The Impact of Coilin Nonsynonymous SNP Variants E121K and V145I on Cell Growth and Cajal Body Formation: The First Characterization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Cell Culture and Transfection
2.3. Immunofluorescence Microscopy
2.4. Microtubule Regrowth Assay
2.5. Cell Cycle Analysis
2.6. Cell Proliferation Assay
2.7. Protein Expression Examination
2.8. Gene Expression Examination
2.9. Protein Structure Prediction and Structural Bioinformatic Analysis
2.10. Clinical Bioinformatic Analysis
2.11. Statistical Analysis
3. Results
3.1. Global Distribution of the Coilin Nonsynonymous SNPs (rs116022828 and rs61731978)
3.2. Establishment of HeLa Cell Lines Expressing Coilin WT or SNP Variants
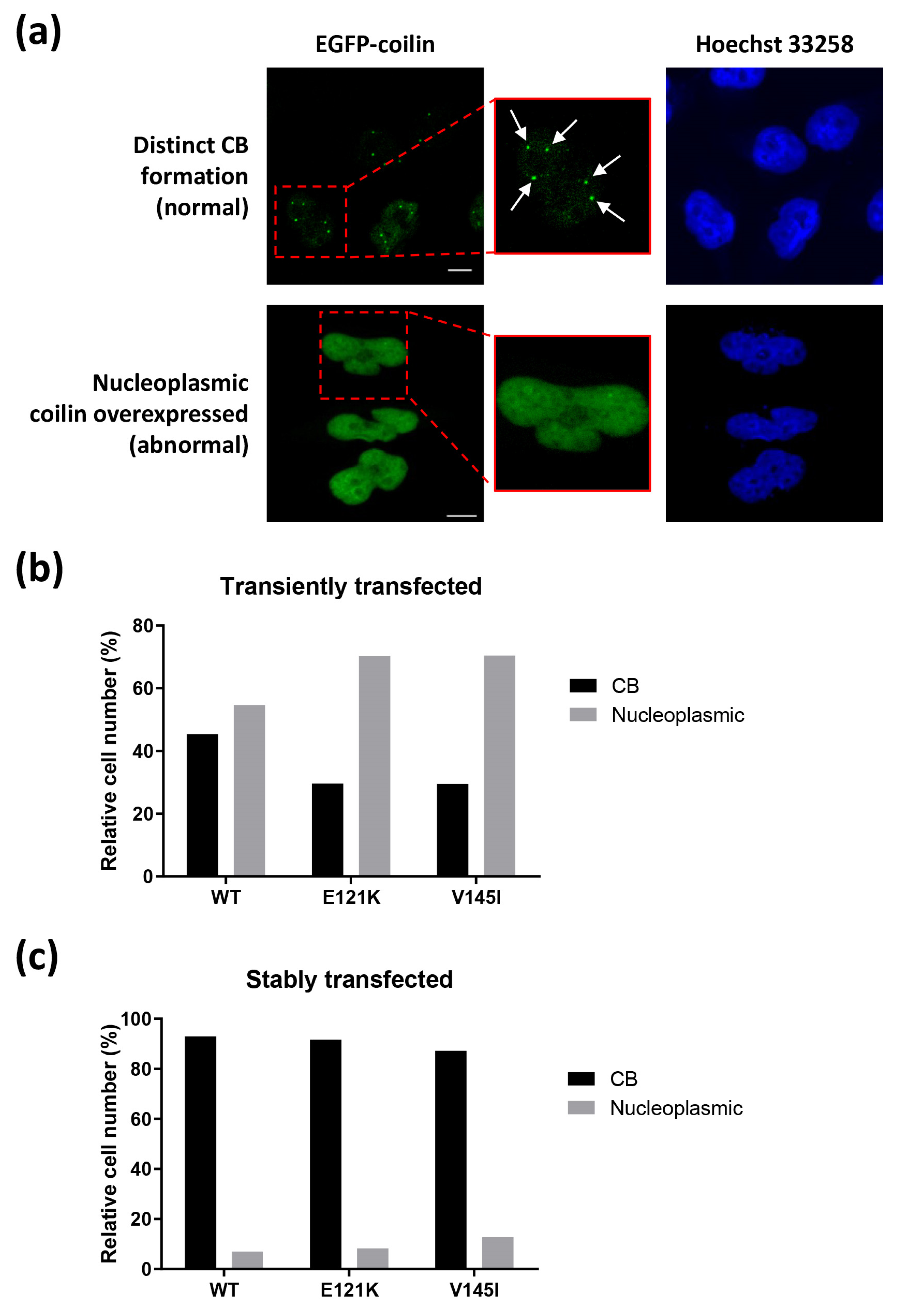
3.3. Effects of Transiently and Stably Transfected Coilin SNP Variant Cells on CB Formation
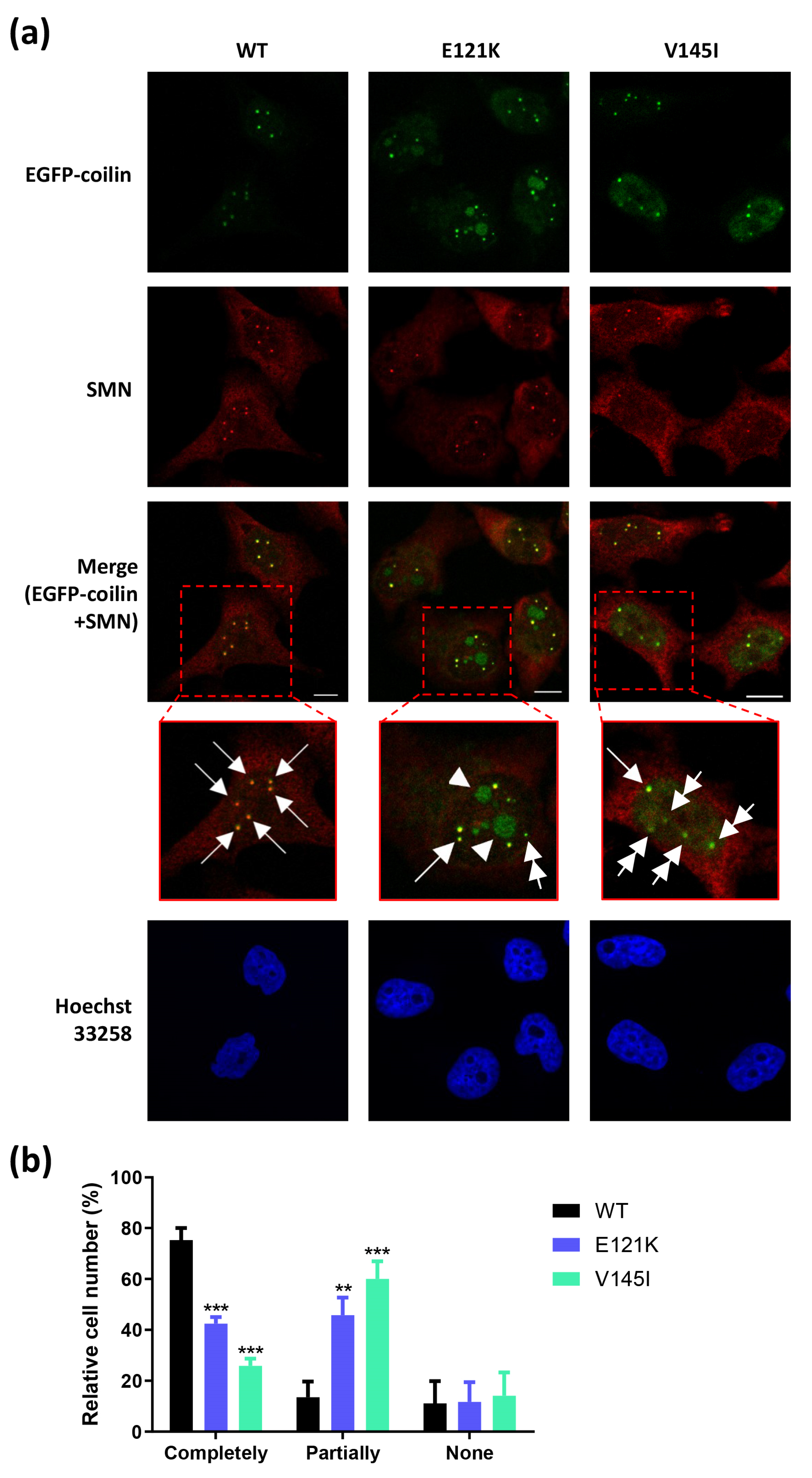
3.4. Effects of Coilin SNP Variants on Subcellular Co-Localization with SMN
3.5. Effects of Coilin SNP Variants on Microtubule Formation, Cell Cycle, and Cell Proliferation
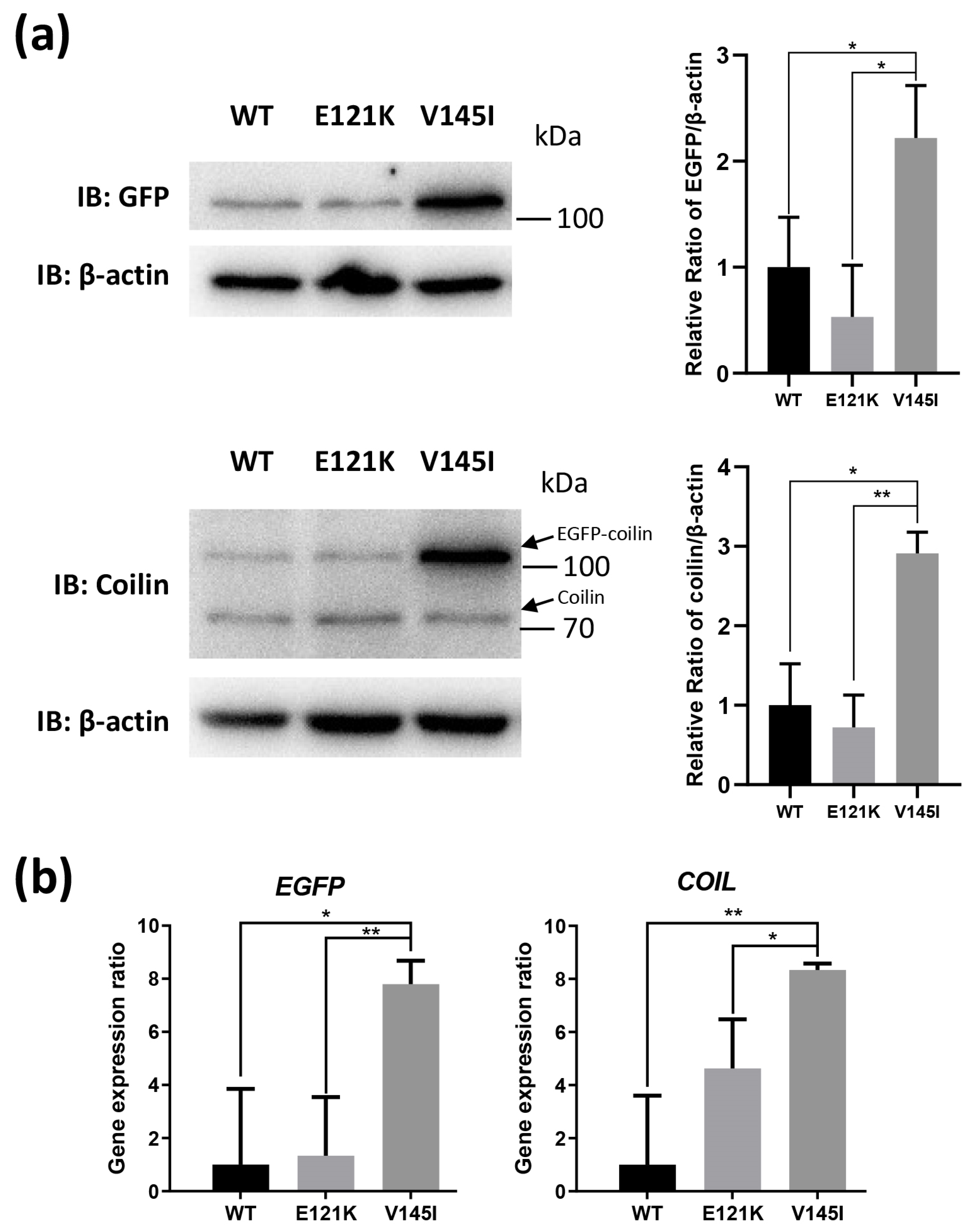
3.6. Effects of Coilin SNP Variants on Coilin Expression
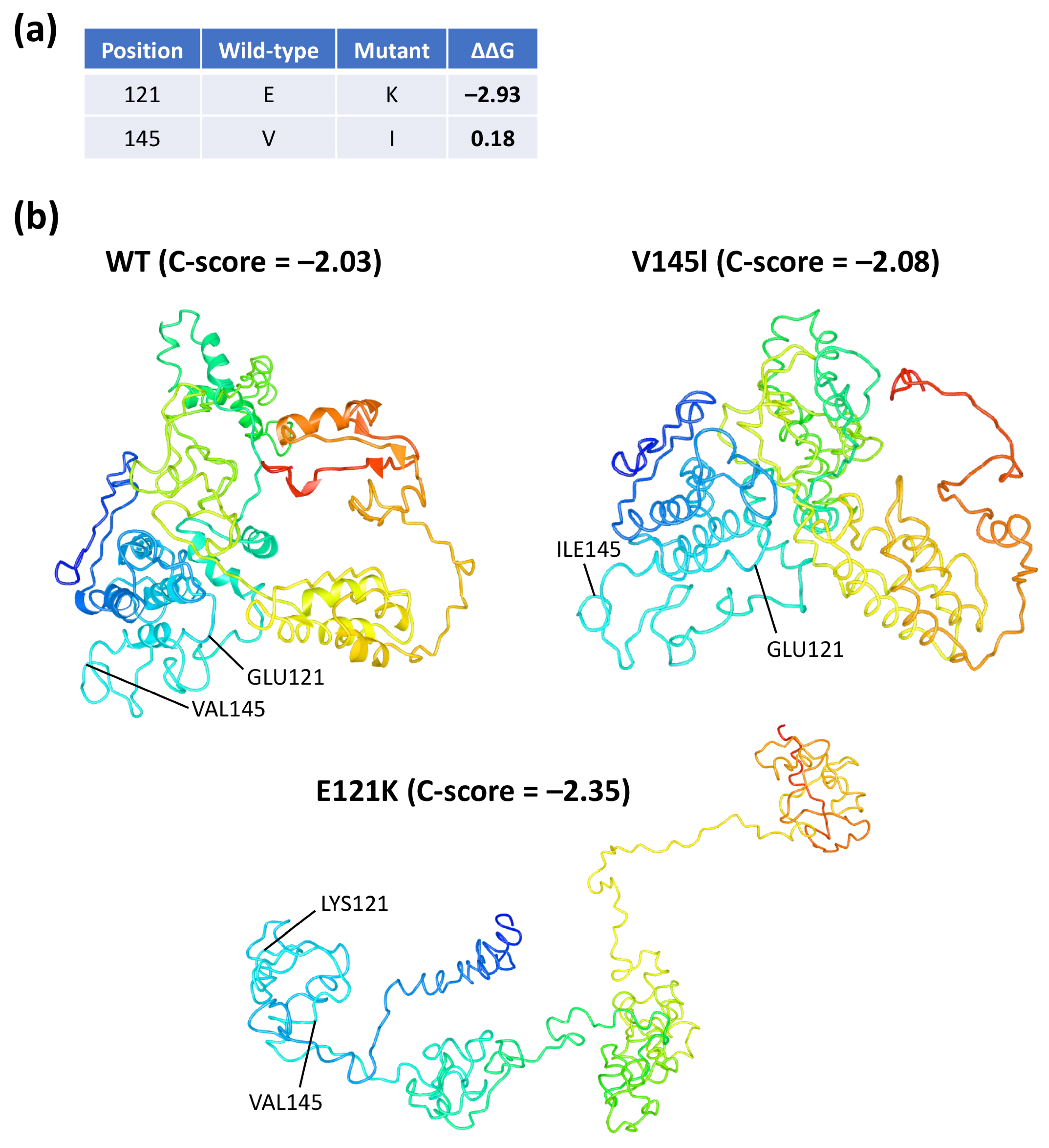
3.7. In Silico Three-Dimensional Modeling and Bioinformatic Analysis of Coilin WT and SNP Variants
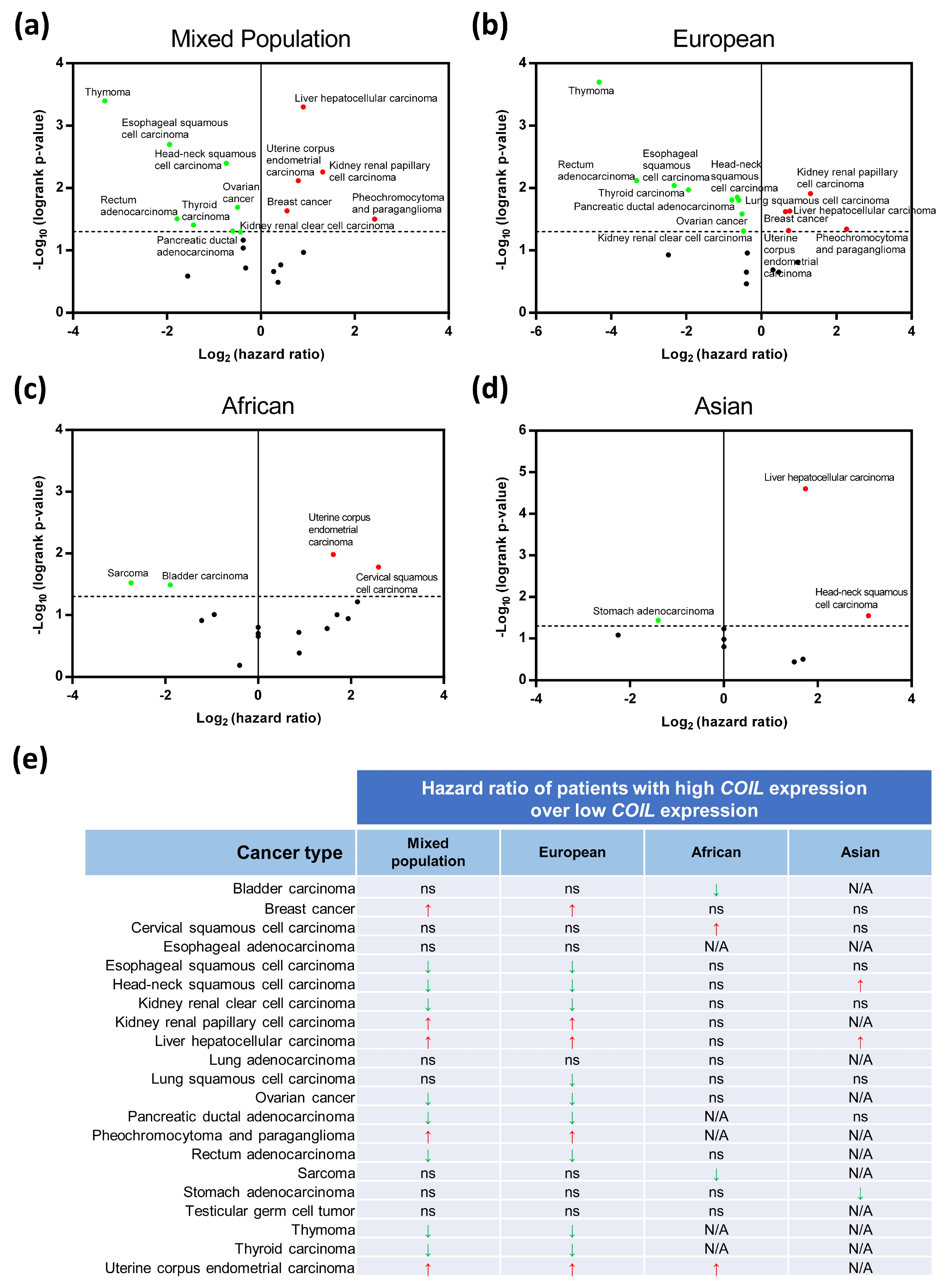
