Identifying Candidate Genes for Hypoxia Adaptation of Tibet Chicken Embryos by Selection Signature Analyses and RNA Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. DNA Samples and Single Nucleotide Polymorphism (SNP) Genotyping
2.3. RNA Samples and RNA Sequencing Data Collection
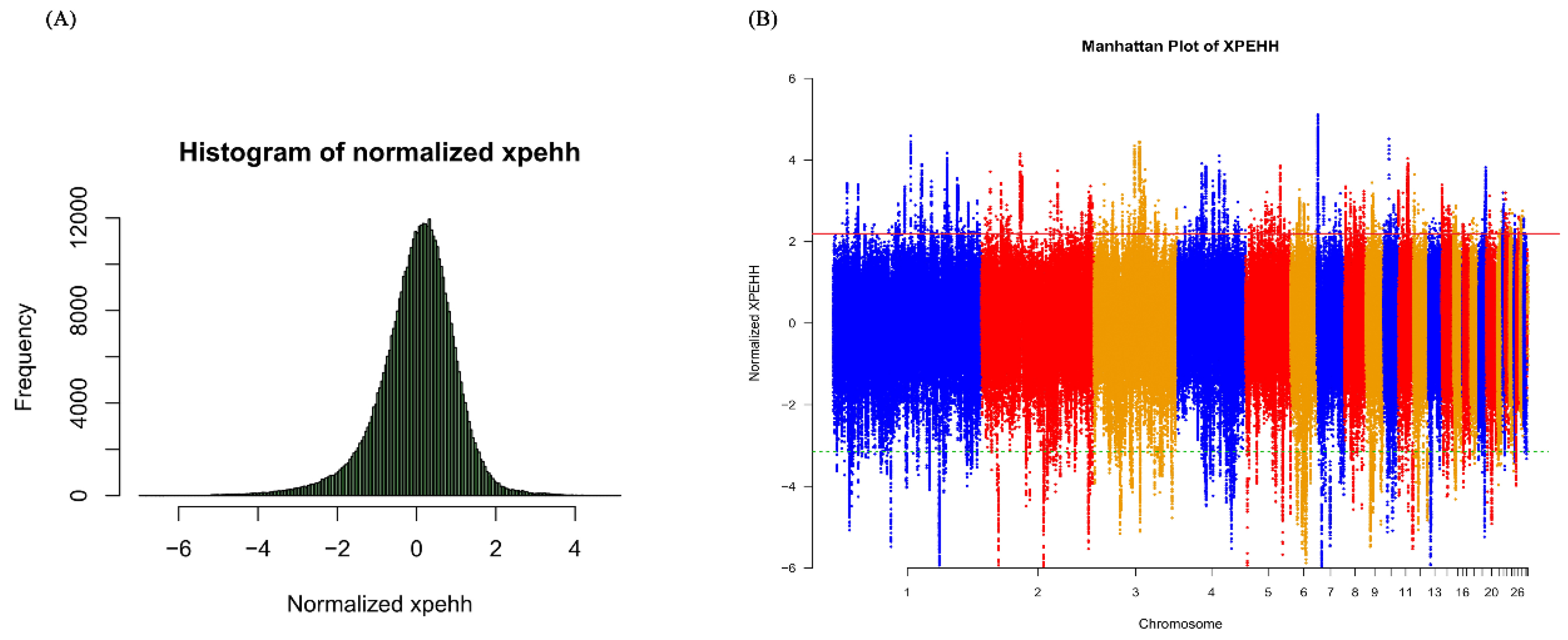
2.4. Identifying Signatures of Positive Selection by XPEHH Test and Fst Statistics
2.5. Screening for Differentially Expressed Transcripts and Genes
2.6. Identifying Candidate Genes for Hypoxia Adaptation of the Tibet Chicken Embryos
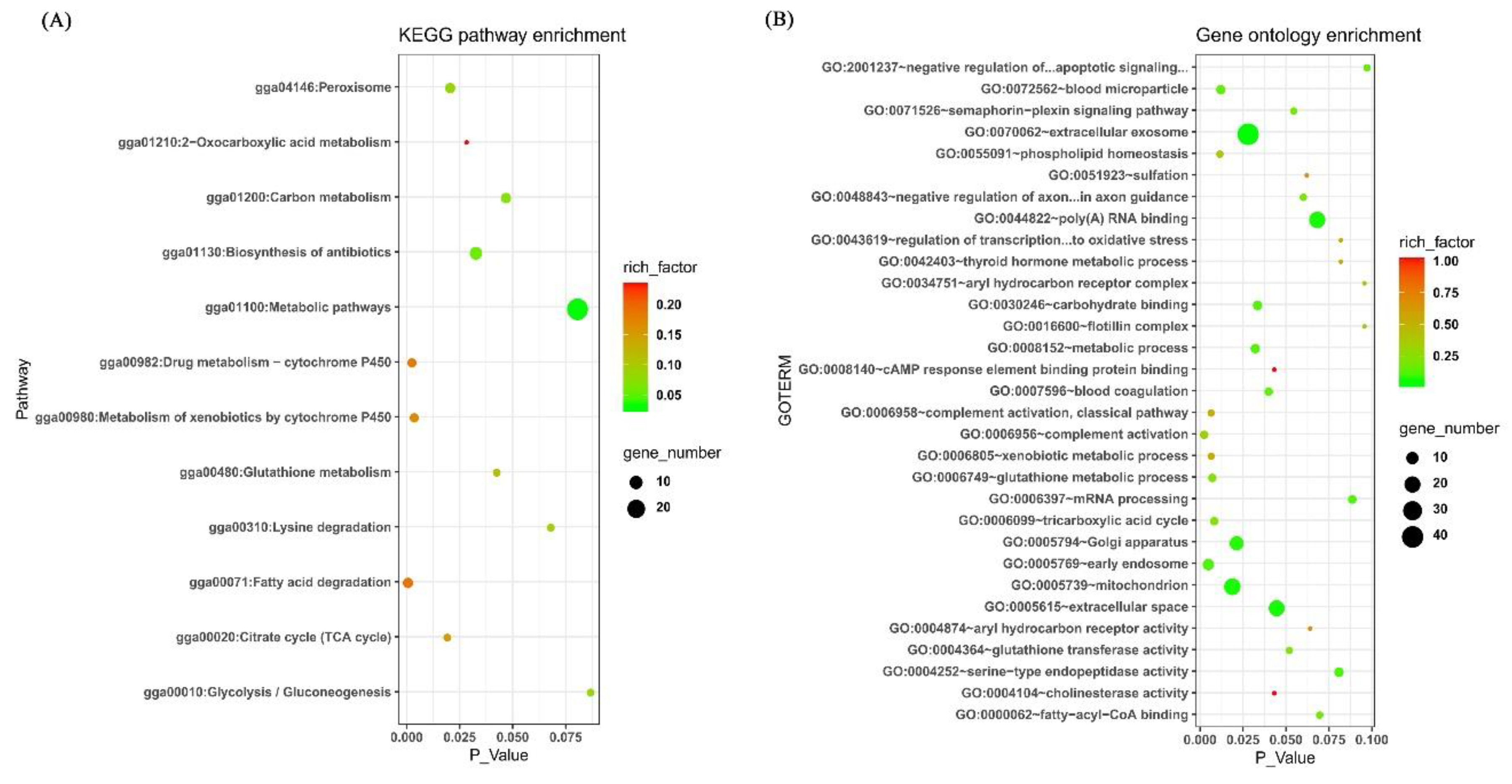
2.7. Functional Annotation of Candidate Genes for Hypoxia Adaptation of the Tibet Chicken Embryos
3. Results and Discussions
3.1. Analyses for Signatures of Selection
3.2. Analysis for RNA Sequencing Data
3.3. Overlapping Genes between the Results of Identification of Signatures of Selection and RNA Sequencing Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, H.; Wang, X.T.; Chamba, Y.; Ling, Y.; Wu, C.X. Influences of hypoxia on hatching performance in chickens with different genetic adaptation to high altitude. Poult. Sci. 2008, 87, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Jaffee, O.C. The effects of moderate hypoxia and moderate hypoxia plus hypercapnea on cardiac development in chick embryos. Teratology 1974, 10, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Visschedijk, A.H. Gas exchange and hatchability of chicken eggs incubated at simulated high altitude. J. Appl. Physiol. 1985, 58, 416–418. [Google Scholar] [CrossRef] [PubMed]
- Meuer, H.J.; Hartmann, V.; Jopp, S. Tissue PO2 and growth rate in early chick embryos. Respir. Physiol. 1992, 90, 227–237. [Google Scholar] [CrossRef]
- Şahan, Ü.; İpek, A.; Yilmaz-Dıkmen, B.; Aydin, C.; Kederlı, E. Effect of oxygen supplementation in the hatcher at high altitude on the incubation results of broiler eggs laid at low altitude. Br. Poult. Sci. 2011, 52, 388–394. [Google Scholar] [CrossRef]
- Mortola, J.P. Gas exchange in avian embryos and hatchlings. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2009, 153, 359–377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, H. Review of hypoxia adaptation in Tibetan chicken. China Poult. 2018, 40, 47–52. [Google Scholar]
- Liu, C.; Zhang, L.F.; Li, N. The specific expression pattern of globin mRNAs in Tibetan chicken during late embryonic stage under hypoxia. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2013, 164, 638–644. [Google Scholar] [CrossRef]
