Guarding the Genome: CENP-A-Chromatin in Health and Cancer
Abstract
1. Introduction
2. The Structure and Composition of CENP-A-Containing Chromatin
3. CENP-B Roles in Centromere Specification and Function
4. The Constitutive Centromere-Associated Network Role in Kinetochore Assembly
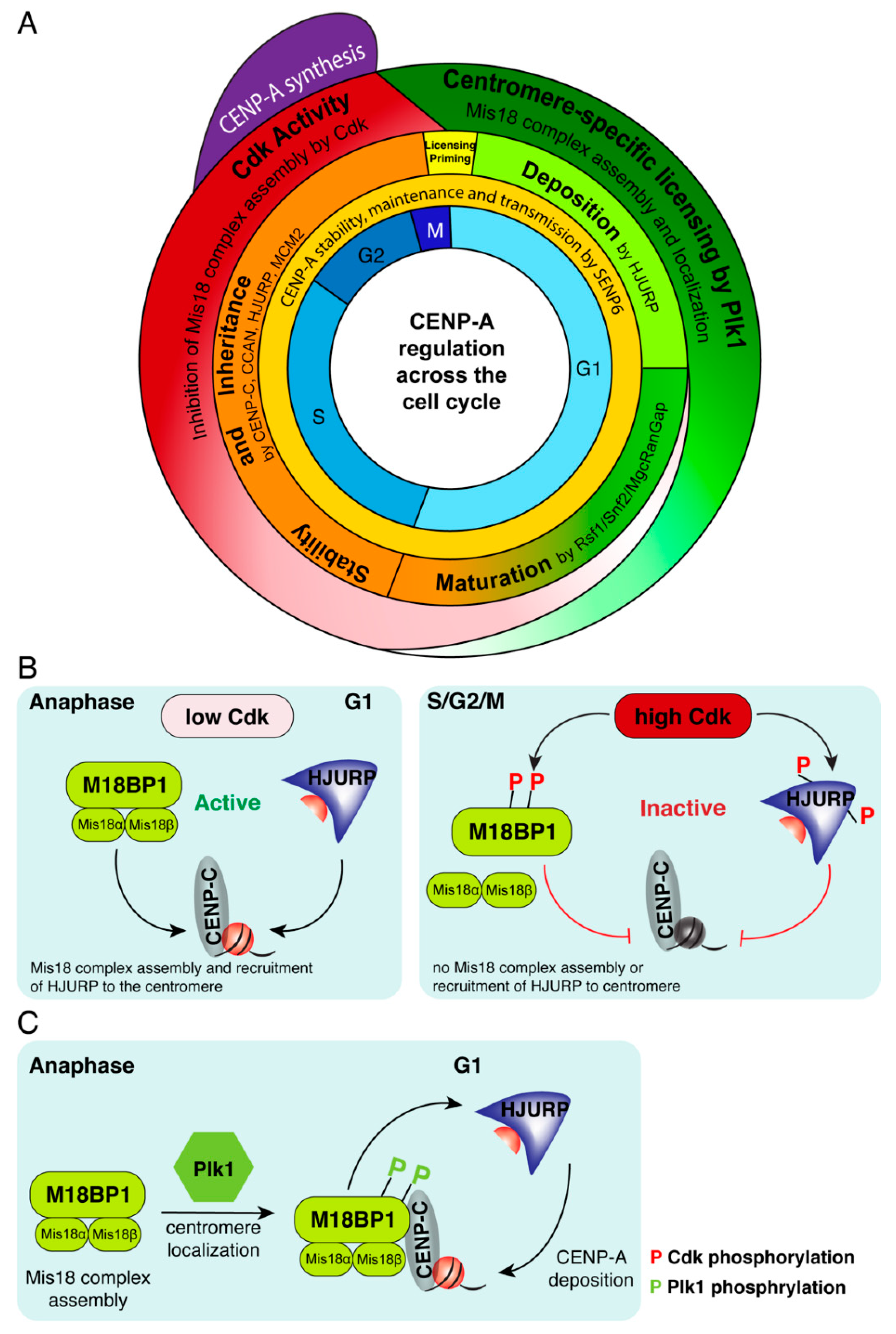
5. Temporal Regulation of CENP-A Deposition: Precision in Action
6. Global Regulation Restricts CENP-A Assembly to Early G1
7. Maturation and Stability of Newly Deposited CENP-A
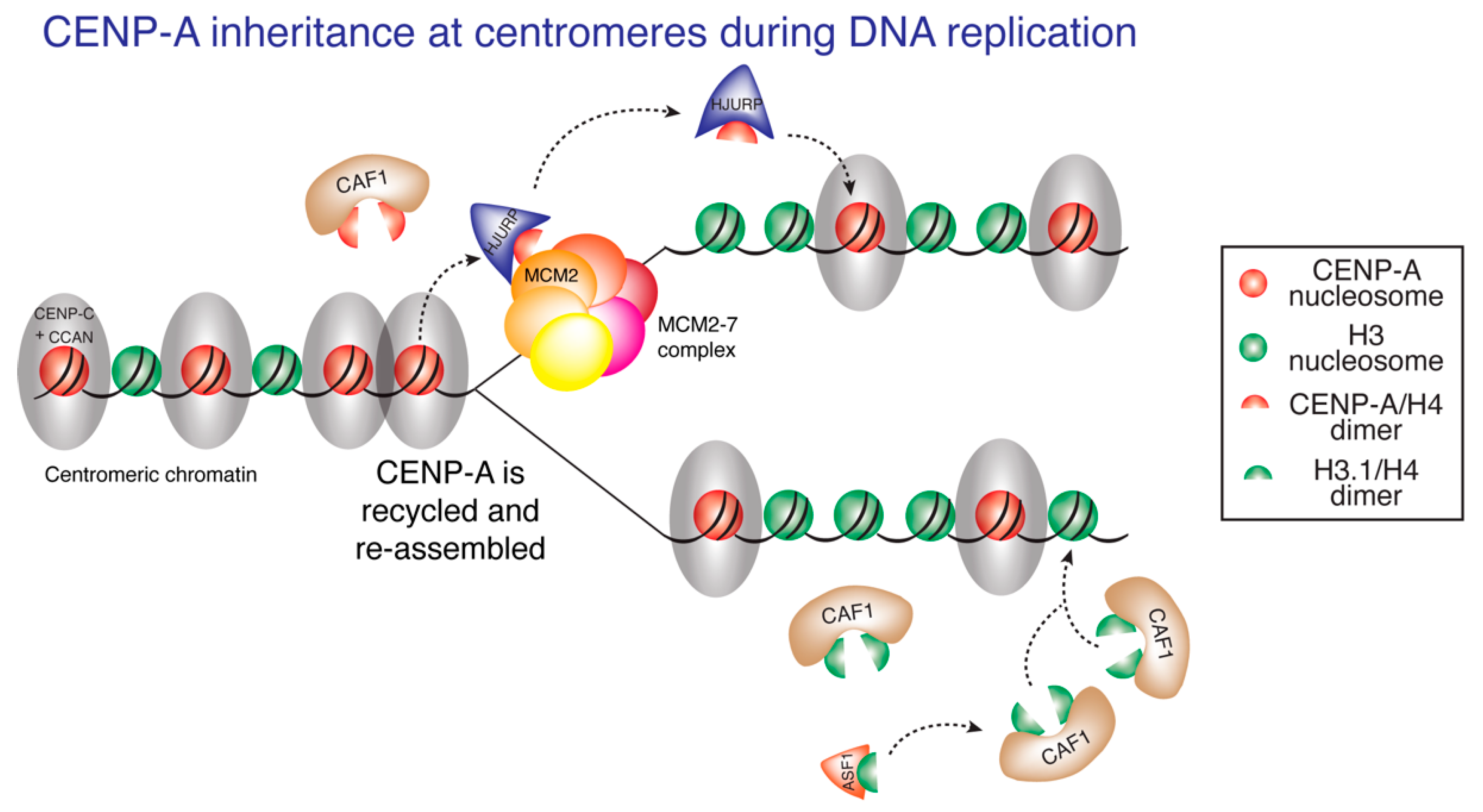
8. CENP-A Inheritance at the Centromeric DNA Replication Fork Crossroad
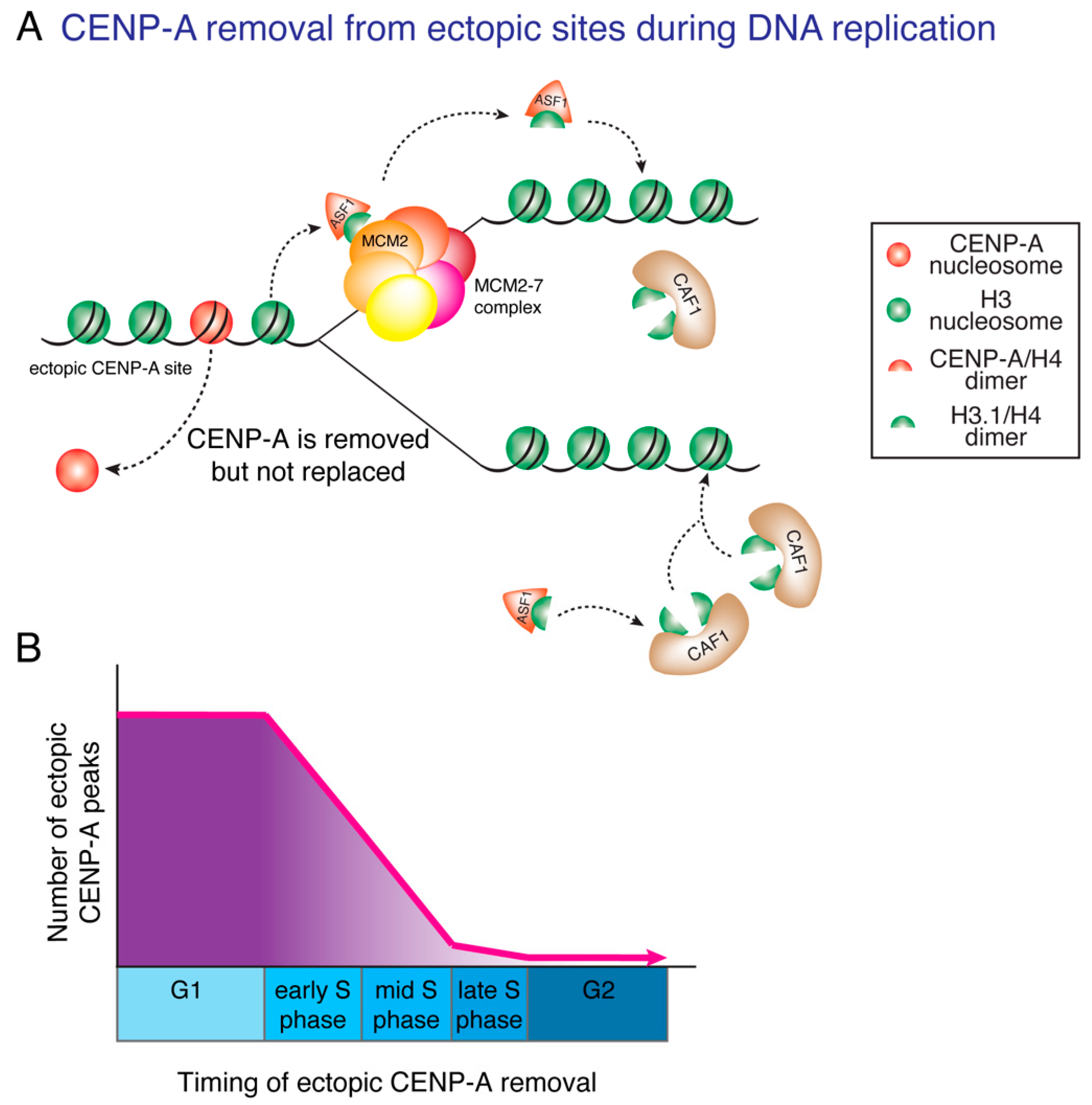
9. DNA Replication Ensures Centromere Specificity
10. Connecting CENP-A, Chromosomal Instability and Cancer
11. Ectopic Deposition of CENP-A and CIN
12. CENP-A Misregulation and Gene Expression
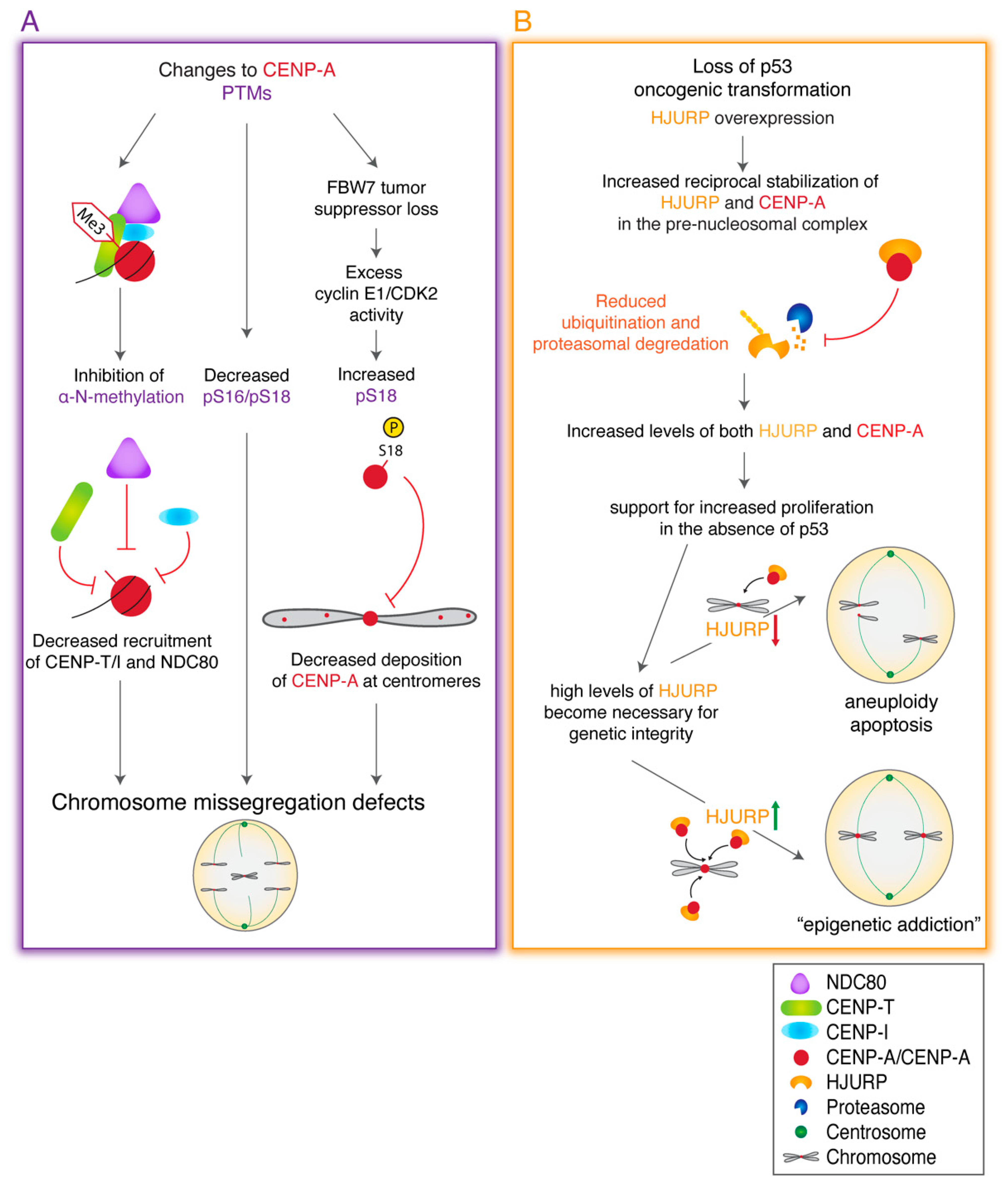
13. Perturbations in CENP-A Posttranslational Modifications are Linked to Cancer
14. CENP-A Overexpression May Be Indispensable for Cancer Progression
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carter, S.L.; Cibulskis, K.; Helman, E.; McKenna, A.; Shen, H.; Zack, T.; Laird, P.W.; Onofrio, R.C.; Winckler, W.; Weir, B.A.; et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012, 30, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instabilities in human cancers. Nature 1998, 396, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Funk, L.C.; Zasadil, L.M.; Weaver, B.A. Living in CIN: Mitotic Infidelity and Its Consequences for Tumor Promotion and Suppression. Dev. Cell 2016, 39, 638–652. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, H.; Lengauer, C. Aneuploidy and cancer. Nature 2004, 432, 338–341. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Cantley, L.C. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 2018, 174, 1347–1360. [Google Scholar] [CrossRef]
- Bolhaqueiro, A.C.F.; Ponsioen, B.; Bakker, B.; Klaasen, S.J.; Kucukkose, E.; van Jaarsveld, R.H.; Vivie, J.; Verlaan-Klink, I.; Hami, N.; Spierings, D.C.J.; et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 2019, 51, 824–834. [Google Scholar] [CrossRef]
- Henikoff, S.; Ahmad, K.; Malik, H.S. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 2001, 293, 1098–1102. [Google Scholar] [CrossRef]
- Cleveland, D.W.; Mao, Y.; Sullivan, K.F. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 2003, 112, 407–421. [Google Scholar] [CrossRef]
- Manuelidis, L.; Wu, J.C. Homology between human and simian repeated DNA. Nature 1978, 276, 92–94. [Google Scholar] [CrossRef]
- Willard, H.F. Chromosome-specific organization of human α satellite DNA. Am. J. Hum. Genet. 1985, 37, 524–532. [Google Scholar]
- Alexandrov, I.; Kazakov, A.; Tumeneva, I.; Shepelev, V.; Yurov, Y. α-satellite DNA of primates: Old and new families. Chromosoma 2001, 110, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Nechemia-Arbely, Y.; Miga, K.H.; Shoshani, O.; Aslanian, A.; McMahon, M.A.; Lee, A.Y.; Fachinetti, D.; Yates, J.R.; Ren, B.; Cleveland, D.W. DNA replication acts as an error correction mechanism to maintain centromere identity by restricting CENP-A to centromeres. Nat. Cell Biol. 2019, 21, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Willard, H.F.; Waye, J.S. Chromosome-specific subsets of human α satellite DNA: Analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J. Mol. Evol. 1987, 25, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Nechemia-Arbely, Y.; Fachinetti, D.; Miga, K.H.; Sekulic, N.; Soni, G.V.; Kim, D.H.; Wong, A.K.; Lee, A.Y.; Nguyen, K.; Dekker, C.; et al. Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points. J. Cell Biol. 2017, 216, 607–621. [Google Scholar] [CrossRef]
- Hayden, K.E.; Strome, E.D.; Merrett, S.L.; Lee, H.R.; Rudd, M.K.; Willard, H.F. Sequences associated with centromere competency in the human genome. Mol. Cell Biol. 2013, 33, 763–772. [Google Scholar] [CrossRef]
- Miga, K.H. Centromeric Satellite DNAs: Hidden Sequence Variation in the Human Population. Genes Basel 2019, 10, 352. [Google Scholar] [CrossRef]
- Stimpson, K.M.; Sullivan, B.A. Epigenomics of centromere assembly and function. Curr. Opin. Cell Biol. 2010, 22, 772–780. [Google Scholar] [CrossRef]
- Marshall, O.J.; Chueh, A.C.; Wong, L.H.; Choo, K.H. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 2008, 82, 261–282. [Google Scholar] [CrossRef]
- Amor, D.J.; Bentley, K.; Ryan, J.; Perry, J.; Wong, L.; Slater, H.; Choo, K.H. Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. USA 2004, 101, 6542–6547. [Google Scholar] [CrossRef]
- Smith, M.M. Centromeres and variant histones: What, where, when and why? Curr. Opin. Cell Biol. 2002, 14, 279–285. [Google Scholar] [CrossRef]
- Earnshaw, W.C.; Rothfield, N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 1985, 91, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.K.; O’Day, K.; Wener, M.H.; Andrews, B.S.; Margolis, R.L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 1987, 104, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Fachinetti, D.; Folco, H.D.; Nechemia-Arbely, Y.; Valente, L.P.; Nguyen, K.; Wong, A.J.; Zhu, Q.; Holland, A.J.; Desai, A.; Jansen, L.E.; et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 2013, 15, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Howman, E.V.; Fowler, K.J.; Newson, A.J.; Redward, S.; MacDonald, A.C.; Kalitsis, P.; Choo, K.H. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 2000, 97, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Gemble, S.; Simon, A.; Pennetier, C.; Dumont, M.; Herve, S.; Meitinger, F.; Oegema, K.; Rodriguez, R.; Almouzni, G.; Fachinetti, D.; et al. Centromere Dysfunction Compromises Mitotic Spindle Pole Integrity. Curr. Biol. 2019, 29, 3072–3080. [Google Scholar] [CrossRef]
