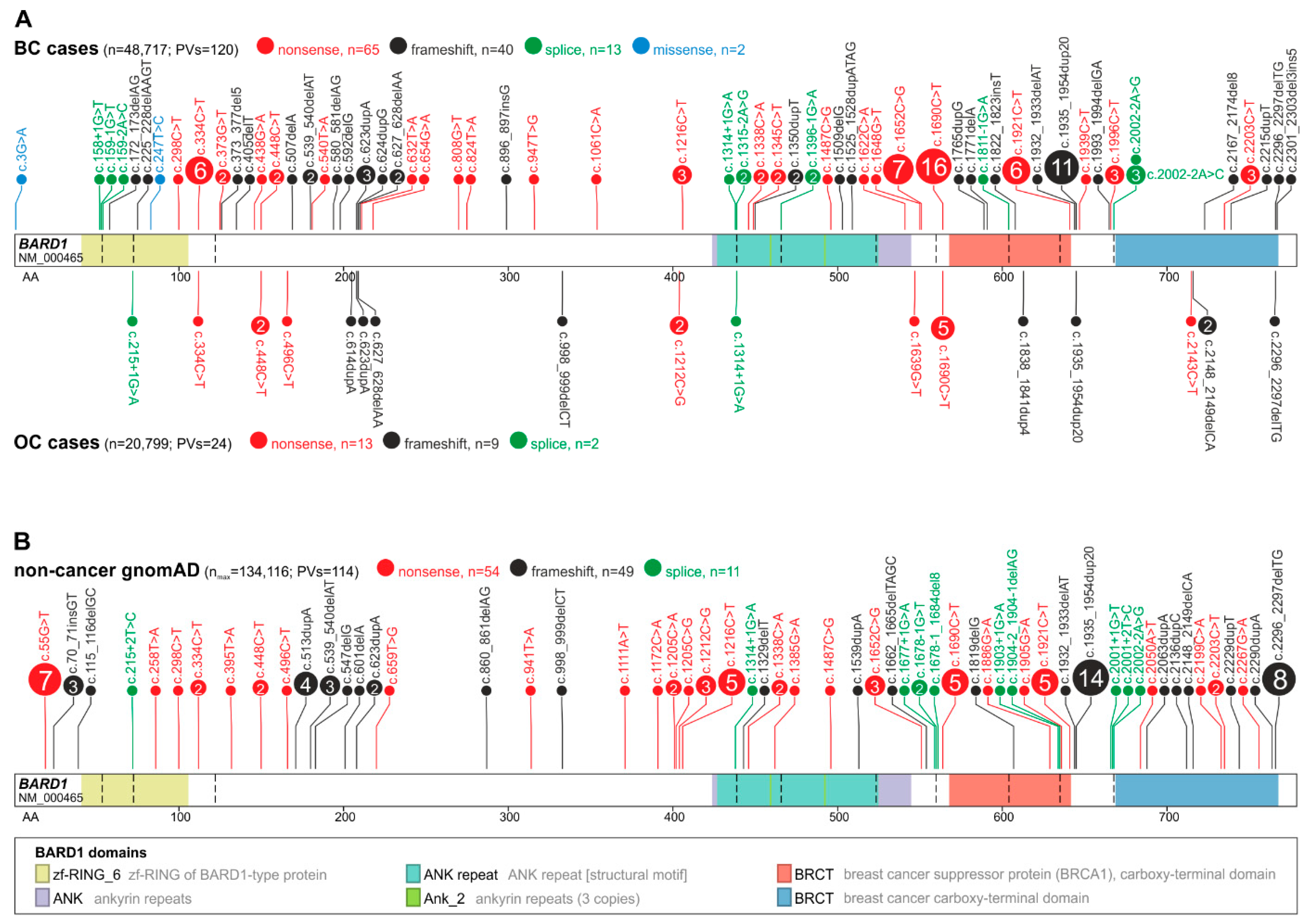
3.2. BARD1 Mutational Spectrum
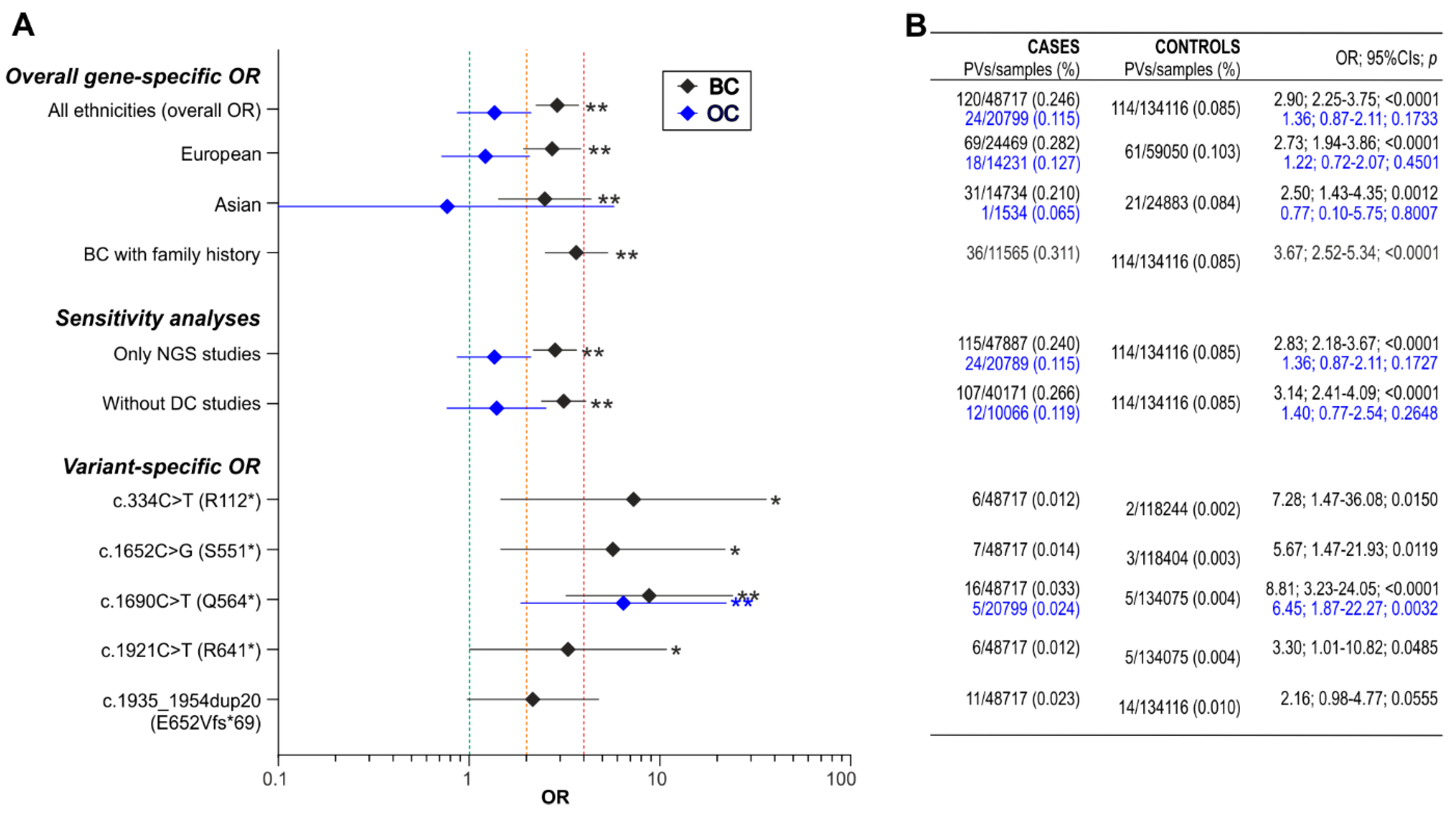
3.3. Association of BARD1 Pathogenic Variants with Breast Cancer
3.4. Association of BARD1 Pathogenic Variants with Ovarian Cancer
3.5. Large Mutations in the BARD1 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Suszynska, M.; Klonowska, K.; Jasinska, A.J.; Kozlowski, P. Large-scale meta-analysis of mutations identified in panels of breast/ovarian cancer-related genes—Providing evidence of cancer predisposition genes. Gynecol. Oncol. 2019, 153, 452–462. [Google Scholar] [CrossRef] [Green Version]
- Walsh, T.; Casadei, S.; Lee, M.K.; Pennil, C.C.; Nord, A.S.; Thornton, A.M.; Roeb, W.; Agnew, K.J.; Stray, S.M.; Wickramanayake, A.; et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 18032–18037. [Google Scholar] [CrossRef] [Green Version]
- Couch, F.J.; Shimelis, H.; Hu, C.; Hart, S.N.; Polley, E.C.; Na, J.; Hallberg, E.; Moore, R.; Thomas, A.; Lilyquist, J.; et al. Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol. 2017, 3, 1190–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, M.B.; Pilarski, R.; Yurgelun, M.B.; Berry, M.P.; Buys, S.S.; Dickson, P.; Domchek, S.M.; Elkhanany, A.; Friedman, S.; Garber, J.E.; et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2020. J. Natl. Compr. Canc. Netw. 2020, 18, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.B.; Pilarski, R.; Berry, M.; Buys, S.S.; Farmer, M.; Friedman, S.; Garber, J.E.; Kauff, N.D.; Khan, S.; Klein, C.; et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast and Ovarian, Version 2.2017. J. Natl. Compr. Canc. Netw. 2017, 15, 9–20. [Google Scholar] [CrossRef]
- Hashizume, R.; Fukuda, M.; Maeda, I.; Nishikawa, H.; Oyake, D.; Yabuki, Y.; Ogata, H.; Ohta, T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 2001, 276, 14537–14540. [Google Scholar] [CrossRef] [Green Version]
- Irminger-Finger, I.; Ratajska, M.; Pilyugin, M. New concepts on BARD1: Regulator of BRCA pathways and beyond. Int. J. Biochem. Cell Biol. 2016, 72, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020, 21, 284–299. [Google Scholar] [CrossRef]
- Irminger-Finger, I.; Leung, W.C.; Li, J.; Dubois-Dauphin, M.; Harb, J.; Feki, A.; Jefford, C.E.; Soriano, J.V.; Jaconi, M.; Montesano, R.; et al. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol. Cell 2001, 8, 1255–1266. [Google Scholar] [CrossRef]
- Cimmino, F.; Formicola, D.; Capasso, M. Dualistic Role of BARD1 in Cancer. Genes 2017, 8, 375. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Ryser, S.; Dizin, E.; Pils, D.; Krainer, M.; Jefford, C.E.; Bertoni, F.; Zeillinger, R.; Irminger-Finger, I. Oncogenic BARD1 isoforms expressed in gynecological cancers. Cancer Res. 2007, 67, 11876–11885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.Q.; Bianco, A.; Malkinson, A.M.; Leoni, V.P.; Frau, G.; De Rosa, N.; Andre, P.A.; Versace, R.; Boulvain, M.; Laurent, G.J.; et al. BARD1: An independent predictor of survival in non-small cell lung cancer. Int. J. Cancer 2012, 131, 83–94. [Google Scholar] [CrossRef]
- Bosse, K.R.; Diskin, S.J.; Cole, K.A.; Wood, A.C.; Schnepp, R.W.; Norris, G.; Nguyen le, B.; Jagannathan, J.; Laquaglia, M.; Winter, C.; et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012, 72, 2068–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suszynska, M.; Kluzniak, W.; Wokolorczyk, D.; Jakubowska, A.; Huzarski, T.; Gronwald, J.; Debniak, T.; Szwiec, M.; Ratajska, M.; Klonowska, K.; et al. BARD1 is A Low/Moderate Breast Cancer Risk Gene: Evidence Based on An Association Study of the Central European p.Q564X Recurrent Mutation. Cancers 2019, 11, 740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber-Lassalle, N.; Borde, J.; Weber-Lassalle, K.; Horvath, J.; Niederacher, D.; Arnold, N.; Kaulfuss, S.; Ernst, C.; Paul, V.G.; Honisch, E.; et al. Germline loss-of-function variants in the BARD1 gene are associated with early-onset familial breast cancer but not ovarian cancer. Breast Cancer Res. 2019, 21, 55. [Google Scholar] [CrossRef] [Green Version]
- Ramus, S.J.; Song, H.; Dicks, E.; Tyrer, J.P.; Rosenthal, A.N.; Intermaggio, M.P.; Fraser, L.; Gentry-Maharaj, A.; Hayward, J.; Philpott, S.; et al. Germline Mutations in the BRIP1, BARD1, PALB2, and NBN Genes in Women With Ovarian Cancer. J. Natl. Cancer Inst. 2015, 107, djv214. [Google Scholar] [CrossRef]
- Lu, H.M.; Li, S.; Black, M.H.; Lee, S.; Hoiness, R.; Wu, S.; Mu, W.; Huether, R.; Chen, J.; Sridhar, S.; et al. Association of Breast and Ovarian Cancers With Predisposition Genes Identified by Large-Scale Sequencing. JAMA Oncol 2019, 5, 51–57. [Google Scholar] [CrossRef]
- Lilyquist, J.; LaDuca, H.; Polley, E.; Davis, B.T.; Shimelis, H.; Hu, C.; Hart, S.N.; Dolinsky, J.S.; Couch, F.J.; Goldgar, D.E. Frequency of mutations in a large series of clinically ascertained ovarian cancer cases tested on multi-gene panels compared to reference controls. Gynecol. Oncol. 2017, 147, 375–380. [Google Scholar] [CrossRef]
- Norquist, B.M.; Harrell, M.I.; Brady, M.F.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Yi, Q.; Burger, R.A.; et al. Inherited Mutations in Women With Ovarian Carcinoma. JAMA Oncol. 2016, 2, 482–490. [Google Scholar] [CrossRef]
- Slavin, T.P.; Maxwell, K.N.; Lilyquist, J.; Vijai, J.; Neuhausen, S.L.; Hart, S.N.; Ravichandran, V.; Thomas, T.; Maria, A.; Villano, D.; et al. The contribution of pathogenic variants in breast cancer susceptibility genes to familial breast cancer risk. NPJ Breast Cancer 2017, 3, 22. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [Green Version]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- De Souza Timoteo, A.R.; Goncalves, A.; Sales, L.A.P.; Albuquerque, B.M.; de Souza, J.E.S.; de Moura, P.C.P.; de Aquino, M.A.A.; Agnez-Lima, L.F.; Lajus, T.B.P. A portrait of germline mutation in Brazilian at-risk for hereditary breast cancer. Breast Cancer Res. Treat. 2018, 172, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, E.S.C.S.; Cury, N.M.; Brotto, D.B.; de Araujo, L.F.; Rosa, R.C.A.; Texeira, L.A.; Placa, J.R.; Marques, A.A.; Peronni, K.C.; Ruy, P.C.; et al. Germline variants in DNA repair genes associated with hereditary breast and ovarian cancer syndrome: Analysis of a 21 gene panel in the Brazilian population. BMC Med. Genom. 2020, 13, 21. [Google Scholar]
