Nuclear Regulation of Wnt/β-Catenin Signaling: It’s a Complex Situation
Abstract
:1. Introduction
2. Nuclear Transport of β-Catenin
3. Nuclear Mediators of Wnt Signaling
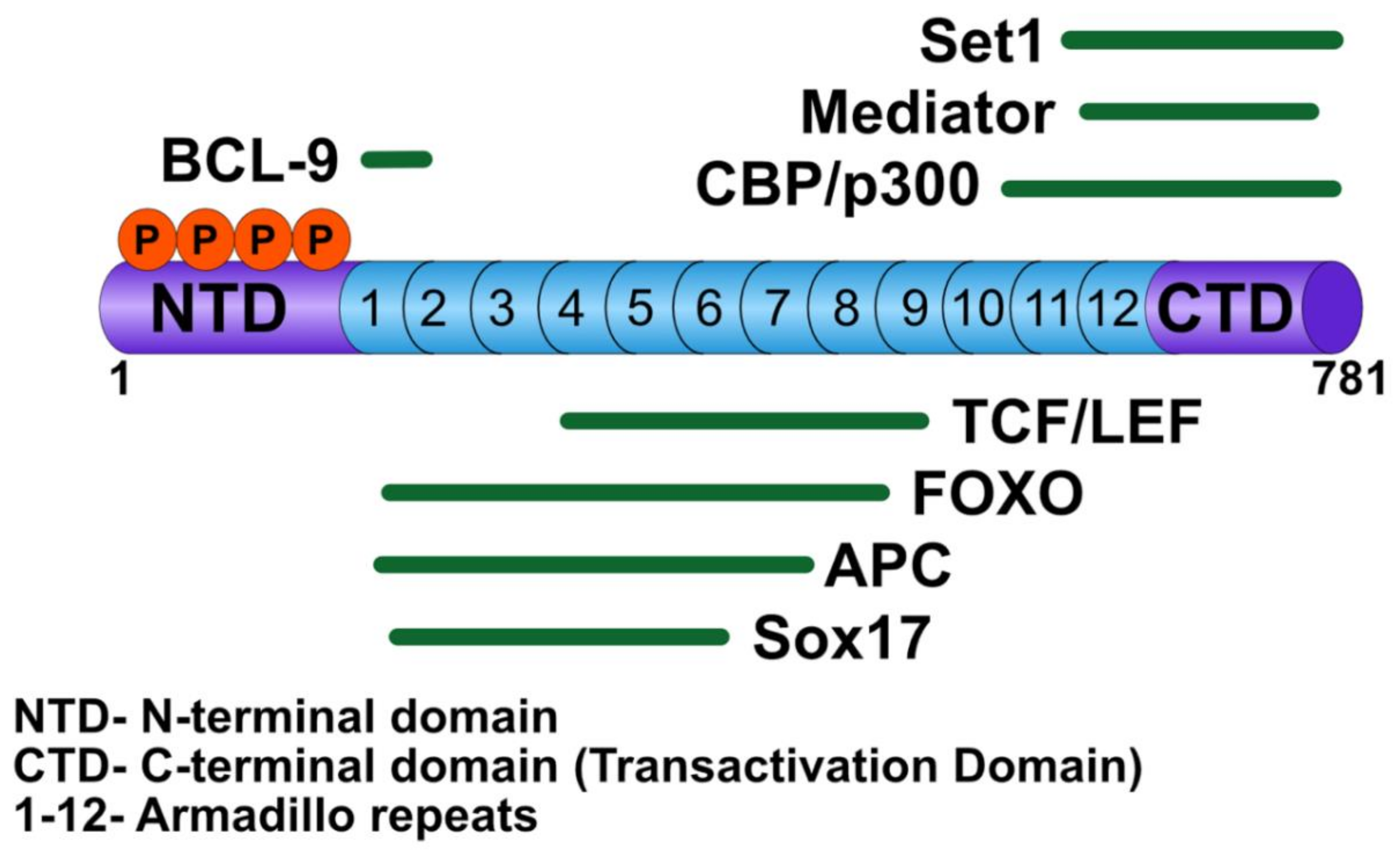
3.1. β-Catenin
3.2. T-Cell Factor/Lymphoid Enhancer Factor
3.3. Groucho/Transducing-Like Enhancer
3.4. Pygopus
3.5. B-cell CLL/Lymphoma 9
3.6. Non-TCF/LEF Transcription Partners of β-catenin
4. Wnt/β-Catenin-Mediated Transcriptional Repression
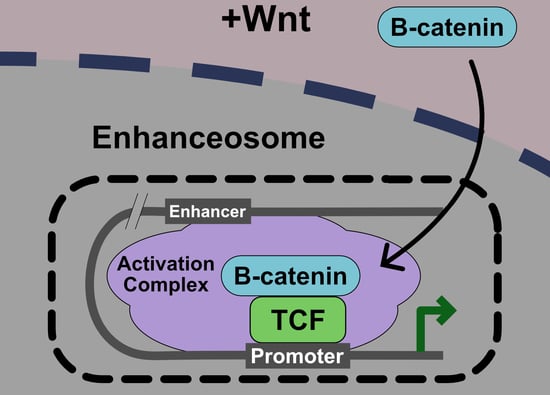
5. Current Model: The Wnt Enhanceosome
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nusse, R.; Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Saito-Diaz, K.; Chen, T.W.; Wang, X.; Thorne, C.A.; Wallace, H.A.; Page-McCaw, A.; Lee, E. The way Wnt works: Components and mechanism. Growth Factors Chur. Switz. 2013, 31, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Bilic, J.; Huang, Y.-L.; Davidson, G.; Zimmermann, T.; Cruciat, C.-M.; Bienz, M.; Niehrs, C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 2007, 316, 1619–1622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, F.; Schweizer, L.; Varmus, H. Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, frizzled and LRP. Dev. Camb. Engl. 2004, 131, 5103–5115. [Google Scholar]
- Fiedler, M.; Graeb, M.; Mieszczanek, J.; Rutherford, T.J.; Johnson, C.M.; Bienz, M. An ancient Pygo-dependent Wnt enhanceosome integrated by Chip/LDB-SSDP. ELife 2015, 4, e09073. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Jamieson, C.; Lui, C.; Henderson, B.R. Distinct hydrophobic “patches” in the N- and C-tails of beta-catenin contribute to nuclear transport. Exp. Cell Res. 2016, 348, 132–145. [Google Scholar] [CrossRef]
- Fagotto, F.; Glück, U.; Gumbiner, B.M. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of β-catenin. Curr. Biol. 1998, 8, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Suh, E.-K.; Gumbiner, B.M. Translocation of beta-catenin into the nucleus independent of interactions with FG-rich nucleoporins. Exp. Cell Res. 2003, 290, 447–456. [Google Scholar] [CrossRef]
- Henderson, B.R. Nuclear-cytoplasmic shuttling of APC regulates beta-catenin subcellular localization and turnover. Nat. Cell Biol. 2000, 2, 653–660. [Google Scholar] [CrossRef]
- Townsley, F.M.; Cliffe, A.; Bienz, M. Pygopus and Legless target Armadillo/beta-catenin to the nucleus to enable its transcriptional co-activator function. Nat. Cell Biol. 2004, 6, 626–633. [Google Scholar] [CrossRef]
- Lilien, J.; Balsamo, J. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of β-catenin. Curr. Opin. Cell Biol. 2005, 17, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.R.; Fagotto, F. The ins and outs of APC and beta-catenin nuclear transport. EMBO Rep. 2002, 3, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.N.; Del Viso, F.; Duncan, A.R.; Robson, A.; Hwang, W.; Kulkarni, S.; Liu, K.J.; Khokha, M.K. RAPGEF5 regulates nuclear translocation of β-catenin. Dev. Cell 2018, 44, 248–260. [Google Scholar] [CrossRef] [Green Version]