- Regnier, V.; Vagnarelli, P.; Fukagawa, T.; Zerjal, T.; Burns, E.; Trouche, D.; Earnshaw, W.; Brown, W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell Biol. 2005, 25, 3967–3981. [Google Scholar] [CrossRef]
- Ho, K.H.; Tsuchiya, D.; Oliger, A.C.; Lacefield, S. Localization and function of budding yeast CENP-A depends upon kinetochore protein interactions and is independent of canonical centromere sequence. Cell Rep. 2014, 9, 2027–2033. [Google Scholar] [CrossRef]
- Mendiburo, M.J.; Padeken, J.; Fulop, S.; Schepers, A.; Heun, P. Drosophila CENH3 is sufficient for centromere formation. Science 2011, 334, 686–690. [Google Scholar] [CrossRef]
- Olszak, A.M.; van Essen, D.; Pereira, A.J.; Diehl, S.; Manke, T.; Maiato, H.; Saccani, S.; Heun, P. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat. Cell Biol. 2011, 13, 799–808. [Google Scholar] [CrossRef]
- Palladino, J.; Chavan, A.; Sposato, A.; Mason, T.D.; Mellone, B.G. Targeted De Novo Centromere Formation in Drosophila Reveals Plasticity and Maintenance Potential of CENP-A Chromatin. Dev. Cell 2020, 52, 379–394. [Google Scholar] [CrossRef]
- Barnhart, M.C.; Kuich, P.H.; Stellfox, M.E.; Ward, J.A.; Bassett, E.A.; Black, B.E.; Foltz, D.R. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011, 194, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Logsdon, G.A.; Gambogi, C.W.; Liskovykh, M.A.; Barrey, E.J.; Larionov, V.; Miga, K.H.; Heun, P.; Black, B.E. Human Artificial Chromosomes that Bypass Centromeric DNA. Cell 2019, 178, 624–639. [Google Scholar] [CrossRef] [PubMed]
- Ohzeki, J.-I.; Nakano, M.; Okada, T.; Masumoto, H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002, 159, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Schueler, M.G.; Higgins, A.W.; Rudd, M.K.; Gustashaw, K.; Willard, H.F. Genomic and Genetic Definition of a Functional Human Centromere. Science 2001, 294, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Ohzeki, J.-I.; Nakano, M.; Yoda, K.; Brinkley, W.R.; Larionov, V.; Masumoto, H. CENP-B Controls Centromere Formation Depending on the Chromatin Context. Cell 2007, 131, 1287–1300. [Google Scholar] [CrossRef]
- Gambogi, C.W.; Dawicki-McKenna, J.M.; Logsdon, G.A.; Black, B.E. The unique kind of human artificial chromosome: Bypassing the requirement for repetitive centromere DNA. Exp. Cell Res. 2020, 391, 111978. [Google Scholar] [CrossRef]
- Bodor, D.L.; Mata, J.F.; Sergeev, M.; David, A.F.; Salimian, K.J.; Panchenko, T.; Cleveland, D.W.; Black, B.E.; Shah, J.V.; Jansen, L.E. The quantitative architecture of centromeric chromatin. eLife 2014, 3, e02137. [Google Scholar] [CrossRef]
- Blower, M.D.; Sullivan, B.A.; Karpen, G.H. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2002, 2, 319–330. [Google Scholar] [CrossRef]
- Sullivan, B.A.; Karpen, G.H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004, 11, 1076–1083. [Google Scholar] [CrossRef]
- Black, B.E.; Cleveland, D.W. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 2011, 144, 471–479. [Google Scholar] [CrossRef]
- Black, B.E.; Brock, M.A.; Bedard, S.; Woods, V.L., Jr.; Cleveland, D.W. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 2007, 104, 5008–5013. [Google Scholar] [CrossRef] [PubMed]
- Falk, S.J.; Lee, J.; Sekulic, N.; Sennett, M.A.; Lee, T.H.; Black, B.E. CENP-C directs a structural transition of CENP-A nucleosomes mainly through sliding of DNA gyres. Nat. Struct. Mol. Biol. 2016, 23, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Tachiwana, H.; Kagawa, W.; Shiga, T.; Osakabe, A.; Miya, Y.; Saito, K.; Hayashi-Takanaka, Y.; Oda, T.; Sato, M.; Park, S.Y.; et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 2011, 476, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Dalal, Y.; Wang, H.; Lindsay, S.; Henikoff, S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007, 5, e218. [Google Scholar] [CrossRef] [PubMed]
- Dimitriadis, E.K.; Weber, C.; Gill, R.K.; Diekmann, S.; Dalal, Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 20317–20322. [Google Scholar] [CrossRef]
- Furuyama, T.; Henikoff, S. Centromeric nucleosomes induce positive DNA supercoils. Cell 2009, 138, 104–113. [Google Scholar] [CrossRef]
- Williams, J.S.; Hayashi, T.; Yanagida, M.; Russell, P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell 2009, 33, 287–298. [Google Scholar] [CrossRef]
- Lacoste, N.; Woolfe, A.; Tachiwana, H.; Garea, A.V.; Barth, T.; Cantaloube, S.; Kurumizaka, H.; Imhof, A.; Almouzni, G. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell 2014, 53, 631–644. [Google Scholar] [CrossRef]
- Arimura, Y.; Shirayama, K.; Horikoshi, N.; Fujita, R.; Taguchi, H.; Kagawa, W.; Fukagawa, T.; Almouzni, G.; Kurumizaka, H. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci. Rep. 2014, 4, 7115. [Google Scholar] [CrossRef]
- Bui, M.; Dimitriadis, E.K.; Hoischen, C.; An, E.; Quenet, D.; Giebe, S.; Nita-Lazar, A.; Diekmann, S.; Dalal, Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell 2012, 150, 317–326. [Google Scholar] [CrossRef]