3.8. Clinical Bioinformatic Analysis: Relationships Between Coilin Expression and Cancer Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cioce, M.; Lamond, A.I. Cajal bodies: A long history of discovery. Annu. Rev. Cell Dev. Biol. 2005, 21, 105–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matera, A.G. Drosophila Cajal bodies: Accessories not included. J. Cell Biol. 2006, 172, 791–793. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, J.E.; Ajuh, P.; Lamond, A.I. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J. Cell Sci. 2001, 114, 4407–4419. [Google Scholar] [PubMed]
- Trinkle-Mulcahy, L.; Sleeman, J.E. The Cajal body and the nucleolus: In a relationship or It‘s complicated? RNA Biol. 2017, 14, 739–751. [Google Scholar] [CrossRef] [Green Version]
- Andrade, L.E.; Tan, E.M.; Chan, E.K. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc. Natl. Acad. Sci. USA 1993, 90, 1947–1951. [Google Scholar] [CrossRef] [Green Version]
- Chan, E.K.; Takano, S.; Andrade, L.E.; Hamel, J.C.; Matera, A.G. Structure, expression and chromosomal localization of human p80-coilin gene. Nucleic Acids Res. 1994, 22, 4462–4469. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, R.; Pena, E.; Navascues, J.; Casafont, I.; Lafarga, M.; Berciano, M.T. cAMP-dependent reorganization of the Cajal bodies and splicing machinery in cultured Schwann cells. Glia 2002, 40, 378–388. [Google Scholar] [CrossRef]
- Hebert, M.D.; Poole, A.R. Towards an understanding of regulating Cajal body activity by protein modification. RNA Biol. 2017, 14, 761–778. [Google Scholar] [CrossRef] [Green Version]