- Zhang, Q.; Gou, W.; Wang, X.; Zhang, Y.; Ma, J.; Zhang, H.; Zhang, Y.; Zhang, H. Genome Resequencing Identifies Unique Adaptations of Tibetan Chickens to Hypoxia and High-Dose Ultraviolet Radiation in High-Altitude Environments. Genome Biol. Evol. 2016, 8, 765–776. [Google Scholar] [CrossRef]
- Nielsen, R. Molecular signatures of natural selection. Annu. Rev. Genet. 2005, 39, 197–218. [Google Scholar] [CrossRef] [Green Version]
- Perez, O.A.; Utsunomiya, Y.T.; Meszaros, G.; Bickhart, D.M.; Liu, G.E.; Van Tassell, C.P.; Sonstegard, T.S.; Da, S.M.; Garcia, J.F.; Solkner, J. Assessing signatures of selection through variation in linkage disequilibrium between taurine and indicine cattle. Genet. Sel. Evol. 2014, 46, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Simoni, G.J.; Da, S.M.; Paiva, S.R.; de Oliveira, S.M. Identification of selection signatures in livestock species. Genet. Mol. Biol. 2014, 37, 330–342. [Google Scholar] [CrossRef] [Green Version]
- Grossman, S.R.; Shlyakhter, I.; Karlsson, E.K.; Byrne, E.H.; Morales, S.; Frieden, G.; Hostetter, E.; Angelino, E.; Garber, M.; Zuk, O.; et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 2010, 327, 883–886. [Google Scholar] [CrossRef] [Green Version]
- Maynard, J.; Haigh, J. The hitch-hiking effect of a favourable gene. Genet. Res. 2007, 89, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B. A hitch-hiking guide to the genome: A commentary on ’The hitch-hiking effect of a favourable gene’ by John Maynard Smith and John Haigh. Genet. Res. 2007, 89, 389–390. [Google Scholar] [CrossRef] [Green Version]
- Weigand, H.; Leese, F. Detecting signatures of positive selection in non-model species using genomic data. Zool. J. Linn. Soc. 2018, 184, 528–583. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Joseph, K.; Pickrell, J.K.; Coop, G. The genetics of human adaptation: Hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 2010, 20, R208–R215. [Google Scholar] [CrossRef] [Green Version]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar]
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R.; et al. Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef]
- Rubin, C.J.; Zody, M.C.; Eriksson, J.; Meadows, J.R.; Sherwood, E.; Webster, M.T.; Jiang, L.; Ingman, M.; Sharpe, T.; Ka, S.; et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 2010, 464, 587–591. [Google Scholar] [CrossRef] [Green Version]