- Padeganeh, A.; Ryan, J.; Boisvert, J.; Ladouceur, A.M.; Dorn, J.F.; Maddox, P.S. Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle. Curr. Biol. 2013, 23, 764–769. [Google Scholar] [CrossRef]
- Miell, M.D.; Fuller, C.J.; Guse, A.; Barysz, H.M.; Downes, A.; Owen-Hughes, T.; Rappsilber, J.; Straight, A.F.; Allshire, R.C. CENP-A confers a reduction in height on octameric nucleosomes. Nat. Struct. Mol. Biol. 2013, 20, 763–765. [Google Scholar] [CrossRef]
- Sekulic, N.; Bassett, E.A.; Rogers, D.J.; Black, B.E. The structure of (CENP-A-H4) (2) reveals physical features that mark centromeres. Nature 2010, 467, 347–351. [Google Scholar] [CrossRef]
- Boopathi, R.; Danev, R.; Khoshouei, M.; Kale, S.; Nahata, S.; Ramos, L.; Angelov, D.; Dimitrov, S.; Hamiche, A.; Petosa, C.; et al. Phase-plate cryo-EM structure of the Widom 601 CENP-A nucleosome core particle reveals differential flexibility of the DNA ends. Nucleic Acids Res. 2020, 48, 5735–5748. [Google Scholar] [CrossRef]
- Roulland, Y.; Ouararhni, K.; Naidenov, M.; Ramos, L.; Shuaib, M.; Syed, S.H.; Lone, I.N.; Boopathi, R.; Fontaine, E.; Papai, G.; et al. The Flexible Ends of CENP-A Nucleosome Are Required for Mitotic Fidelity. Mol. Cell 2016, 63, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Conde e Silva, N.; Black, B.E.; Sivolob, A.; Filipski, J.; Cleveland, D.W.; Prunell, A. CENP-A-containing nucleosomes: Easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 2007, 370, 555–573. [Google Scholar] [CrossRef] [PubMed]
- Hasson, D.; Panchenko, T.; Salimian, K.J.; Salman, M.U.; Sekulic, N.; Alonso, A.; Warburton, P.E.; Black, B.E. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 2013, 20, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Panchenko, T.; Sorensen, T.C.; Woodcock, C.L.; Kan, Z.Y.; Wood, S.; Resch, M.G.; Luger, K.; Englander, S.W.; Hansen, J.C.; Black, B.E. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc. Natl. Acad. Sci. USA 2011, 108, 16588–16593. [Google Scholar] [CrossRef]
- Takizawa, Y.; Ho, C.H.; Tachiwana, H.; Matsunami, H.; Kobayashi, W.; Suzuki, M.; Arimura, Y.; Hori, T.; Fukagawa, T.; Ohi, M.D.; et al. Cryo-EM Structures of Centromeric Tri-nucleosomes Containing a Central CENP-A Nucleosome. Structure 2020, 28, 44–53. [Google Scholar] [CrossRef]
- Zhou, B.R.; Yadav, K.N.S.; Borgnia, M.; Hong, J.; Cao, B.; Olins, A.L.; Olins, D.E.; Bai, Y.; Zhang, P. Atomic resolution cryo-EM structure of a native-like CENP-A nucleosome aided by an antibody fragment. Nat. Commun. 2019, 10, 2301. [Google Scholar] [CrossRef]
- Masumoto, H.; Masukata, H.; Muro, Y.; Nozaki, N.; Okazaki, T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 1989, 109, 1963–1973. [Google Scholar] [CrossRef]
- Muro, Y.; Masumoto, H.; Yoda, K.; Nozaki, N.; Ohashi, M.; Okazaki, T. Centromere protein B assembles human centromeric α-satellite DNA at the 17-bp sequence, CENP-B box. J. Cell Biol. 1992, 116, 585–596. [Google Scholar] [CrossRef]
- Tanaka, Y.; Nureki, O.; Kurumizaka, H.; Fukai, S.; Kawaguchi, S.; Ikuta, M.; Iwahara, J.; Okazaki, T.; Yokoyama, S. Crystal structure of the CENP-B protein-DNA complex: The DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 2001, 20, 6612–6618. [Google Scholar] [CrossRef] [PubMed]
- Fachinetti, D.; Han, J.S.; McMahon, M.A.; Ly, P.; Abdullah, A.; Wong, A.J.; Cleveland, D.W. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev. Cell 2015, 33, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Gamba, R.; Fachinetti, D. From evolution to function: Two sides of the same CENP-B coin? Exp. Cell Res. 2020, 111959. [Google Scholar] [CrossRef]
- Hoffmann, S.; Dumont, M.; Barra, V.; Ly, P.; Nechemia-Arbely, Y.; McMahon, M.A.; Herve, S.; Cleveland, D.W.; Fachinetti, D. CENP-A Is Dispensable for Mitotic Centromere Function after Initial Centromere/Kinetochore Assembly. Cell Rep. 2016, 17, 2394–2404. [Google Scholar] [CrossRef] [PubMed]
- Dumont, M.; Gamba, R.; Gestraud, P.; Klaasen, S.; Worrall, J.T.; De Vries, S.G.; Boudreau, V.; Salinas-Luypaert, C.; Maddox, P.S.; Lens, S.M.; et al. Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features. EMBO J. 2020, 39, e102924. [Google Scholar] [CrossRef] [PubMed]
- Tyler-Smith, C.; Gimelli, G.; Giglio, S.; Floridia, G.; Pandya, A.; Terzoli, G.; Warburton, P.E.; Earnshaw, W.C.; Zuffardi, O. Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet. 1999, 64, 1440–1444. [Google Scholar] [CrossRef]
- Koren, A.; Tsai, H.J.; Tirosh, I.; Burrack, L.S.; Barkai, N.; Berman, J. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet. 2010, 6, e1001068. [Google Scholar] [CrossRef]
- Hori, T.; Fukagawa, T. Artificial generation of centromeres and kinetochores to understand their structure and function. Exp. Cell Res. 2020, 389, 111898. [Google Scholar] [CrossRef]