- Vuong, L.T.; Iomini, C.; Balmer, S.; Esposito, D.; Aaronson, S.A.; Mlodzik, M. Kinesin-2 and IFT-A act as a complex promoting nuclear localization of β-catenin during Wnt signalling. Nat. Commun. 2018, 9, 5304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayat, R.; Leber, B.; Grubac, V.; Wiltshire, L.; Persad, S. O-GlcNAc-glycosylation of beta-catenin regulates its nuclear localization and transcriptional activity. Exp. Cell Res. 2008, 314, 2774–2787. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Xu, X.; Hecht, A.; Boyer, T.G. Mediator is a transducer of Wnt/beta-catenin signaling. J. Biol. Chem. 2006, 281, 14066–14075. [Google Scholar] [CrossRef] [Green Version]
- Takemaru, K.I.; Moon, R.T. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 2000, 149, 249–254. [Google Scholar] [CrossRef] [Green Version]
- Chodaparambil, J.V.; Pate, K.T.; Hepler, M.R.D.; Tsai, B.P.; Muthurajan, U.M.; Luger, K.; Waterman, M.L.; Weis, W.I. Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J. 2014, 33, 719–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmans, R.; Basler, K. BCL9-2 binds Arm/beta-catenin in a Tyr142-independent manner and requires Pygopus for its function in Wg/Wnt signaling. Mech. Dev. 2007, 124, 59–67. [Google Scholar] [CrossRef]
- Sustmann, C.; Flach, H.; Ebert, H.; Eastman, Q.; Grosschedl, R. Cell-type-specific function of BCL9 involves a transcriptional activation domain that synergizes with β-catenin. Mol. Cell. Biol. 2008, 28, 3526–3537. [Google Scholar] [CrossRef] [Green Version]
- Sierra, J.; Yoshida, T.; Joazeiro, C.A.; Jones, K.A. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006, 20, 586–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, B.M. The adjustable nucleosome: An epigenetic signaling module. Trends Genet. TIG 2012, 28, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sutter, C.; Parker, D.S.; Blauwkamp, T.; Fang, M.; Cadigan, K.M. CBP/p300 are bimodal regulators of Wnt signaling. EMBO J. 2007, 26, 2284–2294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cadigan, K.M.; Waterman, M.L. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect. Biol. 2012, 4, a007906. [Google Scholar] [CrossRef]
- van de Wetering, M.; Clevers, H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. EMBO J. 1992, 11, 3039–3044. [Google Scholar] [CrossRef]
- Bhambhani, C.; Ravindranath, A.J.; Mentink, R.A.; Chang, M.V.; Betist, M.C.; Yang, Y.X.; Koushika, S.P.; Korswagen, H.C.; Cadigan, K.M. Distinct DNA binding sites contribute to the TCF transcriptional switch in C. elegans and Drosophila. PLoS Genet. 2014, 10, e1004133. [Google Scholar] [CrossRef] [Green Version]
- Hikasa, H.; Sokol, S.Y. Phosphorylation of TCF proteins by homeodomain-interacting protein kinase 2. J. Biol. Chem. 2011, 286, 12093–12100. [Google Scholar] [CrossRef] [Green Version]