- Hebert, M.D.; Matera, A.G. Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell 2000, 11, 4159–4171. [Google Scholar] [CrossRef] [Green Version]
- Goto, N.; Sugiura, K.; Ogawa, Y.; Watanabe, A.; Onouchi, H.; Tomita, Y.; Muro, Y. Anti-p80 coilin autoantibodies react with a conserved epitope and are associated with anti-DFS70/LEDGF autoantibodies. J. Autoimmun. 2006, 26, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell 2013, 50, 613–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanbhag, R.; Kurabi, A.; Kwan, J.J.; Donaldson, L.W. Solution structure of the carboxy-terminal Tudor domain from human Coilin. FEBS Lett. 2010, 584, 4351–4356. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Bhatia, K.; Kannan, A.; Gangwani, L. Molecular mechanisms of neurodegeneration in spinal muscular atrophy. J. Exp. Neurosci. 2016, 10, JEN-S33122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henriksson, S.; Farnebo, M. On the road with WRAP53β: Guardian of Cajal bodies and genome integrity. Front. Genet. 2015, 6, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coady, T.H.; Lorson, C.L. SMN in spinal muscular atrophy and snRNP biogenesis. WIREs RNA 2011, 2, 546–564. [Google Scholar] [CrossRef]
- Tapia, O.; Bengoechea, R.; Berciano, M.T.; Lafarga, M. Nucleolar targeting of coilin is regulated by its hypomethylation state. Chromosoma 2010, 119, 527–540. [Google Scholar] [CrossRef]