- Hudson, D.F.; Fowler, K.J.; Earle, E.; Saffery, R.; Kalitsis, P.; Trowell, H.; Hill, J.; Wreford, N.G.; de Kretser, D.M.; Cancilla, M.R.; et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 1998, 141, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Montes de Oca Luna, R.; Liu, G.; Lozano, G.; Cummings, C.; Mancini, M.; Ouspenski, I.; Brinkley, B.R.; May, G.S. The cenpB gene is not essential in mice. Chromosoma 1998, 107, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Perez-Castro, A.V.; Shamanski, F.L.; Meneses, J.J.; Lovato, T.L.; Vogel, K.G.; Moyzis, R.K.; Pedersen, R. Centromeric protein B null mice are viable with no apparent abnormalities. Dev. Biol. 1998, 201, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Foltz, D.R.; Jansen, L.E.; Black, B.E.; Bailey, A.O.; Yates, J.R.; Cleveland, D.W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006, 8, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Amano, M.; Suzuki, A.; Backer, C.B.; Welburn, J.P.; Dong, Y.; McEwen, B.F.; Shang, W.H.; Suzuki, E.; Okawa, K.; et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 2008, 135, 1039–1052. [Google Scholar] [CrossRef]
- Black, B.E.; Bassett, E.A. The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 2008, 20, 91–100. [Google Scholar] [CrossRef]
- Allu, P.K.; Dawicki-McKenna, J.M.; Van Eeuwen, T.; Slavin, M.; Braitbard, M.; Xu, C.; Kalisman, N.; Murakami, K.; Black, B.E. Structure of the Human Core Centromeric Nucleosome Complex. Curr. Biol. 2019, 29, 2625–2639. [Google Scholar] [CrossRef]
- Yan, K.; Yang, J.; Zhang, Z.; McLaughlin, S.H.; Chang, L.; Fasci, D.; Ehrenhofer-Murray, A.E.; Heck, A.J.R.; Barford, D. Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature 2019, 574, 278–282. [Google Scholar] [CrossRef]
- Hara, M.; Fukagawa, T. Critical Foundation of the Kinetochore: The Constitutive Centromere-Associated Network (CCAN). Prog. Mol. Subcell Biol. 2017, 56, 29–57. [Google Scholar] [CrossRef]
- Klare, K.; Weir, J.R.; Basilico, F.; Zimniak, T.; Massimiliano, L.; Ludwigs, N.; Herzog, F.; Musacchio, A. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 2015, 210, 11–22. [Google Scholar] [CrossRef]
- Shono, N.; Ohzeki, J.; Otake, K.; Martins, N.M.; Nagase, T.; Kimura, H.; Larionov, V.; Earnshaw, W.C.; Masumoto, H. CENP-C and CENP-I are key connecting factors for kinetochore and CENP-A assembly. J. Cell Sci. 2015, 128, 4572–4587. [Google Scholar] [CrossRef] [PubMed]
- Ali-Ahmad, A.; Bilokapic, S.; Schafer, I.B.; Halic, M.; Sekulic, N. CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail. EMBO Rep. 2019, 20, e48913. [Google Scholar] [CrossRef] [PubMed]
- Falk, S.J.; Guo, L.Y.; Sekulic, N.; Smoak, E.M.; Mani, T.; Logsdon, G.A.; Gupta, K.; Jansen, L.E.T.; Van Duyne, G.D.; Vinogradov, S.A.; et al. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 2015, 348, 699–703. [Google Scholar] [CrossRef]
- Watanabe, R.; Hara, M.; Okumura, E.-I.; Hervé, S.; Fachinetti, D.; Ariyoshi, M.; Fukagawa, T. CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization. J. Cell Biol. 2019, 218, 4042–4062. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.W.; Milks, K.J.; Straight, A.F. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 2010, 189, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Tachiwana, H.; Muller, S.; Blumer, J.; Klare, K.; Musacchio, A.; Almouzni, G. HJURP involvement in de novo CenH3 (CENP-A) and CENP-C recruitment. Cell Rep. 2015, 11, 22–32. [Google Scholar] [CrossRef] [PubMed]
- McKinley, K.L.; Sekulic, N.; Guo, L.Y.; Tsinman, T.; Black, B.E.; Cheeseman, I.M. The CENP-L-N Complex Forms a Critical Node in an Integrated Meshwork of Interactions at the Centromere-Kinetochore Interface. Mol. Cell 2015, 60, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.W.; Silva, M.C.; Godek, K.M.; Jansen, L.E.; Straight, A.F. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 2009, 11, 896–902. [Google Scholar] [CrossRef]
- Weir, J.R.; Faesen, A.C.; Klare, K.; Petrovic, A.; Basilico, F.; Fischbock, J.; Pentakota, S.; Keller, J.; Pesenti, M.E.; Pan, D.; et al. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature 2016, 537, 249–253. [Google Scholar] [CrossRef]
- Giunta, S.; Funabiki, H. Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc. Natl. Acad. Sci. USA 2017, 114, 1928–1933. [Google Scholar] [CrossRef]
- Basilico, F.; Maffini, S.; Weir, J.R.; Prumbaum, D.; Rojas, A.M.; Zimniak, T.; De Antoni, A.; Jeganathan, S.; Voss, B.; van Gerwen, S.; et al. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife 2014, 3, e02978. [Google Scholar] [CrossRef] [PubMed]
- Milks, K.J.; Moree, B.; Straight, A.F. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell 2009, 20, 4246–4255. [Google Scholar] [CrossRef]
- Kwon, M.S.; Hori, T.; Okada, M.; Fukagawa, T. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell 2007, 18, 2155–2168. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, H.; Hori, T.; Furukawa, A.; Sugase, K.; Osakabe, A.; Kurumizaka, H.; Fukagawa, T. Dynamic changes in CCAN organization through CENP-C during cell-cycle progression. Mol. Biol. Cell 2015, 26, 3768–3776. [Google Scholar] [CrossRef] [PubMed]
- Fukagawa, T.; Brown, W.R.A. Efficient Conditional Mutation of the Vertebrate CENP-C Gene. Hum. Mol. Genet. 1997, 6, 2301–2308. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Bodor, D.L.; David, A.F.; Abdul-Zani, I.; Mata, J.F.; Neumann, B.; Reither, S.; Tischer, C.; Jansen, L.E.T. Genetic screening identifies a SUMO protease dynamically maintaining centromeric chromatin. Nat. Commun. 2020, 11, 501. [Google Scholar] [CrossRef]
- Liebelt, F.; Jansen, N.S.; Kumar, S.; Gracheva, E.; Claessens, L.A.; Verlaan-de Vries, M.; Willemstein, E.; Vertegaal, A.C.O. The poly-SUMO2/3 protease SENP6 enables assembly of the constitutive centromere-associated network by group deSUMOylation. Nat. Commun. 2019, 10, 3987. [Google Scholar] [CrossRef]