- Hrckulak, D.; Kolar, M.; Strnad, H.; Korinek, V. TCF/LEF transcription factors: An update from the internet resources. Cancers 2016, 8, 70. [Google Scholar] [CrossRef] [Green Version]
- Cavallo, R.A.; Cox, R.T.; Moline, M.M.; Roose, J.; Polevoy, G.A.; Clevers, H.; Peifer, M.; Bejsovec, A. Drosophila Tcf and groucho interact to repress wingless signalling activity. Nature 1998, 395, 604–608. [Google Scholar] [CrossRef]
- Roose, J.; Molenaar, M.; Peterson, J.; Hurenkamp, J.; Brantjes, H.; Moerer, P.; van de Wetering, M.; Destrée, O.; Clevers, H. The Xenopus Wnt effector XTcf-3 interacts with groucho-related transcriptional repressors. Nature 1998, 395, 608–612. [Google Scholar] [CrossRef]
- Chen, G.; Nguyen, P.H.; Courey, A.J. A role for Groucho tetramerization in transcriptional repression. Mol. Cell. Biol. 1998, 18, 7259–7268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, H.; Hasson, P.; Paroush, Z.; Courey, A.J. Groucho oligomerization is required for repression in vivo. Mol. Cell. Biol. 2004, 24, 4341–4350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Fernandez, J.; Mische, S.; Courey, A.J. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999, 13, 2218–2230. [Google Scholar] [CrossRef]
- Arce, L.; Pate, K.T.; Waterman, M.L. Groucho binds two conserved regions of LEF-1 for HDAC-dependent repression. BMC Cancer 2009, 9, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, D.L.; Weis, W.I. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 2005, 12, 364–371. [Google Scholar] [CrossRef]
- Hanson, A.J.; Wallace, H.A.; Freeman, T.J.; Beauchamp, R.D.; Lee, L.A.; Lee, E. XIAP monoubiquitylates Groucho/TLE to promote canonical Wnt signaling. Mol. Cell 2012, 45, 619–628. [Google Scholar] [CrossRef] [Green Version]
- Flack, J.E.; Mieszczanek, J.; Novcic, N.; Bienz, M. Wnt-dependent inactivation of the groucho/TLE co-repressor by the HECT E3 ubiquitin ligase Hyd/UBR5. Mol. Cell 2017, 67, 181–193. [Google Scholar] [CrossRef]
- Ng, V.H.; Hang, B.I.; Sawyer, L.M.; Neitzel, L.R.; Crispi, E.E.; Rose, K.L.; Popay, T.M.; Zhong, A.; Lee, L.A.; Tansey, W.P.; et al. Phosphorylation of XIAP at threonine 180 controls its activity in Wnt signaling. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [Green Version]
- Belenkaya, T.Y.; Han, C.; Standley, H.J.; Lin, X.; Houston, D.W.; Heasman, J.; Lin, X. Pygopus Encodes a nuclear protein essential for wingless/Wnt signaling. Dev. Camb. Engl. 2002, 129, 4089–4101. [Google Scholar]
- Kramps, T.; Peter, O.; Brunner, E.; Nellen, D.; Froesch, B.; Chatterjee, S.; Murone, M.; Züllig, S.; Basler, K. Wnt/Wingless signaling requires BCL9/legless-mediated recruitment of Pygopus to the nuclear β-catenin-TCF complex. Cell 2002, 109, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Parker, D.S.; Jemison, J.; Cadigan, K.M. Pygopus, a nuclear PHD-finger protein required for wingless signaling in Drosophila. Dev. Camb. Engl. 2002, 129, 2565–2576. [Google Scholar]
