Genomic Landscape of the Mitochondrial Genome in the United Arab Emirates Native Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement, Sample Collection, and DNA Isolation
2.2. Human Mitogenome Enrichment and Sequencing
2.3. Data QC, Assembly, and Variant Identification
2.4. Tree Reconstruction and Haplogroup Prediction
2.5. Population Structure and Differentiation
2.6. Diversity, Kinship, and Selection
2.7. Reconstruction of Demographic History
2.8. Heteroplasmy and Structural Variation Identification
3. Results
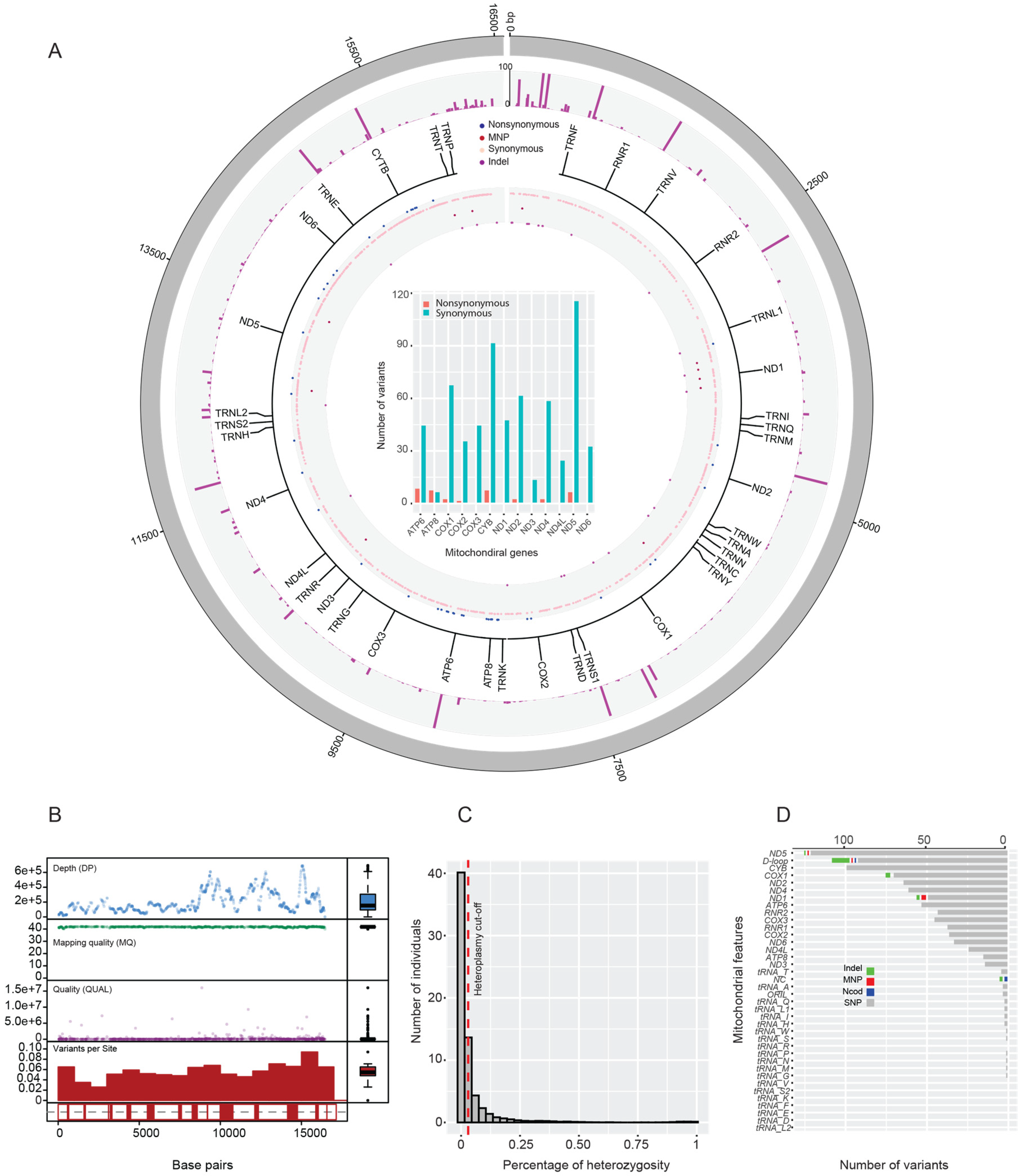
3.1. Mitogenome Assembly and Variant Annotation
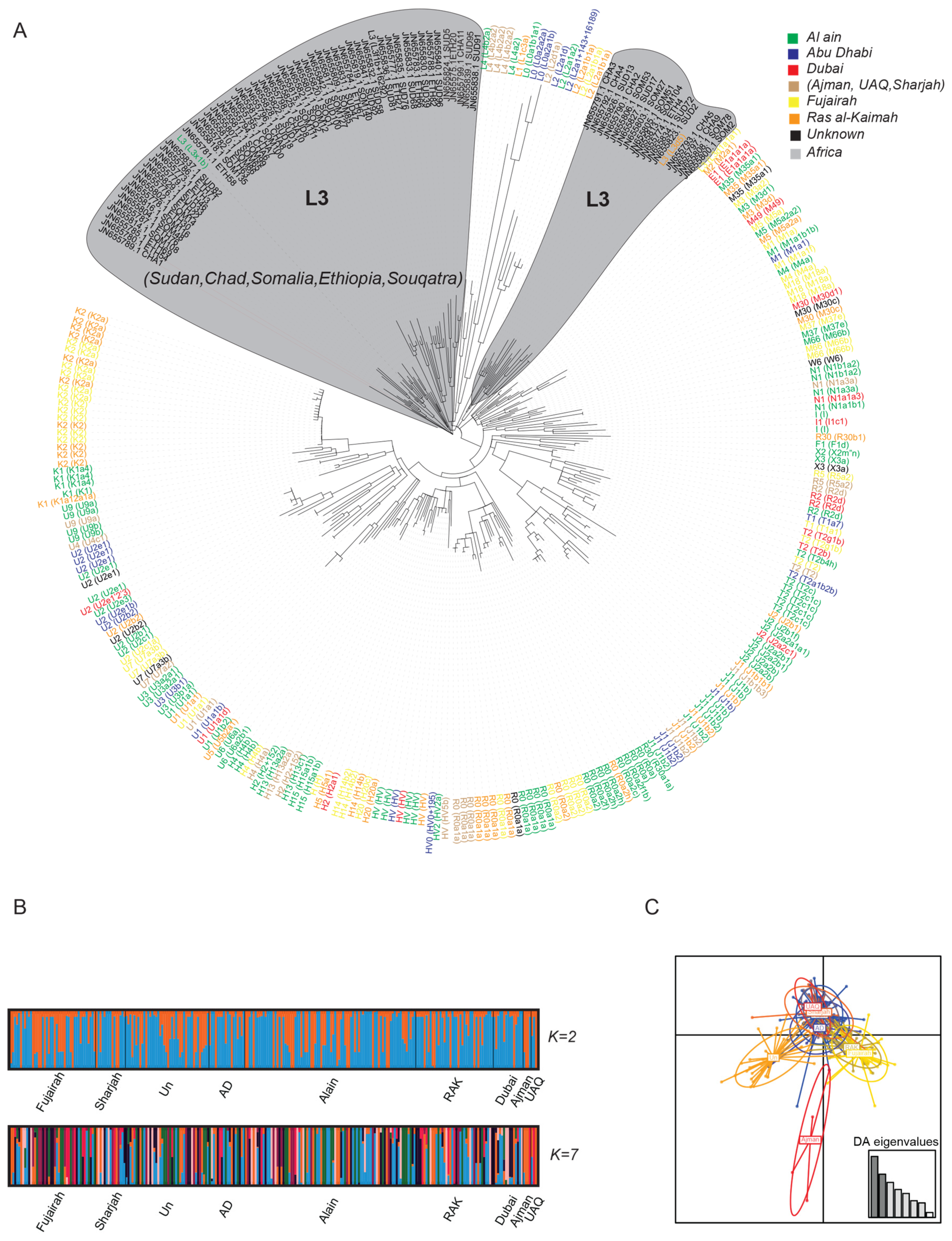
3.2. Haplogroup Identification
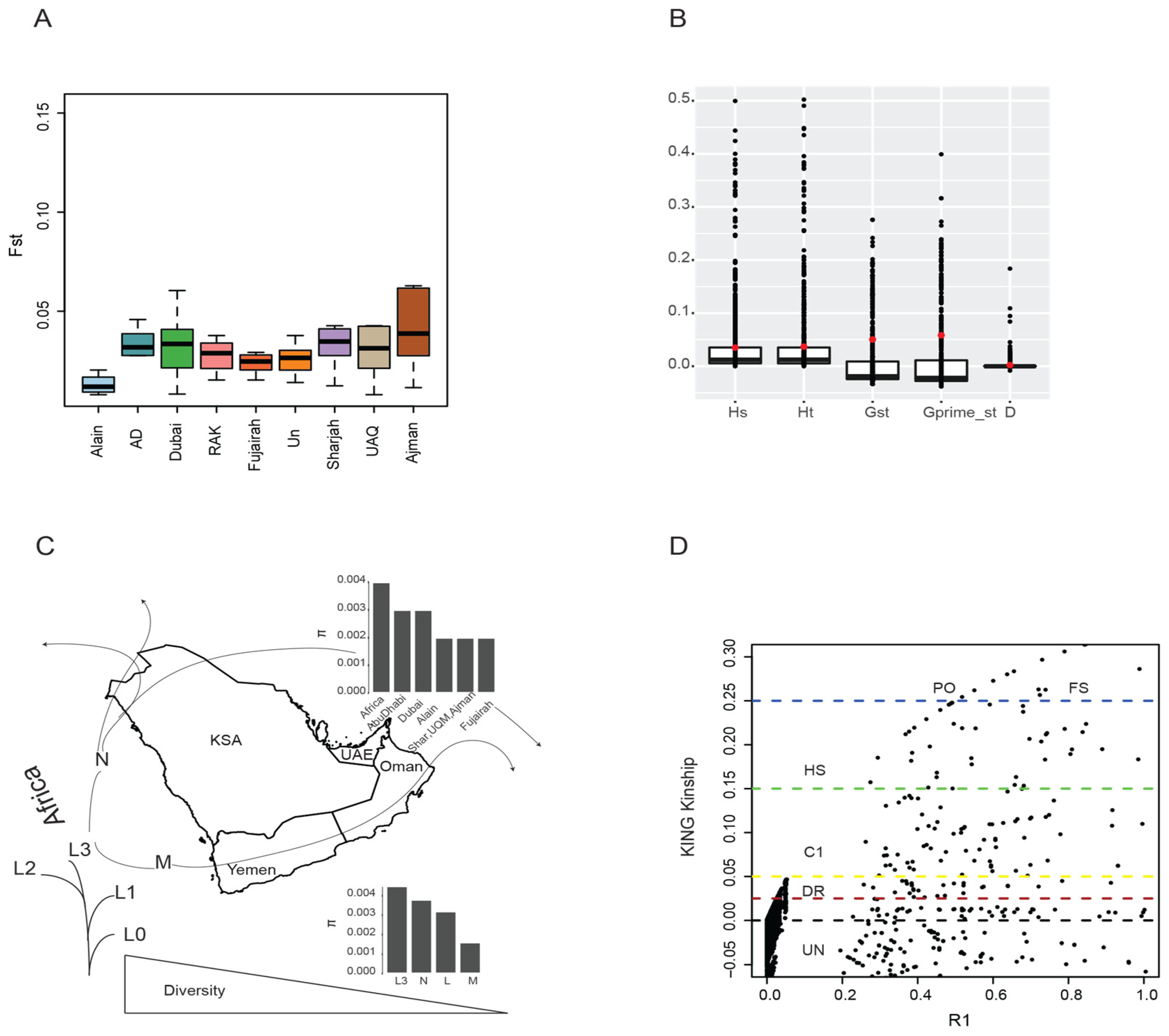
3.3. Population Structure and Differentiation
3.4. Diversity, Kinship, and Selection
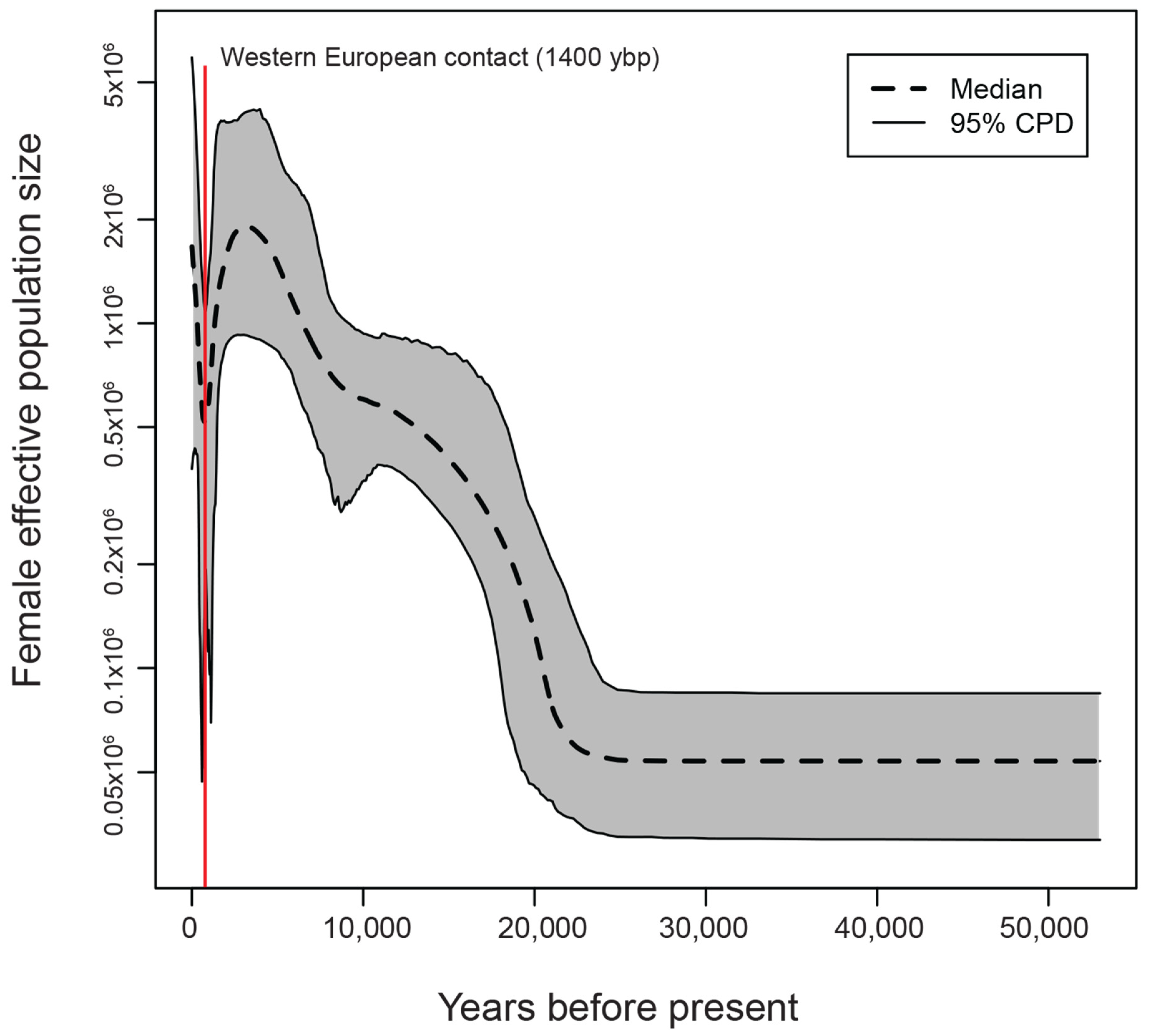
3.5. Demographic History Reconstruction
3.6. Heteroplasmy and Structural Variation Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ingman, M.; Kaessmann, H.; Pääbo, S.; Gyllensten, U. Mitochondrial genome variation and the origin of modern humans. Nature 2000, 408, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Maca-Meyer, N.; González, A.M.; Larruga, J.M.; Flores, C.; Cabrera, V.M. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001, 2, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macaulay, V.; Hill, C.; Achilli, A.; Rengo, C.; Clarke, D.; Meehan, W.; Blackburn, J.; Semino, O.; Scozzari, R.; Cruciani, F. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 2005, 308, 1034–1036. [Google Scholar] [CrossRef]
- Kivisild, T.; Rootsi, S.; Metspalu, M.; Mastana, S.; Kaldma, K.; Parik, J.; Metspalu, E.; Adojaan, M.; Tolk, H.-V.; Stepanov, V. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am. J. Hum. Genet. 2003, 72, 313–332. [Google Scholar] [CrossRef] [Green Version]
- Thangaraj, K.; Chaubey, G.; Singh, V.K.; Vanniarajan, A.; Thanseem, I.; Reddy, A.G.; Singh, L. In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup‘M’in India. BMC Genom. 2006, 7, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Merriwether, D.A.; Hodgson, J.A.; Friedlaender, F.R.; Allaby, R.; Cerchio, S.; Koki, G.; Friedlaender, J.S. Ancient mitochondrial M haplogroups identified in the Southwest Pacific. Proc. Natl. Acad. Sci. USA 2005, 102, 13034–13039. [Google Scholar] [CrossRef] [Green Version]