- Nechemia-Arbely, Y.; Fachinetti, D.; Cleveland, D.W. Replicating centromeric chromatin: Spatial and temporal control of CENP-A assembly. Exp. Cell Res. 2012, 318, 1353–1360. [Google Scholar] [CrossRef]
- Ten Hagen, K.G.; Gilbert, D.M.; Willard, H.F.; Cohen, S.N. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol. Cell Biol. 1990, 10, 6348–6355. [Google Scholar] [CrossRef]
- Jansen, L.E.; Black, B.E.; Foltz, D.R.; Cleveland, D.W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007, 176, 795–805. [Google Scholar] [CrossRef]
- Schuh, M.; Lehner, C.F.; Heidmann, S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007, 17, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Foltz, D.R.; Jansen, L.E.; Bailey, A.O.; Yates, J.R.; Bassett, E.A.; Wood, S.; Black, B.E.; Cleveland, D.W. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 2009, 137, 472–484. [Google Scholar] [CrossRef]
- Dunleavy, E.M.; Roche, D.; Tagami, H.; Lacoste, N.; Ray-Gallet, D.; Nakamura, Y.; Daigo, Y.; Nakatani, Y.; Almouzni-Pettinotti, G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 2009, 137, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Bodor, D.L.; Stellfox, M.E.; Martins, N.M.; Hochegger, H.; Foltz, D.R.; Jansen, L.E. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell 2012, 22, 52–63. [Google Scholar] [CrossRef]
- Stankovic, A.; Guo, L.Y.; Mata, J.F.; Bodor, D.L.; Cao, X.J.; Bailey, A.O.; Shabanowitz, J.; Hunt, D.F.; Garcia, B.A.; Black, B.E.; et al. A Dual Inhibitory Mechanism Sufficient to Maintain Cell-Cycle-Restricted CENP-A Assembly. Mol. Cell 2017, 65, 231–246. [Google Scholar] [CrossRef]
- Fujita, Y.; Hayashi, T.; Kiyomitsu, T.; Toyoda, Y.; Kokubu, A.; Obuse, C.; Yanagida, M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell 2007, 12, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Jansen, L.E. At the right place at the right time: Novel CENP-A binding proteins shed light on centromere assembly. Chromosoma 2009, 118, 567–574. [Google Scholar] [CrossRef]
- Hayashi, T.; Fujita, Y.; Iwasaki, O.; Adachi, Y.; Takahashi, K.; Yanagida, M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 2004, 118, 715–729. [Google Scholar] [CrossRef]
- Maddox, P.S.; Hyndman, F.; Monen, J.; Oegema, K.; Desai, A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 2007, 176, 757–763. [Google Scholar] [CrossRef]
- McKinley, K.L.; Cheeseman, I.M. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell 2014, 158, 397–411. [Google Scholar] [CrossRef]
- Moree, B.; Meyer, C.B.; Fuller, C.J.; Straight, A.F. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 2011, 194, 855–871. [Google Scholar] [CrossRef] [PubMed]
- Stellfox, M.E.; Nardi, I.K.; Knippler, C.M.; Foltz, D.R. Differential Binding Partners of the Mis18α/β YIPPEE Domains Regulate Mis18 Complex Recruitment to Centromeres. Cell Rep. 2016, 15, 2127–2135. [Google Scholar] [CrossRef] [PubMed]
- Nardi, I.K.; Zasadzinska, E.; Stellfox, M.E.; Knippler, C.M.; Foltz, D.R. Licensing of Centromeric Chromatin Assembly through the Mis18α-Mis18β Heterotetramer. Mol. Cell 2016, 61, 774–787. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Klare, K.; Petrovic, A.; Take, A.; Walstein, K.; Singh, P.; Rondelet, A.; Bird, A.W.; Musacchio, A. CDK-regulated dimerization of M18BP1 on a Mis18 hexamer is necessary for CENP-A loading. eLife 2017, 6, e23352. [Google Scholar] [CrossRef] [PubMed]
- Spiller, F.; Medina-Pritchard, B.; Abad, M.A.; Wear, M.A.; Molina, O.; Earnshaw, W.C.; Jeyaprakash, A.A. Molecular basis for Cdk1-regulated timing of Mis18 complex assembly and CENP-A deposition. EMBO Rep. 2017, 18, 894–905. [Google Scholar] [CrossRef]
- Pan, D.; Walstein, K.; Take, A.; Bier, D.; Kaiser, N.; Musacchio, A. Mechanism of centromere recruitment of the CENP-A chaperone HJURP and its implications for centromere licensing. Nat. Commun. 2019, 10, 4046. [Google Scholar] [CrossRef]
- Zasadzinska, E.; Barnhart-Dailey, M.C.; Kuich, P.H.; Foltz, D.R. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 2013, 32, 2113–2124. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Dou, Z.; Chen, L.; Jiang, H.; Fu, C.; Fu, G.; Liu, D.; Zhang, J.; Zhu, T.; et al. Mitotic regulator Mis18beta interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP). J. Biol. Chem. 2014, 289, 8326–8336. [Google Scholar] [CrossRef]
- Pidoux, A.L.; Choi, E.S.; Abbott, J.K.; Liu, X.; Kagansky, A.; Castillo, A.G.; Hamilton, G.L.; Richardson, W.; Rappsilber, J.; He, X.; et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 2009, 33, 299–311. [Google Scholar] [CrossRef]
- Bassett, E.A.; DeNizio, J.; Barnhart-Dailey, M.C.; Panchenko, T.; Sekulic, N.; Rogers, D.J.; Foltz, D.R.; Black, B.E. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev. Cell 2012, 22, 749–762. [Google Scholar] [CrossRef]
- Perpelescu, M.; Nozaki, N.; Obuse, C.; Yang, H.; Yoda, K. Active establishment of centromeric CENP-A chromatin by RSF complex. J. Cell Biol. 2009, 185, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Lagana, A.; Dorn, J.F.; De Rop, V.; Ladouceur, A.M.; Maddox, A.S.; Maddox, P.S. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat. Cell Biol. 2010, 12, 1186–1193. [Google Scholar] [CrossRef] [PubMed]