- Adedokun, B.; Zheng, Y.; Ndom, P.; Gakwaya, A.; Makumbi, T.; Zhou, A.Y.; Yoshimatsu, T.F.; Rodriguez, A.; Madduri, R.K.; Foster, I.T.; et al. Prevalence of Inherited Mutations in Breast Cancer Predisposition Genes among Women in Uganda and Cameroon. Cancer Epidemiol. Prev. Biomark. 2020, 29, 359–367. [Google Scholar] [CrossRef]
- Aloraifi, F.; McDevitt, T.; Martiniano, R.; McGreevy, J.; McLaughlin, R.; Egan, C.M.; Cody, N.; Meany, M.; Kenny, E.; Green, A.J.; et al. Detection of novel germline mutations for breast cancer in non-BRCA1/2 families. FEBS J. 2015, 282, 3424–3437. [Google Scholar] [CrossRef] [PubMed]
- Amemiya, Y.; Bacopulos, S.; Al-Shawarby, M.; Al-Tamimi, D.; Naser, W.; Ahmed, A.; Khalifa, M.; Slodkowska, E.; Seth, A. A Comparative Analysis of Breast and Ovarian Cancer-related Gene Mutations in Canadian and Saudi Arabian Patients with Breast Cancer. Anticancer Res. 2015, 35, 2601–2610. [Google Scholar] [PubMed]
- Bernards, S.S.; Norquist, B.M.; Harrell, M.I.; Agnew, K.J.; Lee, M.K.; Walsh, T.; Swisher, E.M. Genetic characterization of early onset ovarian carcinoma. Gynecol. Oncol. 2016, 140, 221–225. [Google Scholar] [CrossRef] [Green Version]
- Bertelsen, B.; Tuxen, I.V.; Yde, C.W.; Gabrielaite, M.; Torp, M.H.; Kinalis, S.; Oestrup, O.; Rohrberg, K.; Spangaard, I.; Santoni-Rugiu, E.; et al. High frequency of pathogenic germline variants within homologous recombination repair in patients with advanced cancer. NPJ Genom. Med. 2019, 4, 13. [Google Scholar] [CrossRef]
- Bonache, S.; Esteban, I.; Moles-Fernandez, A.; Tenes, A.; Duran-Lozano, L.; Montalban, G.; Bach, V.; Carrasco, E.; Gadea, N.; Lopez-Fernandez, A.; et al. Multigene panel testing beyond BRCA1/2 in breast/ovarian cancer Spanish families and clinical actionability of findings. J. Cancer Res. Clin. Oncol. 2018, 144, 2495–2513. [Google Scholar] [CrossRef]
- Brovkina, O.I.; Shigapova, L.; Chudakova, D.A.; Gordiev, M.G.; Enikeev, R.F.; Druzhkov, M.O.; Khodyrev, D.S.; Shagimardanova, E.I.; Nikitin, A.G.; Gusev, O.A. The Ethnic-Specific Spectrum of Germline Nucleotide Variants in DNA Damage Response and Repair Genes in Hereditary Breast and Ovarian Cancer Patients of Tatar Descent. Front. Oncol. 2018, 8, 421. [Google Scholar] [CrossRef]
- Caminsky, N.G.; Mucaki, E.J.; Perri, A.M.; Lu, R.; Knoll, J.H.; Rogan, P.K. Prioritizing Variants in Complete Hereditary Breast and Ovarian Cancer Genes in Patients Lacking Known BRCA Mutations. Hum. Mutat. 2016, 37, 640–652. [Google Scholar] [CrossRef]
- Carter, N.J.; Marshall, M.L.; Susswein, L.R.; Zorn, K.K.; Hiraki, S.; Arvai, K.J.; Torene, R.I.; McGill, A.K.; Yackowski, L.; Murphy, P.D.; et al. Germline pathogenic variants identified in women with ovarian tumors. Gynecol. Oncol. 2018, 151, 481–488. [Google Scholar] [CrossRef]
- Castera, L.; Krieger, S.; Rousselin, A.; Legros, A.; Baumann, J.J.; Bruet, O.; Brault, B.; Fouillet, R.; Goardon, N.; Letac, O.; et al. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur. J. Hum. Genet. 2014, 22, 1305–1313. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Zhang, G.; Li, X.; Ren, C.; Wang, Y.; Li, K.; Mok, H.; Cao, L.; Wen, L.; Jia, M.; et al. Comparison of BRCA versus non-BRCA germline mutations and associated somatic mutation profiles in patients with unselected breast cancer. Aging (Albany NY) 2020, 12, 3140–3155. [Google Scholar] [CrossRef]
- Chirasophon, S.; Manchana, T.; Teerapakpinyo, C. High-risk epithelial ovarian cancer patients for hereditary ovarian cancer. J. Obs. Gynaecol. Res. 2017, 43, 929–934. [Google Scholar] [CrossRef]
- Choi, M.C.; Hwang, S.; Kim, S.; Jung, S.G.; Park, H.; Joo, W.D.; Song, S.H.; Lee, C.; Kim, T.H.; Kang, H.; et al. Clinical Impact of Somatic Variants in Homologous Recombination Repair-Related Genes in Ovarian High-Grade Serous Carcinoma. Cancer Res. Treat. 2020, 52, 634–644. [Google Scholar] [CrossRef] [Green Version]
- Churpek, J.E.; Marquez, R.; Neistadt, B.; Claussen, K.; Lee, M.K.; Churpek, M.M.; Huo, D.; Weiner, H.; Bannerjee, M.; Godley, L.A.; et al. Inherited mutations in cancer susceptibility genes are common among survivors of breast cancer who develop therapy-related leukemia. Cancer 2016, 122, 304–311. [Google Scholar] [CrossRef]
- Churpek, J.E.; Walsh, T.; Zheng, Y.; Moton, Z.; Thornton, A.M.; Lee, M.K.; Casadei, S.; Watts, A.; Neistadt, B.; Churpek, M.M.; et al. Inherited predisposition to breast cancer among African American women. Breast Cancer Res. Treat. 2015, 149, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Cock-Rada, A.M.; Ossa, C.A.; Garcia, H.I.; Gomez, L.R. A multi-gene panel study in hereditary breast and ovarian cancer in Colombia. Fam. Cancer 2018, 17, 23–30. [Google Scholar] [CrossRef]
- Coppa, A.; Nicolussi, A.; D’Inzeo, S.; Capalbo, C.; Belardinilli, F.; Colicchia, V.; Petroni, M.; Zani, M.; Ferraro, S.; Rinaldi, C.; et al. Optimizing the identification of risk-relevant mutations by multigene panel testing in selected hereditary breast/ovarian cancer families. Cancer Med. 2018, 7, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Couch, F.J.; Hart, S.N.; Sharma, P.; Toland, A.E.; Wang, X.; Miron, P.; Olson, J.E.; Godwin, A.K.; Pankratz, V.S.; Olswold, C.; et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015, 33, 304–311. [Google Scholar] [CrossRef]