- Willsey, A.J. Single-Nucleotide Polymorphism. In Encyclopedia of Autism Spectrum Disorders; Volkmar, F.R., Ed.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Kang, S.; Roh, J.W.; Kim, J.W. Single nucleotide polymorphism: A new risk factor for endometrial cancer? Future Oncol. 2005, 1, 323–330. [Google Scholar] [CrossRef]
- Shastry, B.S. SNPs: Impact on gene function and phenotype. Methods Mol. Biol. 2009, 578, 3–22. [Google Scholar]
- Hua, L.; Yang, Z.; Liu, H. A comparison of contributions to disease phenotype between damaging and benign non-synonymous SNPs. In Proceedings of the 2012 5th International Conference on BioMedical Engineering and Informatics, Chongqing, China, 16–18 October 2012; pp. 1133–1137. [Google Scholar]
- Bessenyei, B.; Marka, M.; Urban, L.; Zeher, M.; Semsei, I. Single nucleotide polymorphisms: Aging and diseases. Biogerontology 2004, 5, 291–303. [Google Scholar] [CrossRef]
- Chang, L.C.; Vural, S.; Sonkin, D. Detection of homozygous deletions in tumor-suppressor genes ranging from dozen to hundreds nucleotides in cancer models. Hum. Mutat. 2017, 38, 1449–1453. [Google Scholar] [CrossRef] [PubMed]
- Samuel, N.; Id Said, B.; Guha, T.; Novokmet, A.; Li, W.; Silwal-Pandit, L.; Borrsen-Dale, A.L.; Langerod, A.; Hudson, T.J.; Malkin, D. Assessment of TP53 polymorphisms and MDM2 SNP309 in premenopausal breast cancer risk. Hum. Mutat. 2017, 38, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Katsonis, P.; Koire, A.; Wilson, S.J.; Hsu, T.K.; Lua, R.C.; Wilkins, A.D.; Lichtarge, O. Single nucleotide variations: Biological impact and theoretical interpretation. Protein Sci. 2014, 23, 1650–1666. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.W.; Liang, Z.L.; Yao, Y.; Wu, D.D.; Mo, H.Y.; Gu, J.; Chiu, J.F.; Xu, Y.M.; Lau, A.T.Y. Lasting DNA damage and aberrant DNA repair gene expression profile are associated with post-chronic cadmium exposure in human bronchial epithelial cells. Cells 2019, 8, 842. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.L.; Wu, D.D.; Yao, Y.; Yu, F.Y.; Yang, L.; Tan, H.W.; Hylkema, M.N.; Rots, M.G.; Xu, Y.M.; Lau, A.T.Y. Epiproteome profiling of cadmium-transformed human bronchial epithelial cells by quantitative histone post-translational modification-enzyme-linked immunosorbent assay. J. Appl. Toxicol. 2018, 38, 888–895. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Quan, L.; Lv, Q.; Zhang, Y. STRUM: Structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics 2016, 32, 2936–2946. [Google Scholar] [CrossRef] [Green Version]
- Györffy, B.; Surowiak, P.; Budczies, J.; Lánczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef] [Green Version]
- Hebert, M.D. Signals controlling Cajal body assembly and function. Int. J. Biochem. Cell Biol. 2013, 45, 1314–1317. [Google Scholar] [CrossRef] [Green Version]
- Hebert, M.D.; Shpargel, K.B.; Ospina, J.K.; Tucker, K.E.; Matera, A.G. Coilin methylation regulates nuclear body formation. Dev. Cell 2002, 3, 329–337. [Google Scholar] [CrossRef]