- Hudjashov, G.; Kivisild, T.; Underhill, P.A.; Endicott, P.; Sanchez, J.J.; Lin, A.A.; Shen, P.; Oefner, P.; Renfrew, C.; Villems, R. Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc. Natl. Acad. Sci. USA 2007, 104, 8726–8730. [Google Scholar] [CrossRef] [Green Version]
- Forster, P. Ice Ages and the mitochondrial DNA chronology of human dispersals: A review. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2004, 359, 255–264. [Google Scholar] [CrossRef]
- Forster, P.; Torroni, A.; Renfrew, C.; Röhl, A. Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol. Biol. Evol. 2001, 18, 1864–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oppenheimer, S. Out of Eden: The Peopling of the World; Jonathan Ball Publishers: Johannesburg, South Africa, 2012. [Google Scholar]
- Kivisild, T.; Reidla, M.; Metspalu, E.; Rosa, A.; Brehm, A.; Pennarun, E.; Parik, J.; Geberhiwot, T.; Usanga, E.; Villems, R. Ethiopian mitochondrial DNA heritage: Tracking gene flow across and around the gate of tears. Am. J. Hum. Genet. 2004, 75, 752–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abu-Amero, K.K.; Gonzalez, A.M.; Larruga, J.M.; Bosley, T.M.; Cabrera, V.M. Eurasian and African mitochondrial DNA influences in the Saudi Arabian population. BMC Evol. Biol. 2007, 7, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowold, D.; Luis, J.; Terreros, M.; Herrera, R.J. Mitochondrial DNA geneflow indicates preferred usage of the Levant Corridor over the Horn of Africa passageway. J. Hum. Genet. 2007, 52, 436–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilkington, M.M.; Wilder, J.A.; Mendez, F.L.; Cox, M.P.; Woerner, A.; Angui, T.; Kingan, S.; Mobasher, Z.; Batini, C.; Destro-Bisol, G. Contrasting signatures of population growth for mitochondrial DNA and Y chromosomes among human populations in Africa. Mol. Biol. Evol. 2008, 25, 517–525. [Google Scholar] [CrossRef] [Green Version]
- Quintana-Murci, L.; Quach, H.; Harmant, C.; Luca, F.; Massonnet, B.; Patin, E.; Sica, L.; Mouguiama-Daouda, P.; Comas, D.; Tzur, S. Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter–gatherers and Bantu-speaking farmers. Proc. Natl. Acad. Sci. USA 2008, 105, 1596–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, F.A.; Tishkoff, S.A. African human diversity, origins and migrations. Curr. Opin. Genet. Dev. 2006, 16, 597–605. [Google Scholar] [CrossRef]