- Crawford, B.; Adams, S.B.; Sittler, T.; van den Akker, J.; Chan, S.; Leitner, O.; Ryan, L.; Gil, E.; van ‘t Veer, L. Multi-gene panel testing for hereditary cancer predisposition in unsolved high-risk breast and ovarian cancer patients. Breast Cancer Res. Treat. 2017, 163, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Cybulski, C.; Kluzniak, W.; Huzarski, T.; Wokolorczyk, D.; Kashyap, A.; Rusak, B.; Stempa, K.; Gronwald, J.; Szymiczek, A.; Bagherzadeh, M.; et al. The spectrum of mutations predisposing to familial breast cancer in Poland. Int. J. Cancer 2019, 145, 3311–3320. [Google Scholar] [CrossRef]
- De Brakeleer, S.; De Greve, J.; Desmedt, C.; Joris, S.; Sotiriou, C.; Piccart, M.; Pauwels, I.; Teugels, E. Frequent incidence of BARD1-truncating mutations in germline DNA from triple-negative breast cancer patients. Clin. Genet. 2016, 89, 336–340. [Google Scholar] [CrossRef]
- De Brakeleer, S.; De Greve, J.; Loris, R.; Janin, N.; Lissens, W.; Sermijn, E.; Teugels, E. Cancer predisposing missense and protein truncating BARD1 mutations in non-BRCA1 or BRCA2 breast cancer families. Hum. Mutat. 2010, 31, E1175–E1185. [Google Scholar] [CrossRef]
- Dominguez-Valentin, M.; Nakken, S.; Tubeuf, H.; Vodak, D.; Ekstrom, P.O.; Nissen, A.M.; Morak, M.; Holinski-Feder, E.; Holth, A.; Capella, G.; et al. Results of multigene panel testing in familial cancer cases without genetic cause demonstrated by single gene testing. Sci. Rep. 2019, 9, 18555. [Google Scholar] [CrossRef]
- Dominguez-Valentin, M.; Nakken, S.; Tubeuf, H.; Vodak, D.; Ekstrom, P.O.; Nissen, A.M.; Morak, M.; Holinski-Feder, E.; Martins, A.; Moller, P.; et al. Potentially pathogenic germline CHEK2 c.319+2T>A among multiple early-onset cancer families. Fam. Cancer 2018, 17, 141–153. [Google Scholar] [CrossRef]
- Dong, L.; Wu, N.; Wang, S.; Cheng, Y.; Han, L.; Zhao, J.; Long, X.; Mu, K.; Li, M.; Wei, L.; et al. Detection of novel germline mutations in six breast cancer predisposition genes by targeted next-generation sequencing. Hum. Mutat. 2018, 39, 1442–1455. [Google Scholar] [CrossRef]
- Dutil, J.; Teer, J.K.; Golubeva, V.; Yoder, S.; Tong, W.L.; Arroyo, N.; Karam, R.; Echenique, M.; Matta, J.L.; Monteiro, A.N. Germline variants in cancer genes in high-risk non-BRCA patients from Puerto Rico. Sci. Rep. 2019, 9, 17769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eliade, M.; Skrzypski, J.; Baurand, A.; Jacquot, C.; Bertolone, G.; Loustalot, C.; Coutant, C.; Guy, F.; Fumoleau, P.; Duffourd, Y.; et al. The transfer of multigene panel testing for hereditary breast and ovarian cancer to healthcare: What are the implications for the management of patients and families? Oncotarget 2017, 8, 1957–1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellingson, M.S.; Hart, S.N.; Kalari, K.R.; Suman, V.; Schahl, K.A.; Dockter, T.J.; Felten, S.J.; Sinnwell, J.P.; Thompson, K.J.; Tang, X.; et al. Exome sequencing reveals frequent deleterious germline variants in cancer susceptibility genes in women with invasive breast cancer undergoing neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2015, 153, 435–443. [Google Scholar] [CrossRef] [Green Version]
- Eoh, K.J.; Kim, J.E.; Park, H.S.; Lee, S.T.; Park, J.S.; Han, J.W.; Lee, J.Y.; Kim, S.; Kim, S.W.; Kim, J.H.; et al. Detection of Germline Mutations in Patients with Epithelial Ovarian Cancer Using Multi-gene Panels: Beyond BRCA1/2. Cancer Res. Treat. 2018, 50, 917–925. [Google Scholar] [CrossRef] [Green Version]
- Feliubadalo, L.; Lopez-Fernandez, A.; Pineda, M.; Diez, O.; Del Valle, J.; Gutierrez-Enriquez, S.; Teule, A.; Gonzalez, S.; Stjepanovic, N.; Salinas, M.; et al. Opportunistic testing of BRCA1, BRCA2 and mismatch repair genes improves the yield of phenotype driven hereditary cancer gene panels. Int. J. Cancer 2019, 145, 2682–2691. [Google Scholar] [CrossRef]
- Frey, M.K.; Sandler, G.; Sobolev, R.; Kim, S.H.; Chambers, R.; Bassett, R.Y.; Martineau, J.; Sapra, K.J.; Boyd, L.; Curtin, J.P.; et al. Multigene panels in Ashkenazi Jewish patients yield high rates of actionable mutations in multiple non-BRCA cancer-associated genes. Gynecol. Oncol. 2017, 146, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Gervas, P.; Klyuch, B.; Denisov, E.; Kiselev, A.; Molokov, A.; Pisareva, L.; Malinovskaya, E.; Choynzonov, E.; Cherdyntseva, N. New germline BRCA2 gene variant in the Tuvinian Mongol breast cancer patients. Mol. Biol. Rep. 2019, 46, 5537–5541. [Google Scholar] [CrossRef]
- Ghimenti, C.; Sensi, E.; Presciuttini, S.; Brunetti, I.M.; Conte, P.; Bevilacqua, G.; Caligo, M.A. Germline mutations of the BRCA1-associated ring domain (BARD1) gene in breast and breast/ovarian families negative for BRCA1 and BRCA2 alterations. Genes Chromosomes Cancer 2002, 33, 235–242. [Google Scholar] [CrossRef]
- Girard, E.; Eon-Marchais, S.; Olaso, R.; Renault, A.L.; Damiola, F.; Dondon, M.G.; Barjhoux, L.; Goidin, D.; Meyer, V.; Le Gal, D.; et al. Familial breast cancer and DNA repair genes: Insights into known and novel susceptibility genes from the GENESIS study, and implications for multigene panel testing. Int. J. Cancer 2019, 144, 1962–1974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glentis, S.; Dimopoulos, A.C.; Rouskas, K.; Ntritsos, G.; Evangelou, E.; Narod, S.A.; Mes-Masson, A.M.; Foulkes, W.D.; Rivera, B.; Tonin, P.N.; et al. Exome Sequencing in BRCA1-and BRCA2-Negative Greek Families Identifies MDM1 and NBEAL1 as Candidate Risk Genes for Hereditary Breast Cancer. Front. Genet. 2019, 10, 1005. [Google Scholar] [CrossRef] [PubMed]