- Lyon, C.E.; Bohmann, K.; Sleeman, J.; Lamond, A.I. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp. Cell Res. 1997, 230, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Morse, R.; Shaw, D.J.; Todd, A.G.; Young, P.J. Targeting of SMN to Cajal bodies is mediated by self-association. Hum. Mol. Genet. 2007, 16, 2349–2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawyer, I.A.; Sturgill, D.; Dundr, M. Membraneless nuclear organelles and the search for phases within phases. WIREs RNA 2019, 10, e1514. [Google Scholar] [CrossRef]
- Tucker, K.E.; Berciano, M.T.; Jacobs, E.Y.; LePage, D.F.; Shpargel, K.B.; Rossire, J.J.; Chan, E.K.; Lafarga, M.; Conlon, R.A.; Matera, A.G. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 2001, 154, 293–307. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gall, J.G. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 2003, 4, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.E. The Cajal body. Mol. Cell Res. 2008, 1783, 2108–2115. [Google Scholar] [CrossRef] [Green Version]
- Selenko, P.; Sprangers, R.; Stier, G.; Buhler, D.; Fischer, U.; Sattler, M. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Mol. Biol. 2001, 8, 27–31. [Google Scholar]
- Lafarga, M.; Tapia, O.; Romero, A.M.; Berciano, M.T. Cajal bodies in neurons. RNA Biol. 2017, 14, 712–725. [Google Scholar] [CrossRef] [Green Version]
- Venteicher, A.S.; Abreu, E.B.; Meng, Z.; McCann, K.E.; Terns, R.M.; Veenstra, T.D.; Terns, M.P.; Artandi, S.E. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 2009, 323, 644–648. [Google Scholar] [CrossRef] [Green Version]