- Stellfox, M.E.; Bailey, A.O.; Foltz, D.R. Putting CENP-A in its place. Cell Mol. Life Sci. 2013, 70, 387–406. [Google Scholar] [CrossRef] [PubMed]
- Shelby, R.D.; Monier, K.; Sullivan, K.F. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 2000, 151, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Shelby, R.D.; Vafa, O.; Sullivan, K.F. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 1997, 136, 501–513. [Google Scholar] [CrossRef]
- Miga, K.H.; Newton, Y.; Jain, M.; Altemose, N.; Willard, H.F.; Kent, W.J. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 2014, 24, 697–707. [Google Scholar] [CrossRef]
- Schneider, V.A.; Graves-Lindsay, T.; Howe, K.; Bouk, N.; Chen, H.C.; Kitts, P.A.; Murphy, T.D.; Pruitt, K.D.; Thibaud-Nissen, F.; Albracht, D.; et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017, 27, 849–864. [Google Scholar] [CrossRef]
- Levy, S.; Sutton, G.; Ng, P.C.; Feuk, L.; Halpern, A.L.; Walenz, B.P.; Axelrod, N.; Huang, J.; Kirkness, E.F.; Denisov, G.; et al. The diploid genome sequence of an individual human. PLoS Biol. 2007, 5, e254. [Google Scholar] [CrossRef]
- Huang, H.; Stromme, C.B.; Saredi, G.; Hodl, M.; Strandsby, A.; Gonzalez-Aguilera, C.; Chen, S.; Groth, A.; Patel, D.J. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat. Struct. Mol. Biol. 2015, 22, 618–626. [Google Scholar] [CrossRef]
- Smith, S.; Stillman, B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991, 10, 971–980. [Google Scholar] [CrossRef]
- Richet, N.; Liu, D.; Legrand, P.; Velours, C.; Corpet, A.; Gaubert, A.; Bakail, M.; Moal-Raisin, G.; Guerois, R.; Compper, C.; et al. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 2015, 43, 1905–1917. [Google Scholar] [CrossRef] [PubMed]
- Zasadzinska, E.; Huang, J.; Bailey, A.O.; Guo, L.Y.; Lee, N.S.; Srivastava, S.; Wong, K.A.; French, B.T.; Black, B.E.; Foltz, D.R. Inheritance of CENP-A Nucleosomes during DNA Replication Requires HJURP. Dev. Cell 2018, 47, 348–362. [Google Scholar] [CrossRef]
- Dunleavy, E.M.; Almouzni, G.; Karpen, G.H. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G (1) phase. Nucleus 2011, 2, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.; Tong, P.; White, S.A.; Singh, P.P.; Reid, A.M.; Catania, S.; Pidoux, A.L.; Allshire, R.C. Centromere DNA Destabilizes H3 Nucleosomes to Promote CENP-A Deposition during the Cell Cycle. Curr. Biol. 2018, 28, 3924–3936. [Google Scholar] [CrossRef]
- Athwal, R.K.; Walkiewicz, M.P.; Baek, S.; Fu, S.; Bui, M.; Camps, J.; Ried, T.; Sung, M.H.; Dalal, Y. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenet. Chromatin 2015, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Van Hooser, A.A.; Ouspenski, I.I.; Gregson, H.C.; Starr, D.A.; Yen, T.J.; Goldberg, M.L.; Yokomori, K.; Earnshaw, W.C.; Sullivan, K.F.; Brinkley, B.R. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001, 114, 3529–3542. [Google Scholar]
- Shrestha, R.L.; Ahn, G.S.; Staples, M.I.; Sathyan, K.M.; Karpova, T.S.; Foltz, D.R.; Basrai, M.A. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget 2017, 8, 46781–46800. [Google Scholar] [CrossRef]
- Filipescu, D.; Naughtin, M.; Podsypanina, K.; Lejour, V.; Wilson, L.; Gurard-Levin, Z.A.; Orsi, G.A.; Simeonova, I.; Toufektchan, E.; Attardi, L.D.; et al. Essential role for centromeric factors following p53 loss and oncogenic transformation. Genes Dev. 2017, 31, 463–480. [Google Scholar] [CrossRef]
- Au, W.-C.; Zhang, T.; Mishra, P.K.; Eisenstatt, J.R.; Walker, R.L.; Ocampo, J.; Dawson, A.; Warren, J.; Costanzo, M.; Baryshnikova, A.; et al. Skp, Cullin, F-box (SCF)-Met30 and SCF-Cdc4-Mediated Proteolysis of CENP-A Prevents Mislocalization of CENP-A for Chromosomal Stability in Budding Yeast. PLOS Genet. 2020, 16, e1008597. [Google Scholar] [CrossRef]
- Hewawasam, G.; Shivaraju, M.; Mattingly, M.; Venkatesh, S.; Martin-Brown, S.; Florens, L.; Workman, J.L.; Gerton, J.L. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 2010, 40, 444–454. [Google Scholar] [CrossRef]
- Ranjitkar, P.; Press, M.O.; Yi, X.; Baker, R.; MacCoss, M.J.; Biggins, S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 2010, 40, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Hewawasam, G.S.; Mattingly, M.; Venkatesh, S.; Zhang, Y.; Florens, L.; Workman, J.L.; Gerton, J.L. Phosphorylation by casein kinase 2 facilitates Psh1 protein-assisted degradation of Cse4 protein. J. Biol. Chem. 2014, 289, 29297–29309. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Bao, X.; Rao, H. The F-box Protein Rcy1 Is Involved in the Degradation of Histone H3 Variant Cse4 and Genome Maintenance. J. Biol. Chem. 2016, 291, 10372–10377. [Google Scholar] [CrossRef] [PubMed]
- Ohkuni, K.; Takahashi, Y.; Fulp, A.; Lawrimore, J.; Au, W.C.; Pasupala, N.; Levy-Myers, R.; Warren, J.; Strunnikov, A.; Baker, R.E.; et al. SUMO-Targeted Ubiquitin Ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin. Mol. Biol. Cell 2016. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, T.; Ishii, K.; Takeda, K.; Matsumoto, T. The 19S proteasome subunit Rpt3 regulates distribution of CENP-A by associating with centromeric chromatin. Nat. Commun. 2014, 5, 3597. [Google Scholar] [CrossRef]
- Moreno-Moreno, O.; Torras-Llort, M.; Azorin, F. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 2006, 34, 6247–6255. [Google Scholar] [CrossRef]