- Thompson, B.; Townsley, F.; Rosin-Arbesfeld, R.; Musisi, H.; Bienz, M. A new nuclear component of the Wnt signalling pathway. Nat. Cell Biol. 2002, 4, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Städeli, R.; Basler, K. Dissecting nuclear Wingless signalling: Recruitment of the transcriptional co-activator Pygopus by a chain of adaptor proteins. Mech. Dev. 2005, 122, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Carrera, I.; Janody, F.; Leeds, N.; Duveau, F.; Treisman, J.E. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc. Natl. Acad. Sci. USA 2008, 105, 6644–6649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, P.G.P.; He, Z.; Popadiuk, C.; Kao, K.R. The transcriptional activity of Pygopus is enhanced by its interaction with cAMP-response-element-binding protein (CREB)-binding protein. Biochem. J. 2009, 422, 493–501. [Google Scholar] [CrossRef]
- Kessler, R.; Hausmann, G.; Basler, K. The PHD domain is required to link drosophila Pygopus to legless/β-catenin and not to histone H3. Mech. Dev. 2009, 126, 752–759. [Google Scholar] [CrossRef]
- Li, B.; Rhéaume, C.; Teng, A.; Bilanchone, V.; Munguia, J.E.; Hu, M.; Jessen, S.; Piccolo, S.; Waterman, M.L.; Dai, X. Developmental phenotypes and reduced Wnt signaling in mice deficient for Pygopus 2. Genes 2007, 45, 318–325. [Google Scholar] [CrossRef]
- Schwab, K.R.; Patterson, L.T.; Hartman, H.A.; Song, N.; Lang, R.A.; Lin, X.; Potter, S.S. Pygo1 and Pygo2 roles in Wnt signaling in mammalian kidney development. BMC Biol. 2007, 5, 15. [Google Scholar] [CrossRef] [Green Version]
- Cantù, C.; Felker, A.; Zimmerli, D.; Prummel, K.D.; Cabello, E.M.; Chiavacci, E.; Méndez-Acevedo, K.M.; Kirchgeorg, L.; Burger, S.; Ripoll, J.; et al. Mutations in Bcl9 and Pygo genes cause congenital heart defects by tissue-specific perturbation of Wnt/β-catenin signaling. Genes Dev. 2018, 32, 1443–1458. [Google Scholar] [CrossRef] [Green Version]
- Deka, J.; Wiedemann, N.; Anderle, P.; Murphy-Seiler, F.; Bultinck, J.; Eyckerman, S.; Stehle, J.C.; André, S.; Vilain, N.; Zilian, O.; et al. Bcl9/Bcl9l are critical for Wnt-mediated regulation of stem cell traits in colon epithelium and adenocarcinomas. Cancer Res. 2010, 70, 6619–6628. [Google Scholar] [CrossRef] [Green Version]
- Van Tienen, L.M.; Mieszczanek, J.; Fiedler, M.; Rutherford, T.J.; Bienz, M. Constitutive scaffolding of multiple Wnt enhanceosome components by Legless/BCL9. ELife 2017, 6, e20882. [Google Scholar] [CrossRef]
- Takada, K.; Zhu, D.; Bird, G.H.; Sukhdeo, K.; Zhao, J.J.; Mani, M.; Lemieux, M.; Carrasco, D.E.; Ryan, J.; Horst, D.; et al. Targeted disruption of the BCL9/β-catenin complex inhibits oncogenic Wnt signaling. Sci. Transl. Med. 2012, 4, 148ra117. [Google Scholar] [CrossRef] [Green Version]