- Tishkoff, S.A.; Gonder, M.K.; Henn, B.M.; Mortensen, H.; Knight, A.; Gignoux, C.; Fernandopulle, N.; Lema, G.; Nyambo, T.B.; Ramakrishnan, U. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol. Biol. Evol. 2007, 24, 2180–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, E.T.; Stover, D.A.; Ehret, C.; Destro-Bisol, G.; Spedini, G.; McLeod, H.; Louie, L.; Bamshad, M.; Strassmann, B.I.; Soodyall, H. Contrasting patterns of Y chromosome and mtDNA variation in Africa: Evidence for sex-biased demographic processes. Eur. J. Hum. Genet. 2005, 13, 867–876. [Google Scholar] [CrossRef]
- Campbell, M.C.; Tishkoff, S.A. African genetic diversity: Implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genom. Hum. Genet. 2008, 9, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Shtolz, N.; Mishmar, D. The mitochondrial genome–on selective constraints and signatures at the organism, cell, and single mitochondrion levels. Front. Ecol. Evol. 2019, 7, 342. [Google Scholar] [CrossRef] [Green Version]
- Elson, J.L.; Andrews, R.M.; Chinnery, P.F.; Lightowlers, R.N.; Turnbull, D.M.; Howell, N. Analysis of European mtDNAs for recombination. Am. J. Hum. Genet. 2001, 68, 145–153. [Google Scholar] [CrossRef] [Green Version]
- De Fanti, S.; Vicario, S.; Lang, M.; Simone, D.; Magli, C.; Luiselli, D.; Gianaroli, L.; Romeo, G. Intra-individual purifying selection on mitochondrial DNA variants during human oogenesis. Hum. Reprod. 2017, 32, 1100–1107. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.B.; Freyer, C.; Elson, J.L.; Wredenberg, A.; Cansu, Z.; Trifunovic, A.; Larsson, N.G. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008, 6, e10. [Google Scholar] [CrossRef] [PubMed]
- Bannwarth, S.; Procaccio, V.; Lebre, A.S.; Jardel, C.; Chaussenot, A.; Hoarau, C.; Maoulida, H.; Charrier, N.; Gai, X.; Xie, H.M.; et al. Prevalence of rare mitochondrial DNA mutations in mitochondrial disorders. J. Med. Genet. 2013, 50, 704–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, D.C.; Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a021220. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Schmitt, E.S.; Landsverk, M.L.; Zhang, V.W.; Li, F.Y.; Graham, B.H.; Craigen, W.J.; Wong, L.J. An integrated approach for classifying mitochondrial DNA variants: One clinical diagnostic laboratory’s experience. Genet. Med. 2012, 14, 620–626. [Google Scholar] [CrossRef] [Green Version]