- Moreno-Moreno, O.; Medina-Giro, S.; Torras-Llort, M.; Azorin, F. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3(CID). Curr. Biol. 2011, 21, 1488–1493. [Google Scholar] [CrossRef]
- Sun, X.; Clermont, P.L.; Jiao, W.; Helgason, C.D.; Gout, P.W.; Wang, Y.; Qu, S. Elevated expression of the centromere protein-A (CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int. J. Cancer 2016, 139, 899–907. [Google Scholar] [CrossRef]
- Gu, X.M.; Fu, J.; Feng, X.J.; Huang, X.; Wang, S.M.; Chen, X.F.; Zhu, M.H.; Zhang, S.H. Expression and prognostic relevance of centromere protein A in primary osteosarcoma. Pathol. Res. Pract. 2014, 210, 228–233. [Google Scholar] [CrossRef]
- Mahler, M.; You, D.; Baron, M.; Taillefer, S.S.; Hudson, M.; Canadian Scleroderma Research, Group; Fritzler, M.J. Anti-centromere antibodies in a large cohort of systemic sclerosis patients: Comparison between immunofluorescence, CENP-A and CENP-B ELISA. Clin Chim Acta 2011, 412, 1937–1943. [Google Scholar] [CrossRef]
- Qiu, J.J.; Guo, J.J.; Lv, T.J.; Jin, H.Y.; Ding, J.X.; Feng, W.W.; Zhang, Y.; Hua, K.Q. Prognostic value of centromere protein-A expression in patients with epithelial ovarian cancer. Tumour Biol. 2013, 34, 2971–2975. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Qian, Y.M.; Zhao, X.L.; Wang, S.M.; Feng, X.J.; Chen, X.F.; Zhang, S.H. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer 2012, 77, 407–414. [Google Scholar] [CrossRef]
- McGovern, S.L.; Qi, Y.; Pusztai, L.; Symmans, W.F.; Buchholz, T.A. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012, 14, R72. [Google Scholar] [CrossRef] [PubMed]
- Ly, P.; Teitz, L.S.; Kim, D.H.; Shoshani, O.; Skaletsky, H.; Fachinetti, D.; Page, D.C.; Cleveland, D.W. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 2017, 19, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Umbreit, N.T.; Zhang, C.Z.; Lynch, L.D.; Blaine, L.J.; Cheng, A.M.; Tourdot, R.; Sun, L.; Almubarak, H.F.; Judge, K.; Mitchell, T.J.; et al. Mechanisms generating cancer genome complexity from a single cell division error. Science 2020, 368. [Google Scholar] [CrossRef] [PubMed]
- Ly, P.; Brunner, S.F.; Shoshani, O.; Kim, D.H.; Lan, W.; Pyntikova, T.; Flanagan, A.M.; Behjati, S.; Page, D.C.; Campbell, P.J.; et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet. 2019, 51, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Ly, P.; Cleveland, D.W. Rebuilding Chromosomes After Catastrophe: Emerging Mechanisms of Chromothripsis. Trends Cell Biol. 2017, 27, 917–930. [Google Scholar] [CrossRef]
- Zhang, W.; Mao, J.H.; Zhu, W.; Jain, A.K.; Liu, K.; Brown, J.B.; Karpen, G.H. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat. Commun. 2016, 7, 12619. [Google Scholar] [CrossRef]
- Cimini, D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta 2008, 1786, 32–40. [Google Scholar] [CrossRef]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018, 33, 676–689. [Google Scholar] [CrossRef]
- Takada, M.; Zhang, W.; Suzuki, A.; Kuroda, T.S.; Yu, Z.; Inuzuka, H.; Gao, D.; Wan, L.; Zhuang, M.; Hu, L.; et al. FBW7 Loss Promotes Chromosomal Instability and Tumorigenesis via Cyclin E1/CDK2-Mediated Phosphorylation of CENP-A. Cancer Res. 2017, 77, 4881–4893. [Google Scholar] [CrossRef] [PubMed]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed]
- Laughney, A.M.; Elizalde, S.; Genovese, G.; Bakhoum, S.F. Dynamics of Tumor Heterogeneity Derived from Clonal Karyotypic Evolution. Cell Rep. 2015, 12, 809–820. [Google Scholar] [CrossRef]
- Shih, I.M.; Zhou, W.; Goodman, S.N.; Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 2001, 61, 818–822. [Google Scholar]
- Turajlic, S.; Swanton, C. Implications of cancer evolution for drug development. Nat. Rev. Drug Discov. 2017, 16, 441–442. [Google Scholar] [CrossRef] [PubMed]
- Cimini, D.; Howell, B.; Maddox, P.; Khodjakov, A.; Degrassi, F.; Salmon, E.D. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 2001, 153, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef]
- Levine, M.S.; Holland, A.J. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 2018, 32, 620–638. [Google Scholar] [CrossRef]
- Elsasser, S.J.; D’Arcy, S. Towards a mechanism for histone chaperones. Biochim. Biophys. Acta 2013, 1819, 211–221. [Google Scholar] [CrossRef]
- Demirdizen, E.; Spiller-Becker, M.; Förtsch, A.; Wilhelm, A.; Corless, S.; Bade, D.; Bergner, A.; Hessling, B.; Erhardt, S. Localization of Drosophila CENP-A to non-centromeric sites depends on the NuRD complex. Nucleic Acids Res. 2019, 47, 11589–11608. [Google Scholar] [CrossRef]
- Nye, J.; Sturgill, D.; Athwal, R.; Dalal, Y. HJURP antagonizes CENP-A mislocalization driven by the H3.3 chaperones HIRA and DAXX. PLoS ONE 2018, 13, e0205948. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Huang, G.; Sadanandam, A.; Gu, S.; Lenburg, M.E.; Pai, M.; Bayani, N.; Blakely, E.A.; Gray, J.W.; Mao, J.H. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010, 12, R18. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Kemp, C.J.; Moore, J.M.; Moser, R.; Bernard, B.; Teater, M.; Smith, L.E.; Rabaia, N.A.; Gurley, K.E.; Guinney, J.; Busch, S.E.; et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014, 7, 1020–1029. [Google Scholar] [CrossRef]
- Katainen, R.; Dave, K.; Pitkanen, E.; Palin, K.; Kivioja, T.; Valimaki, N.; Gylfe, A.E.; Ristolainen, H.; Hanninen, U.A.; Cajuso, T.; et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet. 2015, 47, 818–821. [Google Scholar] [CrossRef]