- Ke, Q.; Costa, M. Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 2006, 70, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Kaidi, A.; Williams, A.C.; Paraskeva, C. Interaction between β-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat. Cell Biol. 2007, 9, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Li, Y.; Zhou, Z.; Wu, C.; Xing, Y.; Zou, X.; Tian, W.; Zhang, C. HIF-1α inhibits Wnt signaling pathway by activating sost expression in osteoblasts. PLoS ONE 2013, 8, e65940. [Google Scholar] [CrossRef] [PubMed]
- Boso, D.; Rampazzo, E.; Zanon, C.; Bresolin, S.; Maule, F.; Porcù, E.; Cani, A.; Della Puppa, A.; Trentin, L.; Basso, G.; et al. HIF-1α/Wnt signaling-dependent control of gene transcription regulates neuronal differentiation of glioblastoma stem cells. Theranostics 2019, 9, 4860–4877. [Google Scholar] [CrossRef] [PubMed]
- Tzivion, G.; Dobson, M.; Ramakrishnan, G. FoxO transcription factors; Regulation by AKT and 14–3–3 proteins. Biochim. Biophys. Acta 2011, 1813, 1938–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essers, M.A.G.; de Vries-Smits, L.M.M.; Barker, N.; Polderman, P.E.; Burgering, B.M.T.; Korswagen, H.C. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science 2005, 308, 1181–1184. [Google Scholar] [CrossRef]
- Hoogeboom, D.; Essers, M.A.G.; Polderman, P.E.; Voets, E.; Smits, L.M.M.; Burgering, B.M.T. Interaction of FOXO with β-catenin inhibits β-catenin/T cell factor activity. J. Biol. Chem. 2008, 283, 9224–9230. [Google Scholar] [CrossRef] [Green Version]
- Nlandu-Khodo, S.; Osaki, Y.; Scarfe, L.; Yang, H.; Phillips-Mignemi, M.; Tonello, J.; Saito-Diaz, K.; Neelisetty, S.; Ivanova, A.; Huffstater, T.; et al. Tubular β-catenin and FoxO3 interactions protect in chronic kidney disease. JCI Insight 2020, 5, e135454. [Google Scholar] [CrossRef]
- Sarkar, A.; Hochedlinger, K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell 2013, 12, 15–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zorn, A.M.; Barish, G.D.; Williams, B.O.; Lavender, P.; Klymkowsky, M.W.; Varmus, H.E. Regulation of Wnt signaling by Sox proteins: XSox17 α/β and XSox3 physically interact with beta-catenin. Mol. Cell 1999, 4, 487–498. [Google Scholar] [CrossRef]
- Sinner, D.; Kordich, J.J.; Spence, J.R.; Opoka, R.; Rankin, S.; Lin, S.C.J.; Jonatan, D.; Zorn, A.M.; Wells, J.M. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol. Cell. Biol. 2007, 27, 7802–7815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franz, A.; Shlyueva, D.; Brunner, E.; Stark, A.; Basler, K. Probing the canonicity of the Wnt/Wingless signaling pathway. PLoS Genet. 2017, 13, e1006700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.U.; Blauwkamp, T.A.; Burby, P.E.; Cadigan, K.M. Wnt-mediated repression via bipartite DNA recognition by TCF in the Drosophila hematopoietic system. PLoS Genet. 2014, 10, e1004509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, Q.; Mongan, M.; Wang, J.; Xia, Y. Repression of MAP3K1 expression and JNK activity by canonical Wnt signaling. Dev. Biol. 2018, 440, 129–136. [Google Scholar] [CrossRef]