- Damas, J.; Samuels, D.C.; Carneiro, J.; Amorim, A.; Pereira, F. Mitochondrial DNA rearrangements in health and disease—A comprehensive study. Hum. Mutat. 2014, 35, 1–14. [Google Scholar] [CrossRef]
- Neiman, M.; Taylor, D.R. The causes of mutation accumulation in mitochondrial genomes. Proc. R. Soc. B Biol. Sci. 2009, 276, 1201–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holt, I.J.; Harding, A.E.; Morgan-Hughes, J.A. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 1988, 331, 717–719. [Google Scholar] [CrossRef]
- Wallace, D.C.; Zheng, X.X.; Lott, M.T.; Shoffner, J.M.; Hodge, J.A.; Kelley, R.I.; Epstein, C.M.; Hopkins, L.C. Familial mitochondrial encephalomyopathy (MERRF): Genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell 1988, 55, 601–610. [Google Scholar] [CrossRef]
- Calloway, C.D.; Reynolds, R.L.; Herrin, G.L., Jr.; Anderson, W.W. The frequency of heteroplasmy in the HVII region of mtDNA differs across tissue types and increases with age. Am. J. Hum. Genet. 2000, 66, 1384–1397. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Wu, J.; Dressman, D.C.; Iacobuzio-Donahue, C.; Markowitz, S.D.; Velculescu, V.E.; Diaz, L.A., Jr.; Kinzler, K.W.; Vogelstein, B.; Papadopoulos, N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 2010, 464, 610–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, C.; Montiel, R.; Sierra, B.; Bettencourt, C.; Fernandez, E.; Alvarez, L.; Lima, M.; Abade, A.; Aluja, M.P. Understanding differences between phylogenetic and pedigree-derived mtDNA mutation rate: A model using families from the Azores Islands (Portugal). Mol. Biol. Evol. 2005, 22, 1490–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, C.; Sierra, B.; Alvarez, L.; Ramos, A.; Fernandez, E.; Nogues, R.; Aluja, M.P. Frequency and pattern of heteroplasmy in the control region of human mitochondrial DNA. J. Mol. Evol. 2008, 67, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Kirches, E.; Michael, M.; Warich-Kirches, M.; Schneider, T.; Weis, S.; Krause, G.; Mawrin, C.; Dietzmann, K. Heterogeneous tissue distribution of a mitochondrial DNA polymorphism in heteroplasmic subjects without mitochondrial disorders. J. Med Genet. 