Efficient CRISPR/Cas9-Mediated Knockout of an Endogenous PHYTOENE DESATURASE Gene in T1 Progeny of Apomictic Hieracium Enables New Strategies for Apomixis Gene Identification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth
2.2. Identification of an Hieracium Phytoene Desaturase (HPDS) Gene Ortholog and Confirmation of Expressed Leaf cDNA Sequence
2.3. Development of a CRISPR/Cas9 Gene Editing Construct Targeting Hieracium PDS
2.4. In Vitro Cleavage Assay
2.5. Plant Transformation
2.6. Identification of Edits in HPDS Genes of Transformed Hieracium Plants by Sequencing
2.7. Analyses of T1 Progeny Derived from Chimeric Apomictic T0 Transformed Plants
3. Results
3.1. PDS Edited Apomictic Hieracium T0 Plants Show Dwarfism, Albinism and Chimeric Phenotypes in Vegetative and Floral Tissues
3.2. CRIPSR/Cas9-Induced Indels and Deletions Can Be Rapidly Detected in Transgenic Hieracium Using PCR and Direct Sequencing
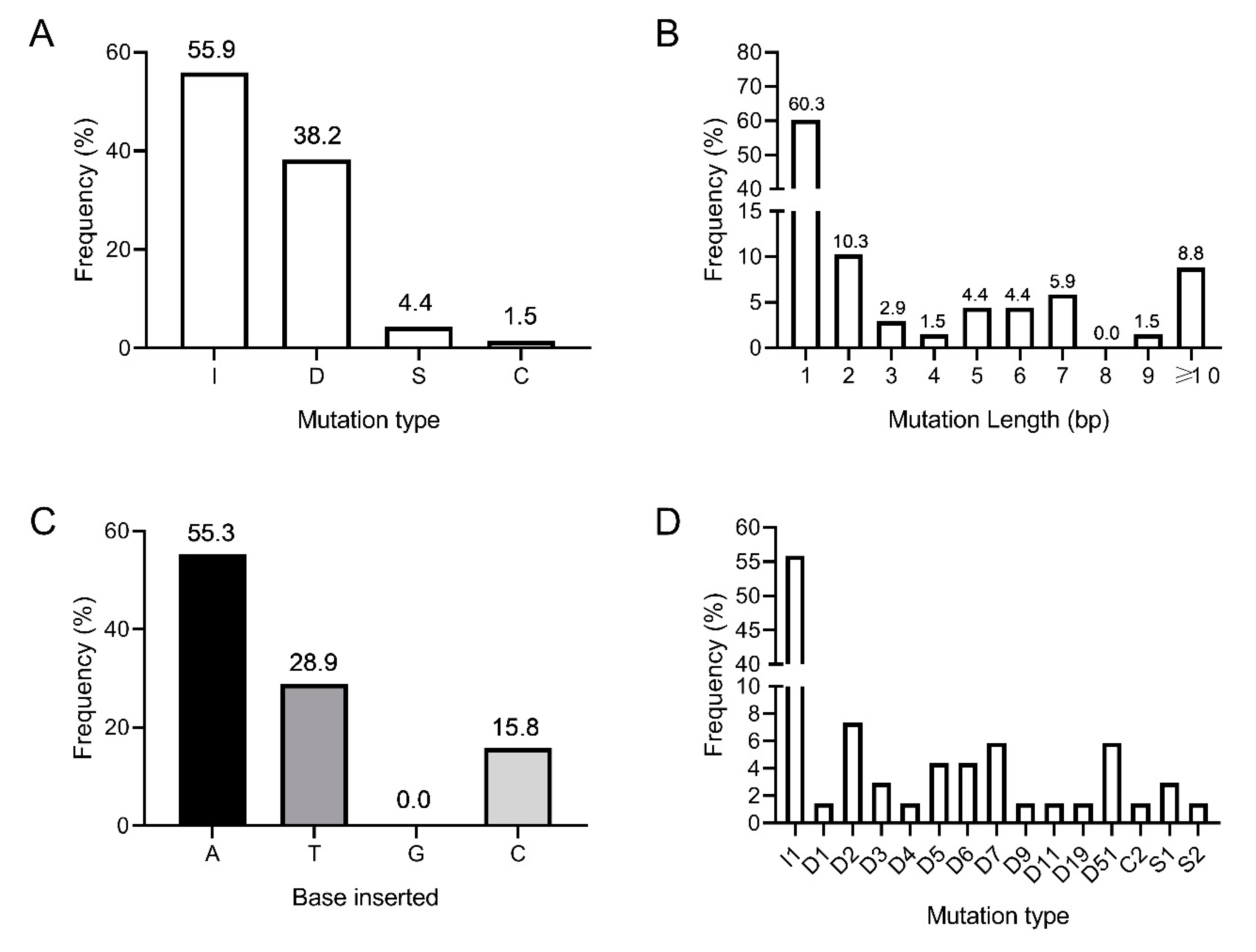
3.3. Amplicon Deep Sequencing Reveals a Wide Range of CRISPR/Cas9-Induced Indels in Hieracium Primary Transformants
3.4. Editing in Chimeric Hieracium Continues through Apomictic Seed Formation and Is Inherited in the Next Generation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Conner, J.A.; Ozias-Akins, P. Apomixis: Engineering the ability to harness hybrid vigor in crop plants. In Plant Germline Development. Methods in Molecular Biology; Schmidt, A., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1669, pp. 17–34. [Google Scholar]
- Hand, M.L.; Koltunow, A.M. The genetic control of apomixis: Asexual seed formation. Genetics 2014, 197, 441–450. [Google Scholar] [CrossRef] [Green Version]
- Koltunow, A.M.; Grossniklaus, U. Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 2003, 54, 547–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sailer, C.; Schmid, B.; Grossniklaus, U. Apomixis allows the transgenerational fixation of phenotypes in hybrid plants. Curr. Biol. 2016, 26, 331–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hörandl, E.; Temsch, E.M. Introgression of apomixis into sexual species is inhibited by mentor effects and ploidy barriers in the Ranunculus auricomus complex. Ann. Bot. 2009, 104, 