- Johansson, H.; Seppa, P.; Helantera, H.; Trontti, K.; Sundstrom, L. ESTIMATING F-STATISTICS FOR THE ANALYSIS OF POPULATION STRUCTURE. PeerJ 2018, 6, e5024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, C.J.; Megens, H.J.; Martinez, B.A.; Maqbool, K.; Sayyab, S.; Schwochow, D.; Wang, C.; Carlborg, O.; Jern, P.; Jorgensen, C.B.; et al. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 2012, 109, 19529–19536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Tian, S.; Jin, L.; Zhou, G.; Li, Y.; Zhang, Y.; Wang, T.; Yeung, C.K.; Chen, L.; Ma, J.; et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 2013, 45, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Fang, Q.; Ma, C.; Zhou, B.; Wan, Y.; Jiang, R. Whole-genome resequencing of Xishuangbanna fighting chicken to identify signatures of selection. Genet. Sel. Evol. 2016, 48, 62. [Google Scholar] [CrossRef] [Green Version]
- Taye, M.; Lee, W.; Jeon, S.; Yoon, J.; Dessie, T.; Hanotte, O.; Mwai, O.A.; Kemp, S.; Cho, S.; Oh, S.J.; et al. Exploring evidence of positive selection signatures in cattle breeds selected for different traits. Mamm. Genome 2017, 28, 528–541. [Google Scholar] [CrossRef]
- Simonson, T.S.; Yang, Y.; Huff, C.D.; Yun, H.; Qin, G.; Witherspoon, D.J.; Bai, Z.; Lorenzo, F.R.; Xing, J.; Jorde, L.B.; et al. Genetic Evidence for High-Altitude Adaptation in Tibet. Science 2010, 329, 72–75. [Google Scholar] [CrossRef] [Green Version]
- Gou, X.; Wang, Z.; Li, N.; Qiu, F.; Xu, Z.; Yan, D.; Yang, S.; Jia, J.; Kong, X.; Wei, Z.; et al. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 2014, 24, 1308–1315. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.; Wang, H.; Liu, G.; Zhao, F.; Kijas, J.W.; Ma, Y.; Lu, J.; Zhang, L.; Cao, J.; Wu, M.; et al. Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep. Sci. Rep. 2016, 6, 26770. [Google Scholar] [CrossRef]
- Wu, H.; Liu, Y.H.; Wang, G.D.; Yang, C.T.; Otecko, N.O.; Liu, F.; Wu, S.F.; Wang, L.; Yu, L.; Zhang, Y.P. Identifying molecular signatures of hypoxia adaptation from sex chromosomes: A case for Tibetan Mastiff based on analyses of X chromosome. Sci. Rep. 2016, 6, 35004. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Li, Y.; Peng, M.; Zhong, L.; Wang, Z.; Li, Q.; Tu, X.; Dong, Y.; Zhu, C.; Wang, L.; et al. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol. Biol. Evol. 2015, 32, 1880–1889. [Google Scholar] [CrossRef]