- Goidescu, I.G.; Caracostea, G.; Eniu, D.T.; Stamatian, F.V. Prevalence of deleterious mutations among patients with breast cancer referred for multigene panel testing in a Romanian population. Clujul Med. 2018, 91, 157–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Rivera, M.; Lobo, M.; Lopez-Tarruella, S.; Jerez, Y.; Del Monte-Millan, M.; Massarrah, T.; Ramos-Medina, R.; Ocana, I.; Picornell, A.; Santillan Garzon, S.; et al. Frequency of germline DNA genetic findings in an unselected prospective cohort of triple-negative breast cancer patients participating in a platinum-based neoadjuvant chemotherapy trial. Breast Cancer Res. Treat. 2016, 156, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Guenard, F.; Labrie, Y.; Ouellette, G.; Beauparlant, C.J.; Durocher, F.; BRCAs, I. Genetic sequence variations of BRCA1-interacting genes AURKA, BAP1, BARD1 and DHX9 in French Canadian families with high risk of breast cancer. J. Hum. Genet. 2009, 54, 152–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guacci, A.; Cordella, A.; Rocco, T.; Giurato, G.; Nassa, G.; Rizzo, F.; Carlomagno, C.; Pepe, S.; Tarallo, R.; Weisz, A. Identification of a novel truncating mutation in PALB2 gene by a multigene sequencing panel for mutational screening of breast cancer risk-associated and related genes. J. Clin. Lab. Anal. 2018, 32, e22418. [Google Scholar] [CrossRef]
- Guan, Y.; Hu, H.; Peng, Y.; Gong, Y.; Yi, Y.; Shao, L.; Liu, T.; Li, G.; Wang, R.; Dai, P.; et al. Detection of inherited mutations for hereditary cancer using target enrichment and next generation sequencing. Fam. Cancer 2015, 14, 9–18. [Google Scholar] [CrossRef]
- Hajkova, N.; Ticha, I.; Hojny, J.; Nemejcova, K.; Bartu, M.; Michalkova, R.; Zikan, M.; Cibula, D.; Laco, J.; Geryk, T.; et al. Synchronous endometrioid endometrial and ovarian carcinomas are biologically related: A clinico-pathological and molecular (next generation sequencing) study of 22 cases. Oncol. Lett. 2019, 17, 2207–2214. [Google Scholar]
- Hahnen, E.; Lederer, B.; Hauke, J.; Loibl, S.; Krober, S.; Schneeweiss, A.; Denkert, C.; Fasching, P.A.; Blohmer, J.U.; Jackisch, C.; et al. Germline Mutation Status, Pathological Complete Response, and Disease-Free Survival in Triple-Negative Breast Cancer: Secondary Analysis of the GeparSixto Randomized Clinical Trial. JAMA Oncol. 2017, 3, 1378–1385. [Google Scholar] [CrossRef]
- Harter, P.; Hauke, J.; Heitz, F.; Reuss, A.; Kommoss, S.; Marme, F.; Heimbach, A.; Prieske, K.; Richters, L.; Burges, A.; et al. Prevalence of deleterious germline variants in risk genes including BRCA1/2 in consecutive ovarian cancer patients (AGO-TR-1). PLoS ONE 2017, 12, e0186043. [Google Scholar] [CrossRef]
- Hata, C.; Nakaoka, H.; Xiang, Y.; Wang, D.; Yang, A.; Liu, D.; Liu, F.; Zou, Q.; Wei, L.; Zheng, K.; et al. Germline mutations of multiple breast cancer-related genes are differentially associated with triple-negative breast cancers and prognostic factors. J. Hum. Genet. 2020, 65, 577–587. [Google Scholar] [CrossRef]
- Hirasawa, A.; Imoto, I.; Naruto, T.; Akahane, T.; Yamagami, W.; Nomura, H.; Masuda, K.; Susumu, N.; Tsuda, H.; Aoki, D. Prevalence of pathogenic germline variants detected by multigene sequencing in unselected Japanese patients with ovarian cancer. Oncotarget 2017, 8, 112258–112267. [Google Scholar] [CrossRef] [Green Version]
- Hirotsu, Y.; Nakagomi, H.; Sakamoto, I.; Amemiya, K.; Oyama, T.; Mochizuki, H.; Omata, M. Multigene panel analysis identified germline mutations of DNA repair genes in breast and ovarian cancer. Mol. Genet. Genom. Med. 2015, 3, 459–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hjortkjaer, M.; Malik Aagaard Jorgensen, M.; Waldstrom, M.; Ornskov, D.; Sogaard-Andersen, E.; Jakobsen, A.; Dahl-Steffensen, K. The clinical importance of BRCAness in a population-based cohort of Danish epithelial ovarian cancer. Int. J. Gynecol. Cancer 2019, 29, 166–173. [Google Scholar] [CrossRef]
- Ishitobi, M.; Miyoshi, Y.; Hasegawa, S.; Egawa, C.; Tamaki, Y.; Monden, M.; Noguchi, S. Mutational analysis of BARD1 in familial breast cancer patients in Japan. Cancer Lett. 2003, 200, 1–7. [Google Scholar] [CrossRef]
- Jalkh, N.; Chouery, E.; Haidar, Z.; Khater, C.; Atallah, D.; Ali, H.; Marafie, M.J.; Al-Mulla, M.R.; Al-Mulla, F.; Megarbane, A. Next-generation sequencing in familial breast cancer patients from Lebanon. BMC Med. Genom. 2017, 10, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jian, W.; Shao, K.; Qin, Q.; Wang, X.; Song, S.; Wang, X. Clinical and genetic characterization of hereditary breast cancer in a Chinese population. Hered Cancer Clin. Pract. 2017, 15, 19. [Google Scholar] [CrossRef] [Green Version]
- Jorge, S.; McFaddin, A.S.; Doll, K.M.; Pennington, K.P.; Norquist, B.M.; Bennett, R.L.; Pritchard, C.C.; Swisher, E.M. Simultaneous germline and somatic sequencing in ovarian carcinoma: Mutation rate and impact on clinical decision-making. Gynecol. Oncol. 2020, 156, 517–522. [Google Scholar] [CrossRef]
- Kanke, Y.; Shimomura, A.; Saito, M.; Honda, T.; Shiraishi, K.; Shimada, Y.; Watanabe, R.; Yoshida, H.; Yoshida, M.; Shimizu, C.; et al. Gene aberration profile of tumors of adolescent and young adult females. Oncotarget 2018, 9, 6228–6237. [Google Scholar] [CrossRef] [Green Version]
- Karppinen, S.M.; Heikkinen, K.; Rapakko, K.; Winqvist, R. Mutation screening of the BARD1 gene: Evidence for involvement of the Cys557Ser allele in hereditary susceptibility to breast cancer. J. Med. Genet. 2004, 41, e114. [Google Scholar] [CrossRef] [Green Version]
- Kiiski, J.I.; Pelttari, L.M.; Khan, S.; Freysteinsdottir, E.S.; Reynisdottir, I.; Hart, S.N.; Shimelis, H.; Vilske, S.; Kallioniemi, A.; Schleutker, J.; et al. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 15172–15177. [Google Scholar] [CrossRef] [Green Version]
- Koczkowska, M.; Krawczynska, N.; Stukan, M.; Kuzniacka, A.; Brozek, I.; Sniadecki, M.; Debniak, J.; Wydra, D.; Biernat, W.; Kozlowski, P.; et al. Spectrum and Prevalence of Pathogenic Variants in Ovarian Cancer Susceptibility Genes in a Group of 333 Patients. Cancers 2018, 10, 442. [Google Scholar] [CrossRef] [Green Version]