81–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conner, J.A.; Mookkan, M.; Huo, H.; Chae, K.; Ozias-Akins, P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. USA 2015, 112, 11205–11210. [Google Scholar] [CrossRef] [Green Version]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Jia, H.; Wang, N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 2014, 9, e93806. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef] [Green Version]
- Hand, M.L.; Vit, P.; Krahulcova, A.; Johnson, S.D.; Oelkers, K.; Siddons, H.; Chrtek, J.; Fehrer, J.; Koltunow, A.M.G. Evolution of apomixis loci in Pilosella and Hieracium (Asteraceae) inferred from the conservation of apomixis-linked markers in natural and experimental populations. Heredity 2015, 114, 17–26. [Google Scholar] [CrossRef]
- Koltunow, A.M.G.; Johnson, S.D.; Rodrigues, J.C.M.; Okada, T.; Hu, Y.; Tsuchiya, T.; Wilson, S.; Fletcher, P.; Ito, K.; Suzuki, G.; et al. Sexual reproduction is the default mode in apomictic Hieracium subgenus Pilosella, in which two dominant loci function to enable apomixis. Plant J. 2011, 66, 890–902. [Google Scholar] [CrossRef] [Green Version]
- Catanach, A.S.; Erasmuson, S.K.; Podivinsky, E.; Jordan, B.R.; Bicknell, R. Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc. Natl. Acad. Sci. USA 2006, 103, 18650–18655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, S.T.; Johnson, S.D.; Eichmann, J.; Koltunow, A.M.G. Genetic analyses of the inheritance and expressivity of autonomous endosperm formation in Hieracium with different modes of embryo sac and seed formation. Ann. Bot. 2017, 119, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, D.; Johnson, S.D.; Henderson, S.T.; Koltunow, A.M. Genetic separation of autonomous endosperm formation (AutE) from the two other components of apomixis in Hieracium. Plant Reprod. 2013, 26, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Ito, K.; Johnson, S.D.; Oelkers, K.; Suzuki, G.; Houben, A.; Mukai, Y.; Koltunow, A.M. Chromosomes carrying meiotic avoidance loci in three apomictic eudicot Hieracium subgenus Pilosella species share structural features with two monocot apomicts. Plant Physiol. 2011, 157, 1327–1341. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, Y.; Conner, J.A.; Goel, S.; Morishige, D.T.; Mullet, J.E.; Hanna, W.W.; Ozias-Akins, P. High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol. 2004, 134, 1733–1741. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, Y.; Hanna, W.W.; Ozias-Akins, P. High-resolution physical mapping reveals that the apospory-specific genomic region (ASGR) in Cenchrus ciliaris is located on a heterochromatic and hemizygous region of a single chromosome. Theor. Appl. Genet. 2005, 111, 1042–1051. [Google Scholar] [CrossRef]
- Bräuning, S.; Catanach, A.; Lord, J.M.; Bicknell, R.; Macknight, R.C. Comparative transcriptome analysis of the wild-type model apomict Hieracium praealtum and its loss of parthenogenesis (lop) mutant. BMC Plant Biol. 2018, 18, 206. [Google Scholar] [CrossRef] [Green Version]
- Rabiger, D.S.; Taylor, J.M.; Spriggs, A.; Hand, M.L.; Henderson, S.T.; Johnson, S.D.; Oelkers, K.; Hrmova, M.; Saito, K.; Suzuki, G.; et al. Generation of an integrated Hieracium genomic and transcriptomic resource enables exploration of small RNA pathways during apomixis initiation. BMC Biol. 2016, 14, 86. [Google Scholar] [CrossRef]
- Juranić, M.; Tucker, M.R.; Schultz, C.J.; Shirley, N.J.; Taylor, J.M.; Spriggs, A.; Johnson, S.D.; Bulone, V.; Koltunow, A.M. Asexual female gametogenesis involves contact with a sexually-fated megaspore in apomictic Hieracium. Plant Physiol. 2018, 177, 1027–1049. [Google Scholar] [CrossRef] [Green Version]
- Okada, T.; Hu, Y.; Tucker, M.R.; Taylor, J.M.; Johnson, S.D.; Spriggs, A.; Tsuchiya, T.; Oelkers, K.; Rodrigues, J.C.M.; Koltunow, A.M.G. Enlarging cells initiating apomixis in Hieracium praealtum transition to an embryo sac program prior to entering mitosis. Plant Physiol. 2013, 163, 216–231. [Google Scholar] [CrossRef] [Green Version]
- Kotani, Y.; Henderson, S.T.; Suzuki, G.; Johnson, S.D.; Okada, T.; Siddons, H.; Mukai, Y.; Koltunow, A.M.G. The LOSS OF APOMEIOSIS (LOA) locus in Hieracium praealtum can function independently of the associated large-scale repetitive chromosomal structure. New Phytol. 2014, 201, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Bicknell, R.A.; Borst, N.K. Agrobacterium-mediated transformation of Hieracium aurantiacum. Int. J. Plant Sci. 1994, 155, 467–470. [Google Scholar] [CrossRef]