- Kharrati-Koopaee, H.; Ebrahimie, E.; Dadpasand, M.; Niazi, A.; Esmailizadeh, A. Genomic analysis reveals variant association with high altitude adaptation in native chickens. Sci. Rep. UK 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Kranis, A.; Gheyas, A.A.; Boschiero, C.; Turner, F.; Yu, L.; Smith, S.; Talbot, R.; Pirani, A.; Brew, F.; Kaiser, P.; et al. Development of a high density 600K SNP genotyping array for chicken. BMC Genom. 2013, 14, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Browning, B.L.; Browning, S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009, 84, 210–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szpiech, Z.A.; Hernandez, R.D. selscan: An efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 2014, 31, 2824–2827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, W.; Lee, W.R.; Abasht, B. Detection of genomic signatures of recent selection in commercial broiler chickens. BMC Genet. 2016, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.V.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Flexible isoform level differential expression analysis with Ballgown. In BioRxiv; The Cold Spring Harbor Laboratory: New York, NY, USA, 2014; p. 3665. [Google Scholar]
- Holsinger, K.E.; Weir, B.S. Genetics in geographically structured populations: Defining, estimating and interpreting F(ST). Nat. Rev. Genet. 2009, 10, 639–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Ding, X.; Qanbari, S.; Weigend, S.; Zhang, Q.; Simianer, H. Properties of different selection signature statistics and a new strategy for combining them. Heredity 2015, 115, 426–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biron-Shental, T.; Schaiff, W.T.; Ratajczak, C.K.; Bildirici, I.; Nelson, D.M.; Sadovsky, Y. Hypoxia regulates the expression of fatty acid-binding proteins in primary term human trophoblasts. Am. J. Obstet. Gynecol. 2007, 197, 511–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodd, M.S.; Fialho, M.L.S.; Aparicio, C.N.M.; Kerr, M.; Timm, K.N.; Griffin, J.L.; Luiken, J.J.F.P.; Glatz, J.F.C.; Tyler, D.J.; Heather, L.C. Fatty acids prevent hypoxia-inducible factor-1α signaling through decreased succinate in diabetes. JACC Basic Transl. Sci. 2018, 3, 485–498. [Google Scholar] [CrossRef]
- Morash, A.J.; Kotwica, A.O.; Murray, A.J. Tissue-specific changes in fatty acid oxidation in hypoxic heart and skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 305, R534–R541. [Google Scholar] [CrossRef]
- Malthankar-Phatak, G.H.; Patel, A.B.; Xia, Y.; Hong, S.; Chowdhury, G.M.I.; Behar, K.L.; Orina, I.A.; Lai, J.C.K. Effects of continuous hypoxia on energy metabolism in cultured cerebro-cortical neurons. Brain Res. 2008, 1229, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Bao, H.G.; Wang, X.Y.; Li, J.Y.; Wu, C.X. Comparison of effects of hypoxia on glutathione and activities of related enzymes in livers of Tibet chicken and Silky chicken. Poult. Sci. 2011, 90, 648–652. [Google Scholar] [CrossRef]