- Kwong, A.; Shin, V.Y.; Chen, J.; Cheuk, I.W.Y.; Ho, C.Y.S.; Au, C.H.; Chan, K.K.L.; Ngan, H.Y.S.; Chan, T.L.; Ford, J.M.; et al. Germline Mutation in 1338 BRCA-Negative Chinese Hereditary Breast and/or Ovarian Cancer Patients: Clinical Testing with a Multigene Test Panel. J. Mol. Diagn. 2020, 22, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Larouche, V.; Akirov, A.; Thain, E.; Kim, R.H.; Ezzat, S. Co-occurrence of breast cancer and neuroendocrine tumours: New genetic insights beyond Multiple Endocrine Neoplasia syndromes. Endocrinol. Diabetes Metab. 2019, 2, e00092. [Google Scholar] [CrossRef] [PubMed]
- Lerner-Ellis, J.; Sopik, V.; Wong, A.; Lazaro, C.; Narod, S.A.; Charames, G.S. Retesting of women who are negative for a BRCA1 and BRCA2 mutation using a 20-gene panel. J. Med. Genet. 2020, 57, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Lhota, F.; Zemankova, P.; Kleiblova, P.; Soukupova, J.; Vocka, M.; Stranecky, V.; Janatova, M.; Hartmannova, H.; Hodanova, K.; Kmoch, S.; et al. Hereditary truncating mutations of DNA repair and other genes in BRCA1/BRCA2/PALB2-negatively tested breast cancer patients. Clin. Genet. 2016, 90, 324–333. [Google Scholar] [CrossRef]
- Li, C.; Bonazzoli, E.; Bellone, S.; Choi, J.; Dong, W.; Menderes, G.; Altwerger, G.; Han, C.; Manzano, A.; Bianchi, A.; et al. Mutational landscape of primary, metastatic, and recurrent ovarian cancer reveals c-MYC gains as potential target for BET inhibitors. Proc. Natl. Acad. Sci. USA 2019, 116, 619–624. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Meeks, H.; Feng, B.J.; Healey, S.; Thorne, H.; Makunin, I.; Ellis, J.; kConFab, I.; Campbell, I.; Southey, M.; et al. Targeted massively parallel sequencing of a panel of putative breast cancer susceptibility genes in a large cohort of multiple-case breast and ovarian cancer families. J. Med. Genet. 2016, 53, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Li, J.Y.; Jing, R.; Wei, H.; Wang, M.; Xiaowei, Q.; Liu, H.; Jian, L.; Ou, J.H.; Jiang, W.H.; Tian, F.G.; et al. Germline mutations in 40 cancer susceptibility genes among Chinese patients with high hereditary risk breast cancer. Int. J. Cancer 2019, 144, 281–289. [Google Scholar] [CrossRef]
- Li, W.; Shao, D.; Li, L.; Wu, M.; Ma, S.; Tan, X.; Zhong, S.; Guo, F.; Wang, Z.; Ye, M. Germline and somatic mutations of multi-gene panel in Chinese patients with epithelial ovarian cancer: A prospective cohort study. J. Ovarian Res. 2019, 12, 80. [Google Scholar] [CrossRef]
- Lin, P.H.; Kuo, W.H.; Huang, A.C.; Lu, Y.S.; Lin, C.H.; Kuo, S.H.; Wang, M.Y.; Liu, C.Y.; Cheng, F.T.; Yeh, M.H.; et al. Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer. Oncotarget 2016, 7, 8310–8320. [Google Scholar] [CrossRef] [Green Version]
- Lolas Hamameh, S.; Renbaum, P.; Kamal, L.; Dweik, D.; Salahat, M.; Jaraysa, T.; Abu Rayyan, A.; Casadei, S.; Mandell, J.B.; Gulsuner, S.; et al. Genomic analysis of inherited breast cancer among Palestinian women: Genetic heterogeneity and a founder mutation in TP53. Int. J. Cancer 2017, 141, 750–756. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Xie, M.; Wendl, M.C.; Wang, J.; McLellan, M.D.; Leiserson, M.D.; Huang, K.L.; Wyczalkowski, M.A.; Jayasinghe, R.; Banerjee, T.; et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat. Commun. 2015, 6, 10086. [Google Scholar] [CrossRef] [PubMed]
- Maksimenko, J.; Irmejs, A.; Trofimovics, G.; Berzina, D.; Skuja, E.; Purkalne, G.; Miklasevics, E.; Gardovskis, J. High frequency of pathogenic non-founder germline mutations in BRCA1 and BRCA2 in families with breast and ovarian cancer in a founder population. Hered Cancer Clin. Pract. 2018, 16, 12. [Google Scholar] [CrossRef] [PubMed]
- Mandelker, D.; Zhang, L.; Kemel, Y.; Stadler, Z.K.; Joseph, V.; Zehir, A.; Pradhan, N.; Arnold, A.; Walsh, M.F.; Li, Y.; et al. Mutation Detection in Patients With Advanced Cancer by Universal Sequencing of Cancer-Related Genes in Tumor and Normal DNA vs Guideline-Based Germline Testing. JAMA 2017, 318, 825–835. [Google Scholar] [CrossRef]
- Maxwell, K.N.; Wubbenhorst, B.; D’Andrea, K.; Garman, B.; Long, J.M.; Powers, J.; Rathbun, K.; Stopfer, J.E.; Zhu, J.; Bradbury, A.R.; et al. Prevalence of mutations in a panel of breast cancer susceptibility genes in BRCA1/2-negative patients with early-onset breast cancer. Genet. Med. 2015, 17, 630–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McVeigh, U.M.; McVeigh, T.P.; Curran, C.; Miller, N.; Morris, D.W.; Kerin, M.J. Diagnostic yield of a custom-designed multi-gene cancer panel in Irish patients with breast cancer. Ir. J. Med. Sci. 2020, 1–16. [Google Scholar] [CrossRef]
- Moran, O.; Nikitina, D.; Royer, R.; Poll, A.; Metcalfe, K.; Narod, S.A.; Akbari, M.R.; Kotsopoulos, J. Revisiting breast cancer patients who previously tested negative for BRCA mutations using a 12-gene panel. Breast Cancer Res. Treat. 2017, 161, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.S.; Wen, W.X.; Fadlullah, M.Z.; Yoon, S.Y.; Lee, S.Y.; Thong, M.K.; Yip, C.H.; Mohd Taib, N.A.; Teo, S.H. Identification of germline alterations in breast cancer predisposition genes among Malaysian breast cancer patients using panel testing. Clin. Genet. 2016, 90, 315–323. [Google Scholar] [CrossRef]
- Nunziato, M.; Esposito, M.V.; Starnone, F.; Diroma, M.A.; Calabrese, A.; Del Monaco, V.; Buono, P.; Frasci, G.; Botti, G.; D’Aiuto, M.; et al. A multi-gene panel beyond BRCA1/BRCA2 to identify new breast cancer-predisposing mutations by a picodroplet PCR followed by a next-generation sequencing strategy: A pilot study. Anal. Chim. Acta 2019, 1046, 154–162. [Google Scholar] [CrossRef]
- Oliver, J.; Quezada Urban, R.; Franco Cortes, C.A.; Diaz Velasquez, C.E.; Montealegre Paez, A.L.; Pacheco-Orozco, R.A.; Castro Rojas, C.; Garcia-Robles, R.; Lopez Rivera, J.J.; Gaitan Chaparro, S.; et al. Latin American Study of Hereditary Breast and Ovarian Cancer LACAM: A Genomic Epidemiology Approach. Front. Oncol. 2019, 9, 1429. [Google Scholar] [CrossRef]