- Freed, E.F.; Bleichert, F.; Dutca, L.M.; Baserga, S.J. When ribosomes go bad: Diseases of ribosome biogenesis. Mol. BioSyst. 2010, 6, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Porokhovnik, L.N.; Lyapunova, N.A. Dosage effects of human ribosomal genes (rDNA) in health and disease. Chromosome Res. 2019, 27, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Isaac, C.; Yang, Y.; Meier, U.T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998, 142, 319–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprung, R.; Chen, Y.; Zhang, K.; Cheng, D.; Zhang, T.; Peng, J.; Zhao, Y. Identification and validation of eukaryotic aspartate and glutamate methylation in proteins. J. Proteome Res. 2008, 7, 1001–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.M.; Du, J.Y.; Lau, A.T. Posttranslational modifications of human histone H3: An update. Proteomics 2014, 14, 2047–2060. [Google Scholar] [CrossRef]
- Boisvert, F.M.; Cote, J.; Boulanger, M.C.; Cleroux, P.; Bachand, F.; Autexier, C.; Richard, S. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J. Cell Biol. 2002, 159, 957–969. [Google Scholar] [CrossRef] [Green Version]
- Machyna, M.; Neugebauer, K.M.; Stanek, D. Coilin: The first 25 years. RNA Biol. 2015, 12, 590–596. [Google Scholar] [CrossRef] [Green Version]
- Carrero, Z.I.; Velma, V.; Douglas, H.E.; Hebert, M.D. Coilin phosphomutants disrupt Cajal body formation, reduce cell proliferation and produce a distinct coilin degradation product. PLoS ONE 2011, 6, e25743. [Google Scholar] [CrossRef] [Green Version]