- Ge, R.L.; Cai, Q.; Shen, Y.Y.; San, A.; Ma, L.; Zhang, Y.; Yi, X.; Chen, Y.; Yang, L.; Huang, Y.; et al. Draft genome sequence of the Tibetan antelope. Nat. Commun. 2013, 4, 1858. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.L.; Jiang, B.H.; Rue, E.A.; Semenza, G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar] [CrossRef] [Green Version]
- Rankin, E.B.; Biju, M.P.; Liu, Q.; Unger, T.L.; Rha, J.; Johnson, R.S.; Simon, M.C.; Keith, B.; Haase, V.H. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J. Clin. Investig. 2007, 117, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Ray, P.S.; Zhu, L.; Otani, H.; Asahara, T.; Maulik, N. Hypoxia/Reoxygenation Promotes Myocardial angiogenesis via anÈNF κ B-dependent mechanism in a rat model of chronic myocardial infarction. J. Mol. Cell. Cardiol. 2001, 33, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Komniski, M.S.; Yakushev, S.; Bogdanov, N.; Gassmann, M.; Bogdanova, A. Interventricular heterogeneity in rat heart responses to hypoxia: The tuning of glucose metabolism, ion gradients, and function. Am. J. Physiol. Heart Circ. Physiol. 2011, 300, H1645–H1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dus-Szachniewicz, K.; Drobczynski, S.; Ziolkowski, P.; Kolodziej, P.; Walaszek, K.M.; Korzeniewska, A.K.; Agrawal, A.; Kupczyk, P.; Wozniak, M. Physiological Hypoxia (Physioxia) Impairs the early adhesion of single lymphoma cell to marrow stromal cell and extracellular matrix. Optical tweezers study. Int. J. Mol. Sci. 2018, 19, 1880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandel, N.S.; McClintock, D.S.; Feliciano, C.E.; Wood, T.M.; Melendez, J.A.; Rodriguez, A.M.; Schumacker, P.T. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: A mechanism of O2 sensing. J. Biol. Chem. 2000, 275, 25130–25138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, C.R.; Billiard, S.M.; Di Giulio, R.T. Hypoxia inhibits induction of aryl hydrocarbon receptor activity in topminnow hepatocarcinoma cells in an ARNT dependent manner. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009, 150, 383–389. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.M.; Kim, J.S.; Yoo, Y.D. A novel protein, Romo1, induces ROS production in the mitochondria. Biochem. Biophys. Res. Commun. 2006, 347, 649–655. [Google Scholar] [CrossRef]