2001, 38, 312–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irwin, J.A.; Saunier, J.L.; Niederstätter, H.; Strouss, K.M.; Sturk, K.A.; Diegoli, T.M.; Brandstätter, A.; Parson, W.; Parsons, T.J. Investigation of heteroplasmy in the human mitochondrial DNA control region: A synthesis of observations from more than 5000 global population samples. J. Mol. Evol. 2009, 68, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Al-Gazali, L.; Ali, B.R. Mutations of a country: A mutation review of single gene disorders in the United Arab Emirates (UAE). Hum. Mutat. 2010, 31, 505–520. [Google Scholar] [CrossRef]
- Al-Jasmi, F.A.; Tawfig, N.; Berniah, A.; Ali, B.R.; Taleb, M.; Hertecant, J.L.; Bastaki, F.; Souid, A.K. Prevalence and Novel Mutations of Lysosomal Storage Disorders in United Arab Emirates: LSD in UAE. JIMD Rep. 2013, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Al-Shamsi, A.; Hertecant, J.L.; Al-Hamad, S.; Souid, A.K.; Al-Jasmi, F. Mutation Spectrum and Birth Prevalence of Inborn Errors of Metabolism among Emiratis: A study from Tawam Hospital Metabolic Center, United Arab Emirates. Sultan Qaboos Univ. Med. J. 2014, 14, e42–e49. [Google Scholar] [CrossRef] [Green Version]
- Fendt, L.; Zimmermann, B.; Daniaux, M.; Parson, W. Sequencing strategy for the whole mitochondrial genome resulting in high quality sequences. BMC Genom. 2009, 10, 139. [Google Scholar] [CrossRef] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bragg, L.M.; Stone, G.; Butler, M.K.; Hugenholtz, P.; Tyson, G.W. Shining a light on dark sequencing: Characterising errors in Ion Torrent PGM data. PLoS Comput. Biol. 2013, 9, e1003031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preste, R.; Clima, R.; Attimonelli, M. Human mitochondrial variant annotation with HmtNote. BioRxiv 2019, 600619. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A. 2006–2012. Fig. Tree. Tree Figure Drawing Tool, Version 1.4. 0. University of Edinburgh: Institute of Evolutionary Biology [on-line]. Available online: http://tree.bio.ed.ac.uk/ (accessed on 30 January 2015).