- Ollier, M.; Radosevic-Robin, N.; Kwiatkowski, F.; Ponelle, F.; Viala, S.; Privat, M.; Uhrhammer, N.; Bernard-Gallon, D.; Penault-Llorca, F.; Bignon, Y.J.; et al. DNA repair genes implicated in triple negative familial non-BRCA1/2 breast cancer predisposition. Am. J. Cancer Res. 2015, 5, 2113–2126. [Google Scholar]
- Ow, S.G.W.; Ong, P.Y.; Lee, S.C. Discoveries beyond BRCA1/2: Multigene testing in an Asian multi-ethnic cohort suspected of hereditary breast cancer syndrome in the real world. PLoS ONE 2019, 14, e0213746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.S.; Lee, S.T.; Nam, E.J.; Han, J.W.; Lee, J.Y.; Kim, J.; Kim, T.I.; Park, H.S. Variants of cancer susceptibility genes in Korean BRCA1/2 mutation-negative patients with high risk for hereditary breast cancer. BMC Cancer 2018, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Pritzlaff, M.; Summerour, P.; McFarland, R.; Li, S.; Reineke, P.; Dolinsky, J.S.; Goldgar, D.E.; Shimelis, H.; Couch, F.J.; Chao, E.C.; et al. Male breast cancer in a multi-gene panel testing cohort: Insights and unexpected results. Breast Cancer Res. Treat. 2017, 161, 575–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quezada Urban, R.; Diaz Velasquez, C.E.; Gitler, R.; Rojo Castillo, M.P.; Sirota Toporek, M.; Figueroa Morales, A.; Moreno Garcia, O.; Garcia Esquivel, L.; Torres Mejia, G.; Dean, M.; et al. Comprehensive Analysis of Germline Variants in Mexican Patients with Hereditary Breast and Ovarian Cancer Susceptibility. Cancers 2018, 10, 361. [Google Scholar] [CrossRef] [Green Version]
- Rajkumar, T.; Meenakumari, B.; Mani, S.; Sridevi, V.; Sundersingh, S. Targeted Resequencing of 30 Genes Improves the Detection of Deleterious Mutations in South Indian Women with Breast and/or Ovarian Cancers. Asian Pac. J. Cancer Prev 2015, 16, 5211–5217. [Google Scholar] [CrossRef] [Green Version]
- Ratajska, M.; Antoszewska, E.; Piskorz, A.; Brozek, I.; Borg, A.; Kusmierek, H.; Biernat, W.; Limon, J. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res. Treat. 2012, 131, 89–97. [Google Scholar] [CrossRef]
- Rizzolo, P.; Zelli, V.; Silvestri, V.; Valentini, V.; Zanna, I.; Bianchi, S.; Masala, G.; Spinelli, A.M.; Tibiletti, M.G.; Russo, A.; et al. Insight into genetic susceptibility to male breast cancer by multigene panel testing: Results from a multicenter study in Italy. Int. J. Cancer 2019, 145, 390–400. [Google Scholar] [CrossRef]
- Rodriguez-Balada, M.; Roig, B.; Mele, M.; Albacar, C.; Serrano, S.; Salvat, M.; Querol, M.; Borras, J.; Martorell, L.; Guma, J. Identification of germline pathogenic variants in DNA damage repair genes by a next-generation sequencing multigene panel in BRCAX patients. Clin. Biochem. 2020, 76, 17–23. [Google Scholar] [CrossRef]
- Rostami, P.; Zendehdel, K.; Shirkoohi, R.; Ebrahimi, E.; Ataei, M.; Imanian, H.; Najmabadi, H.; Akbari, M.R.; Sanati, M.H. Gene Panel Testing in Hereditary Breast Cancer. Arch. Iran. Med. 2020, 23, 155–162. [Google Scholar]
- Scarpitta, R.; Zanna, I.; Aretini, P.; Gambino, G.; Scatena, C.; Mei, B.; Ghilli, M.; Rossetti, E.; Roncella, M.; Congregati, C.; et al. Germline investigation in male breast cancer of DNA repair genes by next-generation sequencing. Breast Cancer Res. Treat. 2019, 178, 557–564. [Google Scholar] [CrossRef]
- Shahi, R.B.; De Brakeleer, S.; Caljon, B.; Pauwels, I.; Bonduelle, M.; Joris, S.; Fontaine, C.; Vanhoeij, M.; Van Dooren, S.; Teugels, E.; et al. Identification of candidate cancer predisposing variants by performing whole-exome sequencing on index patients from BRCA1 and BRCA2-negative breast cancer families. BMC Cancer 2019, 19, 313. [Google Scholar] [CrossRef] [PubMed]
- Shao, D.; Cheng, S.; Guo, F.; Zhu, C.; Yuan, Y.; Hu, K.; Wang, Z.; Meng, X.; Jin, X.; Xiong, Y.; et al. Prevalence of hereditary breast and ovarian cancer (HBOC) predisposition gene mutations among 882 HBOC high-risk Chinese individuals. Cancer Sci. 2020, 111, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.C.; Lee, H.B.; Yoo, T.K.; Lee, E.S.; Kim, R.N.; Park, B.; Yoon, K.A.; Park, C.; Lee, E.S.; Moon, H.G.; et al. Detection of Germline Mutations in Breast Cancer Patients with Clinical Features of Hereditary Cancer Syndrome Using a Multi-Gene Panel Test. Cancer Res. Treat. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirts, B.H.; Casadei, S.; Jacobson, A.L.; Lee, M.K.; Gulsuner, S.; Bennett, R.L.; Miller, M.; Hall, S.A.; Hampel, H.; Hisama, F.M.; et al. Improving performance of multigene panels for genomic analysis of cancer predisposition. Genet. Med. 2016, 18, 974–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siraj, A.K.; Masoodi, T.; Bu, R.; Parvathareddy, S.K.; Al-Badawi, I.A.; Al-Sanea, N.; Ashari, L.H.; Abduljabbar, A.; Alhomoud, S.; Al-Sobhi, S.S.; et al. Expanding the spectrum of germline variants in cancer. Hum. Genet. 2017, 136, 1431–1444. [Google Scholar] [CrossRef]
- Sokolenko, A.P.; Preobrazhenskaya, E.V.; Aleksakhina, S.N.; Iyevleva, A.G.; Mitiushkina, N.V.; Zaitseva, O.A.; Yatsuk, O.S.; Tiurin, V.I.; Strelkova, T.N.; Togo, A.V.; et al. Candidate gene analysis of BRCA1/2 mutation-negative high-risk Russian breast cancer patients. Cancer Lett. 2015, 359, 259–261. [Google Scholar] [CrossRef]