- Ye, L.; Mao, S.; Fang, S.; Zhang, J.; Tan, Y.; Gu, W. Increased serum romo1 was correlated with lung function, inflammation, and oxidative Stress in chronic obstructive pulmonary disease. Inflammation 2019, 42, 1555–1560. [Google Scholar] [CrossRef]
- Itsumi, M.; Inoue, S.; Elia, A.J.; Murakami, K.; Sasaki, M.; Lind, E.F.; Brenner, D.; Harris, I.S.; Chio, I.I.; Afzal, S.; et al. Idh1 protects murine hepatocytes from endotoxin-induced oxidative stress by regulating the intracellular NADP(+)/NADPH ratio. Cell Death Differ. 2015, 22, 1837–1845. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Liebermann, D.A.; Holtzman, E.J.; Jeronis, S.; Hoffman, B.; Geifman-Holtzman, O. Preeclampsia-associated stresses activate Gadd45a signaling and sFlt-1 in placental explants. J. Cell. Physiol. 2013, 228, 362–370. [Google Scholar] [CrossRef]
- Li, F.; Han, N.; Wang, Y.; Xu, Q. Gadd45a knockdown alleviates oxidative stress through suppressing the p38 MAPK signaling pathway in the pathogenesis of preeclampsia. Placenta 2018, 65, 20–28. [Google Scholar] [CrossRef]
- Holroyd, E.W.; Delacroix, S.; Larsen, K.; Harbuzariu, A.; Psaltis, P.J.; Wang, L.; Pan, S.; White, T.A.; Witt, T.A.; Kleppe, L.S.; et al. Tissue factor pathway inhibitor blocks angiogenesis via its carboxyl terminus. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 704–711. [Google Scholar] [CrossRef] [Green Version]
- Casazza, A.; Laoui, D.; Wenes, M.; Rizzolio, S.; Bassani, N.; Mambretti, M.; Deschoemaeker, S.; Van Ginderachter, J.A.; Tamagnone, L.; Mazzone, M. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell 2013, 24, 695–709. [Google Scholar] [CrossRef] [Green Version]
- Beischlag, T.V.; Luis, M.J.; Hollingshead, B.D.; Perdew, G.H. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008, 18, 207–250. [Google Scholar] [CrossRef] [Green Version]
- Ziello, J.E.; Jovin, I.S.; Huang, Y. Hypoxia-Inducible Factor (HIF)-1 regulatory pathway and its potential for therapeutic intervention in malignancy and ischemia. Yale J. Biol. Med. 2007, 80, 51–60. [Google Scholar]
- Wang, C.F.; Wu, C.X.; Li, N. Differential gene expression of hypoxia inducible factor-1alpha and hypoxic adaptation in chicken. Yi Chuan 2007, 29, 75–80. [Google Scholar] [CrossRef]
- Tazawa, H.; Andrewartha, S.J.; Burggren, W.W. Acute regulation of hematocrit and blood acid-base balance during severe hypoxic challenges in late chicken embryos (Gallus gallus). Respir. Physiol. Neurobiol. 2012, 184, 86–96. [Google Scholar] [CrossRef]