- Whittom, A.A.; Xu, H.; Hebert, M.D. Coilin levels and modifications influence artificial reporter splicing. Cell Mol. Life Sci. 2008, 65, 1256–1271. [Google Scholar] [CrossRef]
- Walker, M.P.; Tian, L.; Matera, A.G. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS ONE 2009, 4, e6171. [Google Scholar] [CrossRef]
- Strzelecka, M.; Trowitzsch, S.; Weber, G.; Luhrmann, R.; Oates, A.C.; Neugebauer, K.M. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat. Struct. Mol. Biol. 2010, 17, 403–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcus, J.H.; Novembre, J. Visualizing the geography of genetic variants. Bioinformatics 2017, 33, 594–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Y.; Tan, H.W.; Liang, Z.-L.; Wu, G.-Q.; Xu, Y.-M.; Lau, A.T.Y. The Impact of Coilin Nonsynonymous SNP Variants E121K and V145I on Cell Growth and Cajal Body Formation: The First Characterization. Genes 2020, 11, 895. https://doi.org/10.3390/genes11080895
Yao Y, Tan HW, Liang Z-L, Wu G-Q, Xu Y-M, Lau ATY. The Impact of Coilin Nonsynonymous SNP Variants E121K and V145I on Cell Growth and Cajal Body Formation: The First Characterization. Genes. 2020; 11(8):895. https://doi.org/10.3390/genes11080895
Chicago/Turabian StyleYao, Yue, Heng Wee Tan, Zhan-Ling Liang, Gao-Qi Wu, Yan-Ming Xu, and Andy T. Y. Lau. 2020. "The Impact of Coilin Nonsynonymous SNP Variants E121K and V145I on Cell Growth and Cajal Body Formation: The First Characterization" Genes 11, no. 8: 895. https://doi.org/10.3390/genes11080895