- Weissensteiner, H.; Pacher, D.; Kloss-Brandstatter, A.; Forer, L.; Specht, G.; Bandelt, H.J.; Kronenberg, F.; Salas, A.; Schonherr, S. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016, 44, W58–W63. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [Green Version]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [Green Version]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef] [Green Version]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heled, J.; Drummond, A.J. Bayesian inference of population size history from multiple loci. BMC Evol. Biol. 2008, 8, 289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weissensteiner, H.; Forer, L.; Fuchsberger, C.; Schopf, B.; Kloss-Brandstatter, A.; Specht, G.; Kronenberg, F.; Schonherr, S. mtDNA-Server: Next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic Acids Res. 2016, 44, W64–W69. [Google Scholar] [CrossRef]
- Goudenège, D.; Bris, C.; Hoffmann, V.; Desquiret-Dumas, V.; Jardel, C.; Rucheton, B.; Bannwarth, S.; Paquis-Flucklinger, V.; Lebre, A.S.; Colin, E. eKLIPse: A sensitive tool for the detection and quantification of mitochondrial DNA deletions from next-generation sequencing data. Genet. Med. 2019, 21, 1407–1416. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, V.; Brucato, N.; Ferreira, J.C.; Pedro, N.; Cavadas, B.; Ricaut, F.X.; Alshamali, F.; Pereira, L. Genome-Wide Characterization of Arabian Peninsula Populations: Shedding Light on the History of a Fundamental Bridge between Continents. Mol. Biol. Evol. 2019, 36, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Černý, V.; Mulligan, C.J.; Fernandes, V.; Silva, N.M.; Alshamali, F.; Non, A.; Harich, N.; Cherni, L.; El Gaaied, A.B.A.; Al-Meeri, A. Internal diversification of mitochondrial haplogroup R0a reveals post-last glacial maximum demographic expansions in South Arabia. Mol. Biol. Evol. 2011, 28, 71–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Abri, A.; Podgorná, E.; Rose, J.I.; Pereira, L.; Mulligan, C.J.; Silva, N.M.; Bayoumi, R.; Soares, P.; Černý, V. Pleistocene-Holocene boundary in Southern Arabia from the perspective of human mtDNA variation. Am. J. Phys. Anthropol. 2012, 149, 291–298. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, V.; Triska, P.; Pereira, J.B.; Alshamali, F.; Rito, T.; Machado, A.; Fajkošová, Z.; Cavadas, B.; Černý, V.; Soares, P. Genetic stratigraphy of key demographic events in Arabia. PLoS ONE 2015, 10, e0118625. [Google Scholar] [CrossRef] [Green Version]
- Vyas, D.N.; Kitchen, A.; Miro-Herrans, A.T.; Pearson, L.N.; Al-Meeri, A.; Mulligan, C.J. Bayesian analyses of Yemeni mitochondrial genomes suggest multiple migration events with Africa and Western Eurasia. Am. J. Phys. Anthropol. 2016, 159, 382–393. [Google Scholar] [CrossRef]
- Fernandes, V.; Alshamali, F.; Alves, M.; Costa, M.D.; Pereira, J.B.; Silva, N.M.; Cherni, L.; Harich, N.; Cerny, V.; Soares, P.; et al. The Arabian cradle: Mitochondrial relicts of the first steps along the southern route out of Africa. Am. J. Hum. Genet. 2012, 90, 347–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macaulay, V.; Richards, M.; Hickey, E.; Vega, E.; Cruciani, F.; Guida, V.; Scozzari, R.; Bonne-Tamir, B.; Sykes, B.; Torroni, A. The emerging tree of West Eurasian mtDNAs: A synthesis of control-region sequences and RFLPs. Am. J. Hum. Genet. 1999, 64, 232–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, M.; Macaulay, V.; Hickey, E.; Vega, E.; Sykes, B.; Guida, V.; Rengo, C.; Sellitto, D.; Cruciani, F.; Kivisild, T.; et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 2000, 67, 1251–1276. [Google Scholar] [CrossRef]
- Al-Zahery, N.; Semino, O.; Benuzzi, G.; Magri, C.; Passarino, G.; Torroni, A.; Santachiara-Benerecetti, A.S. Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol. Phylogenet. Evol. 2003, 28, 458–472. [Google Scholar] [CrossRef]