- Bertier, L.D.; Ron, M.; Huo, H.; Bradford, K.J.; Britt, A.B.; Michelmore, R.W. High-resolution analysis of the efficiency, heritability, and editing outcomes of CRISPR/Cas9-induced modifications of NCED4 in lettuce (Lactuca sativa). G3: Genes|Genomes|Genet. 2018, 8, 1513–1521. [Google Scholar] [CrossRef] [Green Version]
- Shan, S.; Mavrodiev, E.V.; Li, R.; Zhang, Z.; Hauser, B.A.; Soltis, P.S.; Soltis, D.E.; Yang, B. Application of CRISPR/Cas9 to Tragopogon (Asteraceae), an evolutionary model for the study of polyploidy. Mol. Ecol. Resour. 2018, 18, 1427–1443. [Google Scholar] [CrossRef] [PubMed]
- Kishi-Kaboshi, M.; Aida, R.; Sasaki, K. Generation of gene-edited Chrysanthemum morifolium using multicopy transgenes as targets and markers. Plant Cell Physiol. 2017, 58, 216–226. [Google Scholar] [CrossRef] [Green Version]
- Iaffaldano, B.; Zhang, Y.; Cornish, K. CRISPR/Cas9 genome editing of rubber producing dandelion Taraxacum kok-saghyz using Agrobacterium rhizogenes without selection. Ind. Crop. Prod. 2016, 89, 356–362. [Google Scholar] [CrossRef] [Green Version]
- Qin, G.; Gu, H.; Ma, L.; Peng, Y.; Deng, X.W.; Chen, Z.; Qu, L.J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 17, 471–482. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Odipio, J.; Alicai, T.; Ingelbrecht, I.; Nusinow, D.A.; Bart, R.; Taylor, N.J. Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Howells, R.M.; Craze, M.; Bowden, S.; Wallington, E.J. Efficient generation of stable, heritable gene edits in wheat using CRISPR/Cas9. BMC Plant Biol. 2018, 18, 215. [Google Scholar] [CrossRef]
- Naim, F.; Dugdale, B.; Kleidon, J.; Brinin, A.; Shand, K.; Waterhouse, P.; Dale, J. Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 2018. [Google Scholar] [CrossRef] [Green Version]
- Shirasawa, K.; Hand, M.L.; Henderson, S.T.; Okada, T.; Johnson, S.D.; Taylor, J.M.; Spriggs, A.; Siddons, H.; Hirakawa, H.; Isobe, S.; et al. A reference genetic linkage map of apomictic Hieracium species based on expressed markers derived from developing ovule transcripts. Ann. Bot. 2015, 115, 567–580. [Google Scholar] [CrossRef]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arab. Thaliana Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef]