- Spugnesi, L.; Gabriele, M.; Scarpitta, R.; Tancredi, M.; Maresca, L.; Gambino, G.; Collavoli, A.; Aretini, P.; Bertolini, I.; Salvadori, B.; et al. Germline mutations in DNA repair genes may predict neoadjuvant therapy response in triple negative breast patients. Genes Chromosomes Cancer 2016, 55, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Stafford, J.L.; Dyson, G.; Levin, N.K.; Chaudhry, S.; Rosati, R.; Kalpage, H.; Wernette, C.; Petrucelli, N.; Simon, M.S.; Tainsky, M.A. Reanalysis of BRCA1/2 negative high risk ovarian cancer patients reveals novel germline risk loci and insights into missing heritability. PLoS ONE 2017, 12, e0178450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugino, K.; Tamura, R.; Nakaoka, H.; Yachida, N.; Yamaguchi, M.; Mori, Y.; Yamawaki, K.; Suda, K.; Ishiguro, T.; Adachi, S.; et al. Germline and somatic mutations of homologous recombination-associated genes in Japanese ovarian cancer patients. Sci. Rep. 2019, 9, 17808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Meng, H.; Yao, L.; Lv, M.; Bai, J.; Zhang, J.; Wang, L.; Ouyang, T.; Li, J.; Wang, T.; et al. Germline Mutations in Cancer Susceptibility Genes in a Large Series of Unselected Breast Cancer Patients. Clin. Cancer Res. 2017, 23, 6113–6119. [Google Scholar] [CrossRef] [Green Version]
- alo, P.L.; Wen, K.C.; Chen, Y.J.; Chao, T.C.; Tsai, Y.F.; Tseng, L.M.; Qiu, J.T.; Chao, K.C.; Wu, H.H.; Chuang, C.M.; et al. The frequency of cancer predisposition gene mutations in hereditary breast and ovarian cancer patients in Taiwan: From BRCA1/2 to multi-gene panels. PLoS ONE 2017, 12, e0185615. [Google Scholar]
- Thompson, E.R.; Rowley, S.M.; Li, N.; McInerny, S.; Devereux, L.; Wong-Brown, M.W.; Trainer, A.H.; Mitchell, G.; Scott, R.J.; James, P.A.; et al. Panel Testing for Familial Breast Cancer: Calibrating the Tension Between Research and Clinical Care. J. Clin. Oncol. 2016, 34, 1455–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torrezan, G.T.; de Almeida, F.; Figueiredo, M.C.P.; Barros, B.D.F.; de Paula, C.A.A.; Valieris, R.; de Souza, J.E.S.; Ramalho, R.F.; da Silva, F.C.C.; Ferreira, E.N.; et al. Complex Landscape of Germline Variants in Brazilian Patients With Hereditary and Early Onset Breast Cancer. Front. Genet. 2018, 9, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsaousis, G.N.; Papadopoulou, E.; Apessos, A.; Agiannitopoulos, K.; Pepe, G.; Kampouri, S.; Diamantopoulos, N.; Floros, T.; Iosifidou, R.; Katopodi, O.; et al. Analysis of hereditary cancer syndromes by using a panel of genes: Novel and multiple pathogenic mutations. BMC Cancer 2019, 19, 535. [Google Scholar] [CrossRef] [Green Version]
- Tung, N.; Battelli, C.; Allen, B.; Kaldate, R.; Bhatnagar, S.; Bowles, K.; Timms, K.; Garber, J.E.; Herold, C.; Ellisen, L.; et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer 2015, 121, 25–33. [Google Scholar] [CrossRef]
- Tung, N.; Lin, N.U.; Kidd, J.; Allen, B.A.; Singh, N.; Wenstrup, R.J.; Hartman, A.R.; Winer, E.P.; Garber, J.E. Frequency of Germline Mutations in 25 Cancer Susceptibility Genes in a Sequential Series of Patients With Breast Cancer. J. Clin. Oncol. 2016, 34, 1460–1468. [Google Scholar] [CrossRef] [Green Version]
- Vahteristo, P.; Syrjakoski, K.; Heikkinen, T.; Eerola, H.; Aittomaki, K.; von Smitten, K.; Holli, K.; Blomqvist, C.; Kallioniemi, O.P.; Nevanlinna, H. BARD1 variants Cys557Ser and Val507Met in breast cancer predisposition. Eur. J. Hum. Genet. 2006, 14, 167–172. [Google Scholar] [CrossRef] [Green Version]
- Walsh, T.; Mandell, J.B.; Norquist, B.M.; Casadei, S.; Gulsuner, S.; Lee, M.K.; King, M.C. Genetic Predisposition to Breast Cancer Due to Mutations Other Than BRCA1 and BRCA2 Founder Alleles Among Ashkenazi Jewish Women. JAMA Oncol. 2017, 3, 1647–1653. [Google Scholar] [CrossRef]
- Wang, J.; Li, W.; Shi, Y.; Huang, Y.; Sun, T.; Tang, L.; Lu, Q.; Lei, Q.; Liao, N.; Jin, F.; et al. Germline mutation landscape of Chinese patients with familial breast/ovarian cancer in a panel of 22 susceptibility genes. Cancer Med. 2019, 8, 2074–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.A.; Jian, J.W.; Hung, C.F.; Peng, H.P.; Yang, C.F.; Cheng, H.S.; Yang, A.S. Germline breast cancer susceptibility gene mutations and breast cancer outcomes. BMC Cancer 2018, 18, 315. [Google Scholar] [CrossRef] [Green Version]
- Wong, E.S.Y.; Shekar, S.; Met-Domestici, M.; Chan, C.; Sze, M.; Yap, Y.S.; Rozen, S.G.; Tan, M.H.; Ang, P.; Ngeow, J.; et al. Inherited breast cancer predisposition in Asians: Multigene panel testing outcomes from Singapore. NPJ Genom. Med. 2016, 1, 15003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yablonski-Peretz, T.; Paluch-Shimon, S.; Gutman, L.S.; Kaplan, Y.; Dvir, A.; Barnes-Kedar, I.; Kadouri, L.; Semenisty, V.; Efrat, N.; Neiman, V.; et al. Screening for germline mutations in breast/ovarian cancer susceptibility genes in high-risk families in Israel. Breast Cancer Res. Treat. 2016, 155, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, J.; Lu, J.; Liu, G.; Di, G.; Chen, C.; Hou, Y.; Sun, M.; Yang, W.; Xu, X.; et al. Identification of a comprehensive spectrum of genetic factors for hereditary breast cancer in a Chinese population by next-generation sequencing. PLoS ONE 2015, 10, e0125571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, D.; Xu, L.; Luo, J.; You, X.; Huang, T.; Zi, Y.; Li, X.; Wang, R.; Zhong, Z.; Tang, X.; et al. Germline TP53 and MSH6 mutations implicated in sporadic triple-negative breast cancer (TNBC): A preliminary study. Hum. Genom. 2019, 13, 4. [Google Scholar] [CrossRef] [Green Version]
- Yoo, J.; Lee, G.D.; Kim, J.H.; Lee, S.N.; Chae, H.; Han, E.; Kim, Y.; Kim, M. Clinical Validity of Next-Generation Sequencing Multi-Gene Panel Testing for Detecting Pathogenic Variants in Patients With Hereditary Breast-Ovarian Cancer Syndrome. Ann. Lab. Med. 2020, 40, 148–154. [Google Scholar] [CrossRef]
- Zhao, Q.; Yang, J.; Li, L.; Cao, D.; Yu, M.; Shen, K.; Group, B.G.I. Germline and somatic mutations in homologous recombination genes among Chinese ovarian cancer patients detected using next-generation sequencing. J. Gynecol. Oncol. 2017, 28, e39. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Walsh, T.; Gulsuner, S.; Casadei, S.; Lee, M.K.; Ogundiran, T.O.; Ademola, A.; Falusi, A.G.; Adebamowo, C.A.; Oluwasola, A.O.; et al. Inherited Breast Cancer in Nigerian Women. J. Clin. Oncol. 2018, 36, 2820–2825. [Google Scholar] [CrossRef]