- Achilli, A.; Rengo, C.; Magri, C.; Battaglia, V.; Olivieri, A.; Scozzari, R.; Cruciani, F.; Zeviani, M.; Briem, E.; Carelli, V. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am. J. Hum. Genet. 2004, 75, 910–918. [Google Scholar] [CrossRef]
- Luis, J.R.; Rowold, D.J.; Regueiro, M.; Caeiro, B.; Cinnioglu, C.; Roseman, C.; Underhill, P.A.; Cavalli-Sforza, L.L.; Herrera, R.J. The Levant versus the Horn of Africa: Evidence for bidirectional corridors of human migrations. Am. J. Hum. Genet. 2004, 74, 532–544. [Google Scholar] [CrossRef] [Green Version]
- Salas, A.; Richards, M.; De la Fe, T.; Lareu, M.V.; Sobrino, B.; Sanchez-Diz, P.; Macaulay, V.; Carracedo, A. The making of the African mtDNA landscape. Am. J. Hum. Genet. 2002, 71, 1082–1111. [Google Scholar] [CrossRef] [Green Version]
- Richards, M.; Rengo, C.; Cruciani, F.; Gratrix, F.; Wilson, J.F.; Scozzari, R.; Macaulay, V.; Torroni, A. Extensive female-mediated gene flow from sub-Saharan Africa into near eastern Arab populations. Am. J. Hum. Genet. 2003, 72, 1058–1064. [Google Scholar] [CrossRef] [Green Version]
- Just, R.S.; Irwin, J.A.; Parson, W. Mitochondrial DNA heteroplasmy in the emerging field of massively parallel sequencing. Forensic Sci. Int. Genet. 2015, 18, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Rogell, B.; Dean, R.; Lemos, B.; Dowling, D.K. Mito-nuclear interactions as drivers of gene movement on and off the X-chromosome. BMC Genom. 2014, 15, 330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaidi, A.A.; Wilton, P.R.; Su, M.S.; Paul, I.M.; Arbeithuber, B.; Anthony, K.; Nekrutenko, A.; Nielsen, R.; Makova, K.D. Bottleneck and selection in the germline and maternal age influence transmission of mitochondrial DNA in human pedigrees. Proc. Natl. Acad. Sci. USA 2019, 116, 25172–25178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Chen, Z.J.; Zhang, Y.K.; Le, H.B. The role of mitochondrial tRNA mutations in lung cancer. Int. J. Clin. Exp. Med. 2015, 8, 13341–13346. [Google Scholar] [PubMed]
- Al-Gazali, L.I.; Dawodu, A.H.; Sabarinathan, K.; Varghese, M. The profile of major congenital abnormalities in the United Arab Emirates (UAE) population. J. Med. Genet. 1995, 32, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Fahmy, N.; Benson, P.; Al-Garrah, D. Consanguinity in UAE: Prevalence and analysis of some risk factors. Emirates Med. J. 1993, 1, 39–41. [Google Scholar]
- Mishmar, D.; Ruiz-Pesini, E.; Golik, P.; Macaulay, V.; Clark, A.G.; Hosseini, S.; Brandon, M.; Easley, K.; Chen, E.; Brown, M.D.; et al. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA 2003, 100, 171–176. [Google Scholar] [CrossRef] [Green Version]
- Tay, G.K.; Henschel, A.; Daw Elbait, G.; Al Safar, H.S. Genetic Diversity and Low Stratification of the Population of the United Arab Emirates. Front. Genet. 2020, 11, 608. [Google Scholar] [CrossRef]
- Merriwether, D.A.; Clark, A.G.; Ballinger, S.W.; Schurr, T.G.; Soodyall, H.; Jenkins, T.; Sherry, S.T.; Wallace, D.C. The structure of human mitochondrial DNA variation. J. Mol. Evol. 1991, 33, 543–555. [Google Scholar] [CrossRef]
- Schurr, T.G.; Wallace, D.C. Mitochondrial DNA diversity in Southeast Asian populations. Hum. Biol. 2002, 74, 431–452. [Google Scholar] [CrossRef]
- Zalloua, P.A.; Xue, Y.; Khalife, J.; Makhoul, N.; Debiane, L.; Platt, D.E.; Royyuru, A.K.; Herrera, R.J.; Hernanz, D.F.S.; Blue-Smith, J. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am. J. Hum. Genet. 2008, 82, 873–882. [Google Scholar] [CrossRef]

| NE ** | NW ** | Mid-NW ** | SW ** | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group a | F b | F b% | Unknown | Fujairah | RAK * | UAQ * | Dubai | Sharjah | Ajman | Al Ain | Abu Dhabi |
| E | 2 | 0.86 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| F | 1 | 0.43 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| H | 19 | 8.18 | 0 | 5 | 3 | 0 | 1 | 3 | 0 | 7 | 0 |