- Hill, T.P.; Später, D.; Taketo, M.M.; Birchmeier, W.; Hartmann, C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev. Cell 2005, 8, 727–738. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.C.; Kim, K. Identification of MYCBP as a β-catenin/LEF-1 target using DNA microarray analysis. Life Sci. 2005, 77, 1249–1262. [Google Scholar] [CrossRef]
- Kahler, R.A.; Westendorf, J.J. Lymphoid enhancer factor-1 and β-catenin inhibit Runx2-dependent transcriptional activation of the osteocalcin promoter. J. Biol. Chem. 2003, 278, 11937–11944. [Google Scholar] [CrossRef] [Green Version]
- Smartt, H.J.M.; Greenhough, A.; Ordóñez-Morán, P.; Talero, E.; Cherry, C.A.; Wallam, C.A.; Parry, L.; Al Kharusi, M.; Roberts, H.R.; Mariadason, J.M.; et al. β-catenin represses expression of the tumour suppressor 15-prostaglandin dehydrogenase in the normal intestinal epithelium and colorectal tumour cells. Gut 2012, 61, 1306–1314. [Google Scholar] [CrossRef]
- Benchabane, H.; Xin, N.; Tian, A.; Hafler, B.P.; Nguyen, K.; Ahmed, A.; Ahmed, Y. Jerky/Earthbound facilitates cell-specific Wnt/Wingless signalling by modulating β-catenin-TCF activity. EMBO J. 2011, 30, 1444–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, A.; Benchabane, H.; Wang, Z.; Zimmerman, C.; Xin, N.; Perochon, J.; Kalna, G.; Sansom, O.J.; Cheng, C.; Cordero, J.B.; et al. Intestinal stem cell overproliferation resulting from inactivation of the APC tumor suppressor requires the transcription cofactors Earthbound and Erect wing. PLoS Genet. 2017, 13, e1006870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, N.; Benchabane, H.; Tian, A.; Nguyen, K.; Klofas, L.; Ahmed, Y. Erect Wing facilitates context-dependent Wnt/Wingless signaling by recruiting the cell-specific Armadillo-TCF adaptor Earthbound to chromatin. Dev. Camb. Engl. 2011, 138, 4955–4967. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anthony, C.C.; Robbins, D.J.; Ahmed, Y.; Lee, E. Nuclear Regulation of Wnt/β-Catenin Signaling: It’s a Complex Situation. Genes 2020, 11, 886. https://doi.org/10.3390/genes11080886
Anthony CC, Robbins DJ, Ahmed Y, Lee E. Nuclear Regulation of Wnt/β-Catenin Signaling: It’s a Complex Situation. Genes. 2020; 11(8):886. https://doi.org/10.3390/genes11080886
Chicago/Turabian StyleAnthony, Christin C., David J. Robbins, Yashi Ahmed, and Ethan Lee. 2020. "Nuclear Regulation of Wnt/β-Catenin Signaling: It’s a Complex Situation" Genes 11, no. 8: 886. https://doi.org/10.3390/genes11080886
APA StyleAnthony, C. C., Robbins, D. J., Ahmed, Y., & Lee, E. (2020). Nuclear Regulation of Wnt/β-Catenin Signaling: It’s a Complex Situation. Genes, 11(8), 886. https://doi.org/10.3390/genes11080886