- Pinello, L.; Canver, M.C.; Hoban, M.D.; Orkin, S.H.; Kohn, D.B.; Bauer, D.E.; Yuan, G.-C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016, 34, 695. [Google Scholar] [CrossRef] [Green Version]
- Clement, K.; Rees, H.; Canver, M.C.; Gehrke, J.M.; Farouni, R.; Hsu, J.Y.; Cole, M.A.; Liu, D.R.; Joung, J.K.; Bauer, D.E.; et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019, 37, 224–226. [Google Scholar] [CrossRef]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991, 19, 1349. [Google Scholar] [CrossRef]
- Hsiau, T.; Maures, T.; Waite, K.; Yang, J.; Kelso, R.; Holden, K.; Stoner, R. Inference of CRISPR edits from Sanger trace data. bioRxiv 2018. [Google Scholar] [CrossRef]
- Yang, H.; Wu, J.-J.; Tang, T.; Liu, K.-D.; Dai, C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017, 7, 7489. [Google Scholar] [CrossRef] [Green Version]
- Bicknell, R.A.; Lambie, S.C.; Butler, R.C. Quantification of progeny classes in two facultatively apomictic accessions of Hieracium. Hereditas 2003, 138, 11–20. [Google Scholar] [CrossRef]
- Bicknell, R.A.; Koltunow, A.M. Understanding apomixis: Recent advances and remaining conundrums. Plant Cell 2004, 16, S228–S245. [Google Scholar] [CrossRef] [Green Version]
- Juranić, M.; Johnson, S.D.; Koltunow, A.M. Phenotypic plasticity of aposporous embryo sac development in Hieracium praealtum. Plant Signal. Behav. 2019, 1–4. [Google Scholar] [CrossRef]
- Koltunow, A.M.; Johnson, S.D.; Bicknell, R.A. Apomixis is not developmentally conserved in related, genetically characterized Hieracium plants of varying ploidy. Sex Plant Reprod. 2000, 12, 253–266. [Google Scholar] [CrossRef]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 2016, 6, 31481. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef]
- Koltunow, A.M.G.; Johnson, S.D.; Okada, T. Apomixis in hawkweed: Mendel’s experimental nemesis. J. Exp. Bot. 2011, 62, 1699–1707. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Zhang, J.; Sun, L.; Ma, Y.; Xu, J.; Liang, S.; Deng, J.; Tan, J.; Zhang, Q.; Tu, L.; et al. High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 2018, 16, 137–150. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Shan, S.; Mavrodiev, E.V.; Li, R.; Zhang, Z.; Hauser, B.A.; Soltis, P.S.; Soltis, D.E.; Yang, B. Data from: Application of CRISPR/Cas9 to Tragopogon (Asteraceae), an evolutionary model for the study of polyploidy. Dryad Digit. Repository 2018. [Google Scholar] [CrossRef]