- Zidan, J.; Zhou, A.Y.; van den Akker, J.; Laitman, Y.; Schayek, H.; Schnaider, J.; Friedman, E. Inherited predisposition to breast and ovarian cancer in non-Jewish populations in Israel. Breast Cancer Res. Treat. 2017, 166, 881–885. [Google Scholar] [CrossRef]
- Susswein, L.R.; Marshall, M.L.; Nusbaum, R.; Vogel Postula, K.J.; Weissman, S.M.; Yackowski, L.; Vaccari, E.M.; Bissonnette, J.; Booker, J.K.; Cremona, M.L.; et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet. Med. 2016, 18, 823–832. [Google Scholar] [CrossRef] [Green Version]
- Teer, J.K.; Yoder, S.; Gjyshi, A.; Nicosia, S.V.; Zhang, C.; Monteiro, A.N.A. Mutational heterogeneity in non-serous ovarian cancers. Sci. Rep. 2017, 7, 9728. [Google Scholar] [CrossRef]
- LaDuca, H.; Stuenkel, A.J.; Dolinsky, J.S.; Keiles, S.; Tandy, S.; Pesaran, T.; Chen, E.; Gau, C.L.; Palmaer, E.; Shoaepour, K.; et al. Utilization of multigene panels in hereditary cancer predisposition testing: Analysis of more than 2,000 patients. Genet. Med. 2014, 16, 830–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, S.; Reeves, A.; Campian, S.; Paine, A.; Zakalik, D. Outcomes of retesting BRCA negative patients using multigene panels. Fam. Cancer 2017, 16, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Klonowska, K.; Ratajska, M.; Czubak, K.; Kuzniacka, A.; Brozek, I.; Koczkowska, M.; Sniadecki, M.; Debniak, J.; Wydra, D.; Balut, M.; et al. Analysis of large mutations in BARD1 in patients with breast and/or ovarian cancer: The Polish population as an example. Sci. Rep. 2015, 5, 10424. [Google Scholar] [CrossRef]
- Taylor, A.; Brady, A.F.; Frayling, I.M.; Hanson, H.; Tischkowitz, M.; Turnbull, C.; Side, L.; Group, U.K.C.G. Consensus for genes to be included on cancer panel tests offered by UK genetics services: Guidelines of the UK Cancer Genetics Group. J. Med. Genet. 2018, 55, 372–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castera, L.; Harter, V.; Muller, E.; Krieger, S.; Goardon, N.; Ricou, A.; Rousselin, A.; Paimparay, G.; Legros, A.; Bruet, O.; et al. Landscape of pathogenic variations in a panel of 34 genes and cancer risk estimation from 5131 HBOC families. Genet. Med. 2018, 20, 1677–1686. [Google Scholar] [CrossRef] [Green Version]
- Buys, S.S.; Sandbach, J.F.; Gammon, A.; Patel, G.; Kidd, J.; Brown, K.L.; Sharma, L.; Saam, J.; Lancaster, J.; Daly, M.B. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 2017, 123, 1721–1730. [Google Scholar] [CrossRef] [Green Version]
- Shimelis, H.; LaDuca, H.; Hu, C.; Hart, S.N.; Na, J.; Thomas, A.; Akinhanmi, M.; Moore, R.M.; Brauch, H.; Cox, A.; et al. Triple-Negative Breast Cancer Risk Genes Identified by Multigene Hereditary Cancer Panel Testing. J. Natl. Cancer Inst. 2018, 110, 855–862. [Google Scholar] [CrossRef]
- Southey, M.C.; Goldgar, D.E.; Winqvist, R.; Pylkas, K.; Couch, F.; Tischkowitz, M.; Foulkes, W.D.; Dennis, J.; Michailidou, K.; van Rensburg, E.J.; et al. PALB2, CHEK2 and ATM rare variants and cancer risk: Data from COGS. J. Med. Genet. 2016, 53, 800–811. [Google Scholar] [CrossRef] [Green Version]
- Adamovich, A.I.; Banerjee, T.; Wingo, M.; Duncan, K.; Ning, J.; Martins Rodrigues, F.; Huang, K.L.; Lee, C.; Chen, F.; Ding, L.; et al. Functional analysis of BARD1 missense variants in homology-directed repair and damage sensitivity. PLoS Genet. 2019, 15, e1008049. [Google Scholar] [CrossRef]
- Laufer, M.; Nandula, S.V.; Modi, A.P.; Wang, S.; Jasin, M.; Murty, V.V.; Ludwig, T.; Baer, R. Structural requirements for the BARD1 tumor suppressor in chromosomal stability and homology-directed DNA repair. J. Biol. Chem. 2007, 282, 34325–34333. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, K.; Saredi, G.; Becker, J.R.; Foster, B.M.; Nguyen, N.V.; Beyer, T.E.; Cesa, L.C.; Faull, P.A.; Lukauskas, S.; Frimurer, T.; et al. H4K20me0 recognition by BRCA1-BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 2019, 21, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Ratajska, M.; Matusiak, M.; Kuzniacka, A.; Wasag, B.; Brozek, I.; Biernat, W.; Koczkowska, M.; Debniak, J.; Sniadecki, M.; Kozlowski, P.; et al. Cancer predisposing BARD1 mutations affect exon skipping and are associated with overexpression of specific BARD1 isoforms. Oncol. Rep. 2015, 34, 2609–2617. [Google Scholar] [CrossRef] [Green Version]
- Rudd, M.F.; Webb, E.L.; Matakidou, A.; Sellick, G.S.; Williams, R.D.; Bridle, H.; Eisen, T.; Houlston, R.S.; Consortium, G. Variants in the GH-IGF axis confer susceptibility to lung cancer. Genome Res. 2006, 16, 693–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capasso, M.; Devoto, M.; Hou, C.; Asgharzadeh, S.; Glessner, J.T.; Attiyeh, E.F.; Mosse, Y.P.; Kim, C.; Diskin, S.J.; Cole, K.A.; et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat. Genet. 2009, 41, 718–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suszynska, M.; Kozlowski, P. Summary of BARD1 Mutations and Precise Estimation of Breast and Ovarian Cancer Risks Associated with the Mutations. Genes 2020, 11, 798. https://doi.org/10.3390/genes11070798
Suszynska M, Kozlowski P. Summary of BARD1 Mutations and Precise Estimation of Breast and Ovarian Cancer Risks Associated with the Mutations. Genes. 2020; 11(7):798. https://doi.org/10.3390/genes11070798
Chicago/Turabian StyleSuszynska, Malwina, and Piotr Kozlowski. 2020. "Summary of BARD1 Mutations and Precise Estimation of Breast and Ovarian Cancer Risks Associated with the Mutations" Genes 11, no. 7: 798. https://doi.org/10.3390/genes11070798
APA StyleSuszynska, M., & Kozlowski, P. (2020). Summary of BARD1 Mutations and Precise Estimation of Breast and Ovarian Cancer Risks Associated with the Mutations. Genes, 11(7), 798. https://doi.org/10.3390/genes11070798