- Cori, C.F. The glucose lactic acid cycle and gluconeogenesis. Curr. Top. Cell. Regul. 1981, 18, 377–387. [Google Scholar] [PubMed]
- Diani-Moore, S.; Zhang, S.; Ram, P.; Rifkind, A.B. Aryl Hydrocarbon Receptor Activation by Dioxin Targets Phosphoenolpyruvate Carboxykinase (PEPCK) for ADP-ribosylation via 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD)-inducible Poly(ADP-ribose) Polymerase (TiPARP). J. Biol. Chem. 2013, 288, 21514–21525. [Google Scholar] [CrossRef] [Green Version]
- Jin, N.; Hatton, N.; Swartz, D.R.; Xia, X.; Harrington, M.A.; Larsen, S.H.; Rhoades, R.A. Hypoxia activates Jun-N-Terminal kinase, extracellular signal-regulated protein kinase, and p38 kinase in pulmonary arteries. Am. J. Respir. Cell Mol. Biol. 2000, 23, 593–601. [Google Scholar] [CrossRef]
- Sala, M.A.; Chen, C.; Zhang, Q.; Do-Umehara, H.C.; Wu, W.; Misharin, A.V.; Waypa, G.B.; Fang, D.; Budinger, G.R.S.; Liu, S.; et al. JNK2 up-regulates hypoxia-inducible factors and contributes to hypoxia-induced erythropoiesis and pulmonary hypertension. J. Biol. Chem. 2018, 293, 271–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Gou, W.; Zhang, Y.; Zhang, H.; Wu, C. Insights into hypoxic adaptation in Tibetan chicken embryos from comparative proteomics. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 31, 100602. [Google Scholar] [CrossRef] [PubMed]
- Sayal, K.K.; Higgins, G.S.; Hammond, E.M. Uncovering the influence of the FGFR1 pathway on glioblastoma radiosensitivity. Ann. Transl. Med. 2016, 4, 538. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Wang, X.; Liu, J.; Wang, X.; Bao, H. Identifying Candidate Genes for Hypoxia Adaptation of Tibet Chicken Embryos by Selection Signature Analyses and RNA Sequencing. Genes 2020, 11, 823. https://doi.org/10.3390/genes11070823
Liu X, Wang X, Liu J, Wang X, Bao H. Identifying Candidate Genes for Hypoxia Adaptation of Tibet Chicken Embryos by Selection Signature Analyses and RNA Sequencing. Genes. 2020; 11(7):823. https://doi.org/10.3390/genes11070823
Chicago/Turabian StyleLiu, Xiayi, Xiaochen Wang, Jing Liu, Xiangyu Wang, and Haigang Bao. 2020. "Identifying Candidate Genes for Hypoxia Adaptation of Tibet Chicken Embryos by Selection Signature Analyses and RNA Sequencing" Genes 11, no. 7: 823. https://doi.org/10.3390/genes11070823
APA StyleLiu, X., Wang, X., Liu, J., Wang, X., & Bao, H. (2020). Identifying Candidate Genes for Hypoxia Adaptation of Tibet Chicken Embryos by Selection Signature Analyses and RNA Sequencing. Genes, 11(7), 823. https://doi.org/10.3390/genes11070823