- Kumar, S.; Steche, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]

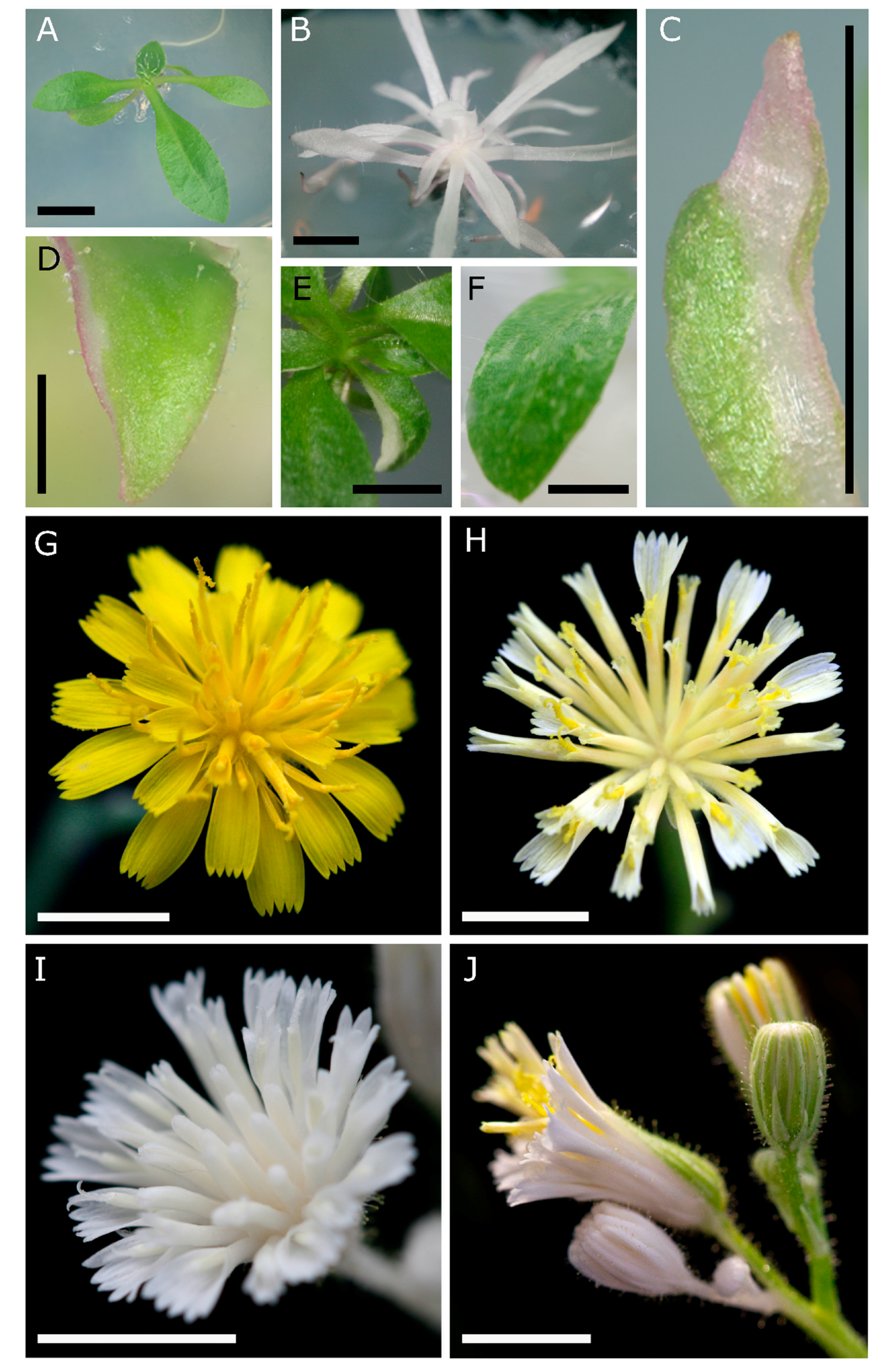



| Experiment 1 | Experiment 2 | Experiment 3 | Combined Total | |||||
|---|---|---|---|---|---|---|---|---|
| No. Plants | % Total | No. Plants | % Total | No. Plants | % Total | No. Plants | % Total | |
| Green | 9 | 64.3 | 6 | 50.0 | 5 | 55. 6 | 20 | 57.1 |
| Albino | 1 | 7.1 | 2 | 16.7 | 1 | 11.1 | 4 | 11.4 |
| Chimera | 4 | 28.6 | 4 | 33.3 | 3 | 33.3 | 11 | 31.4 |
| HPDS Line 1 | HPDS Line 2 | |||
|---|---|---|---|---|
| Progeny | % | Progeny | % | |
| Albino | 95 | 62.9 | 104 | 84.6 |
| Chimera | 56 | 37.1 | 19 | 15.4 |
| Green | 0 | 0 | 0 | 0 |
| Polyembryony | 2 | 1.3 | 7 | 5.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henderson, S.W.; Henderson, S.T.; Goetz, M.; Koltunow, A.M.G. Efficient CRISPR/Cas9-Mediated Knockout of an Endogenous PHYTOENE DESATURASE Gene in T1 Progeny of Apomictic Hieracium Enables New Strategies for Apomixis Gene Identification. Genes 2020, 11, 1064. https://doi.org/10.3390/genes11091064
Henderson SW, Henderson ST, Goetz M, Koltunow AMG. Efficient CRISPR/Cas9-Mediated Knockout of an Endogenous PHYTOENE DESATURASE Gene in T1 Progeny of Apomictic Hieracium Enables New Strategies for Apomixis Gene Identification. Genes. 2020; 11(9):1064. https://doi.org/10.3390/genes11091064
Chicago/Turabian StyleHenderson, Sam W., Steven T. Henderson, Marc Goetz, and Anna M. G. Koltunow. 2020. "Efficient CRISPR/Cas9-Mediated Knockout of an Endogenous PHYTOENE DESATURASE Gene in T1 Progeny of Apomictic Hieracium Enables New Strategies for Apomixis Gene Identification" Genes 11, no. 9: 1064. https://doi.org/10.3390/genes11091064
APA StyleHenderson, S. W., Henderson, S. T., Goetz, M., & Koltunow, A. M. G. (2020). Efficient CRISPR/Cas9-Mediated Knockout of an Endogenous PHYTOENE DESATURASE Gene in T1 Progeny of Apomictic Hieracium Enables New Strategies for Apomixis Gene Identification. Genes, 11(9), 1064. https://doi.org/10.3390/genes11091064