| HV | 10 | 4.31 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 5 | 2 |
| I | 3 | 1.29 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 |
| J | 25 | 10.77 | 0 | 0 | 5 | 0 | 1 | 2 | 1 | 13 | 3 |
| K | 26 | 11.20 | 0 | 11 | 11 | 0 | 0 | 0 | 0 | 4 | 0 |
| L | 20 | 8.62 | 1 | 3 | 4 | 0 | 0 | 4 | 0 | 5 | 3 |
| M | 28 | 12.06 | 2 | 11 | 5 | 0 | 2 | 0 | 0 | 7 | 1 |
| N | 6 | 2.58 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 4 | 0 |
| R | 35 | 15.08 | 1 | 6 | 7 | 2 | 2 | 2 | 1 | 14 | 0 |
| T | 14 | 6.03 | 0 | 3 | 0 | 0 | 2 | 1 | 0 | 6 | 2 |
| U | 39 | 16.81 | 3 | 4 | 3 | 1 | 2 | 2 | 1 | 16 | 7 |
| W | 1 | 0.43 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| X | 3 | 1.29 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Total | 232 | 100 | 9 | 43 | 39 | 3 | 15 | 15 | 4 | 86 | 18 |
| Genes | M | S | Ps | θ | π | D | dN/dS |
|---|---|---|---|---|---|---|---|
| ATP6 | 232 | 45 | 0.067265 | 0.011244 | 0.002034 | −2.391682 | 0.29 |
| ATP8 | 232 | 17 | 0.087179 | 0.014573 | 0.001415 | −2.325058 | 1.03 |
| COX1 | 232 | 75 | 0.050302 | 0.008409 | 0.001305 | −2.556932 | 0.10 |
| COX2 | 232 | 34 | 0.050595 | 0.008458 | 0.001166 | −2.449534 | 0.15 |
| COX3 | 232 | 40 | 0.053763 | 0.008987 | 0.001757 | −2.323988 | 0.12 |
| CYTB | 232 | 95 | 0.085818 | 0.014346 | 0.002837 | −2.457115 | 0.19 |
| ND1 | 232 | 56 | 0.06041 | 0.010098 | 0.002071 | −2.361573 | 0.12 |
| ND2 | 232 | 63 | 0.0625 | 0.010448 | 0.00161 | −2.533238 | 0.12 |
| ND3 | 232 | 16 | 0.048048 | 0.008032 | 0.003149 | −1.548463 | 0.15 |
| ND4 | 232 | 74 | 0.055306 | 0.009245 | 0.002533 | −2.195525 | 0.04 |
| ND4L | 232 | 31 | 0.050542 | 0.016855 | 0.001867 | −2.440526 | 0.06 |
| ND5 | 232 | 121 | 0.068789 | 0.011499 | 0.002293 | −2.476299 | 0.14 |
| ND6 | 232 | 31 | 0.060429 | 0.010102 | 0.001778 | −2.31697 | 0.08 |
| City/Haplogroup | π | D |
|---|---|---|
| Abu Dhabi | 0.0033 ± 0.00310 | −0.9003 ± 0.67193 |
| Al Ain | 0.0022 ± 0.00241 | −1.0783 ± 0.59466 |
| Dubai, Ajman, UQA | 0.0029 ± 0.00264 | −0.8337 ± 0.54551 |
| Fujairah | 0.0023 ± 0.00258 | −1.0999 ± 0.74124 |
| RAK | 0.0021 ± 0.00237 | −0.8034 ± 0.62341 |
| Sharjah | 0.0024 ± 0.00211 | −0.9012 ± 0.53421 |
| UAQ | 0.0021 ± 0.0012 | −0.5231 ± 0.32310 |
| Africa (L3) | 0.004 ± 0.00279 | −1.7485 ± 0.38859 |
| L | 0.0037 ± 0.00302 | −0.8079 ± 0.70818 |
| M | 0.0016 ± 0.00162 | −1.0248 ± 0.56632 |
| N | 0.0021 ± 0.00221 | −0.9080 ± 0.85152 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aljasmi, F.A.; Vijayan, R.; Sudalaimuthuasari, N.; Souid, A.-K.; Karuvantevida, N.; Almaskari, R.; Mohammed Abdul Kader, H.; Kundu, B.; Michel Hazzouri, K.; Amiri, K.M.A. Genomic Landscape of the Mitochondrial Genome in the United Arab Emirates Native Population. Genes 2020, 11, 876. https://doi.org/10.3390/genes11080876
Aljasmi FA, Vijayan R, Sudalaimuthuasari N, Souid A-K, Karuvantevida N, Almaskari R, Mohammed Abdul Kader H, Kundu B, Michel Hazzouri K, Amiri KMA. Genomic Landscape of the Mitochondrial Genome in the United Arab Emirates Native Population. Genes. 2020; 11(8):876. https://doi.org/10.3390/genes11080876
Chicago/Turabian StyleAljasmi, Fatma A., Ranjit Vijayan, Naganeeswaran Sudalaimuthuasari, Abdul-Kader Souid, Noushad Karuvantevida, Raja Almaskari, Hidaya Mohammed Abdul Kader, Biduth Kundu, Khaled Michel Hazzouri, and Khaled M. A. Amiri. 2020. "Genomic Landscape of the Mitochondrial Genome in the United Arab Emirates Native Population" Genes 11, no. 8: 876. https://doi.org/10.3390/genes11080876
APA StyleAljasmi, F. A., Vijayan, R., Sudalaimuthuasari, N., Souid, A.-K., Karuvantevida, N., Almaskari, R., Mohammed Abdul Kader, H., Kundu, B., Michel Hazzouri, K., & Amiri, K. M. A. (2020). Genomic Landscape of the Mitochondrial Genome in the United Arab Emirates Native Population. Genes, 11(8), 876. https://doi.org/10.3390/genes11080876