- Aitken, S.J.; Ibarra-Soria, X.; Kentepozidou, E.; Flicek, P.; Feig, C.; Marioni, J.C.; Odom, D.T. CTCF maintains regulatory homeostasis of cancer pathways. Genome Biol. 2018, 19, 106. [Google Scholar] [CrossRef]
- Docquier, F.; Farrar, D.; D’Arcy, V.; Chernukhin, I.; Robinson, A.F.; Loukinov, D.; Vatolin, S.; Pack, S.; Mackay, A.; Harris, R.A.; et al. Heightened expression of CTCF in breast cancer cells is associated with resistance to apoptosis. Cancer Res. 2005, 65, 5112–5122. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Z.; Zhang, S.; Yu, D.; Yu, H.; Liu, L.; Cao, X.; Wang, L.; Gao, H.; Zhu, M. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS ONE 2011, 6, e17794. [Google Scholar] [CrossRef]
- Sathyan, K.M.; Fachinetti, D.; Foltz, D.R. α-amino trimethylation of CENP-A by NRMT is required for full recruitment of the centromere. Nat. Commun. 2017, 8, 14678. [Google Scholar] [CrossRef]
- Zeitlin, S.G.; Shelby, R.D.; Sullivan, K.F. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 2001, 155, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Barra, V.; Logsdon, G.A.; Scelfo, A.; Hoffmann, S.; Herve, S.; Aslanian, A.; Nechemia-Arbely, Y.; Cleveland, D.W.; Black, B.E.; Fachinetti, D. Phosphorylation of CENP-A on serine 7 does not control centromere function. Nat. Commun. 2019, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.O.; Panchenko, T.; Sathyan, K.M.; Petkowski, J.J.; Pai, P.J.; Bai, D.L.; Russell, D.H.; Macara, I.G.; Shabanowitz, J.; Hunt, D.F.; et al. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc. Natl. Acad Sci. USA 2013, 110, 11827–11832. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhou, X.; Wang, W.; Deng, W.; Fang, J.; Hu, H.; Wang, Z.; Li, S.; Cui, L.; Shen, J.; et al. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev. Cell 2015, 32, 68–81. [Google Scholar] [CrossRef]
- Fachinetti, D.; Logsdon, G.A.; Abdullah, A.; Selzer, E.B.; Cleveland, D.W.; Black, B.E. CENP-A Modifications on Ser68 and Lys124 Are Dispensable for Establishment, Maintenance, and Long-Term Function of Human Centromeres. Dev. Cell 2017, 40, 104–113. [Google Scholar] [CrossRef]
- Niikura, Y.; Kitagawa, R.; Ogi, H.; Abdulle, R.; Pagala, V.; Kitagawa, K. CENP-A K124 Ubiquitylation Is Required for CENP-A Deposition at the Centromere. Dev. Cell 2015, 32, 589–603. [Google Scholar] [CrossRef]
- Niikura, Y.; Kitagawa, R.; Kitagawa, K. CENP-A Ubiquitylation Contributes to Maintaining the Chromosomal Location of the Centromere. Molecules 2019, 24, 402. [Google Scholar] [CrossRef]
- Niikura, Y.; Kitagawa, R.; Fang, L.; Kitagawa, K. CENP-A Ubiquitylation Is Indispensable to Cell Viability. Dev. Cell 2019, 50, 683–689. [Google Scholar] [CrossRef]
- Bui, M.; Pitman, M.; Nuccio, A.; Roque, S.; Donlin-Asp, P.G.; Nita-Lazar, A.; Papoian, G.A.; Dalal, Y. Internal modifications in the CENP-A nucleosome modulate centromeric dynamics. Epigenet. Chromatin 2017, 10, 17. [Google Scholar] [CrossRef]
- Srivastava, S.; Foltz, D.R. Posttranslational modifications of CENP-A: Marks of distinction. Chromosoma 2018, 127, 279–290. [Google Scholar] [CrossRef]
- Davis, R.J.; Welcker, M.; Clurman, B.E. Tumor suppression by the Fbw7 ubiquitin ligase: Mechanisms and opportunities. Cancer Cell 2014, 26, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.-C.; Chen, Y.-J.; Hughes, D.; Petrovic, V.; Major, M.L.; Park, H.J.; Tan, Y.; Ackerson, T.; Costa, R.H. Forkhead Box M1 Regulates the Transcriptional Network of Genes Essential for Mitotic Progression and Genes Encoding the SCF (Skp2-Cks1) Ubiquitin Ligase. Mol. Cell. Biol. 2005, 25, 10875–10894. [Google Scholar] [CrossRef] [PubMed]
- Wonsey, D.R.; Follettie, M.T. Loss of the Forkhead Transcription Factor FoxM1 Causes Centrosome Amplification and Mitotic Catastrophe. Cancer Res. 2005, 65, 5181–5189. [Google Scholar] [CrossRef] [PubMed]
- Bade, D.; Pauleau, A.L.; Wendler, A.; Erhardt, S. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev. Cell 2014, 28, 508–519. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, Q.; Chen, Y.F.; Fu, J.; You, Q.H.; Wang, S.M.; Huang, X.; Feng, X.J.; Zhang, S.H. Short hairpin RNA-mediated down-regulation of CENP-A attenuates the aggressive phenotype of lung adenocarcinoma cells. Cell Oncol. 2014, 37, 399–407. [Google Scholar] [CrossRef]
- Cao, R.; Wang, G.; Qian, K.; Chen, L.; Qian, G.; Xie, C.; Dan, H.C.; Jiang, W.; Wu, M.; Wu, C.L.; et al. Silencing of HJURP induces dysregulation of cell cycle and ROS metabolism in bladder cancer cells via PPARgamma-SIRT1 feedback loop. J. Cancer 2017, 8, 2282–2295. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahlke, M.A.; Nechemia-Arbely, Y. Guarding the Genome: CENP-A-Chromatin in Health and Cancer. Genes 2020, 11, 810. https://doi.org/10.3390/genes11070810
Mahlke MA, Nechemia-Arbely Y. Guarding the Genome: CENP-A-Chromatin in Health and Cancer. Genes. 2020; 11(7):810. https://doi.org/10.3390/genes11070810
Chicago/Turabian StyleMahlke, Megan A., and Yael Nechemia-Arbely. 2020. "Guarding the Genome: CENP-A-Chromatin in Health and Cancer" Genes 11, no. 7: 810. https://doi.org/10.3390/genes11070810
APA StyleMahlke, M. A., & Nechemia-Arbely, Y. (2020). Guarding the Genome: CENP-A-Chromatin in Health and Cancer. Genes, 11(7), 810. https://doi.org/10.3390/genes11070810




