Global Profiling of Dynamic Alternative Splicing Modulation in Arabidopsis Root upon Ralstonia solanacearum Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and R. solanacearum Infection
2.2. RNA Extraction and Sequencing
2.3. Data Quality Control and Read Mapping
2.4. Analysis of Differential Alternative Splicing Events and Expression Genes
2.5. Function Annotation of RNA Sequence Data
2.6. Splicing Factors/RNA Bindng Poteins(SF/RBPs) Analysis
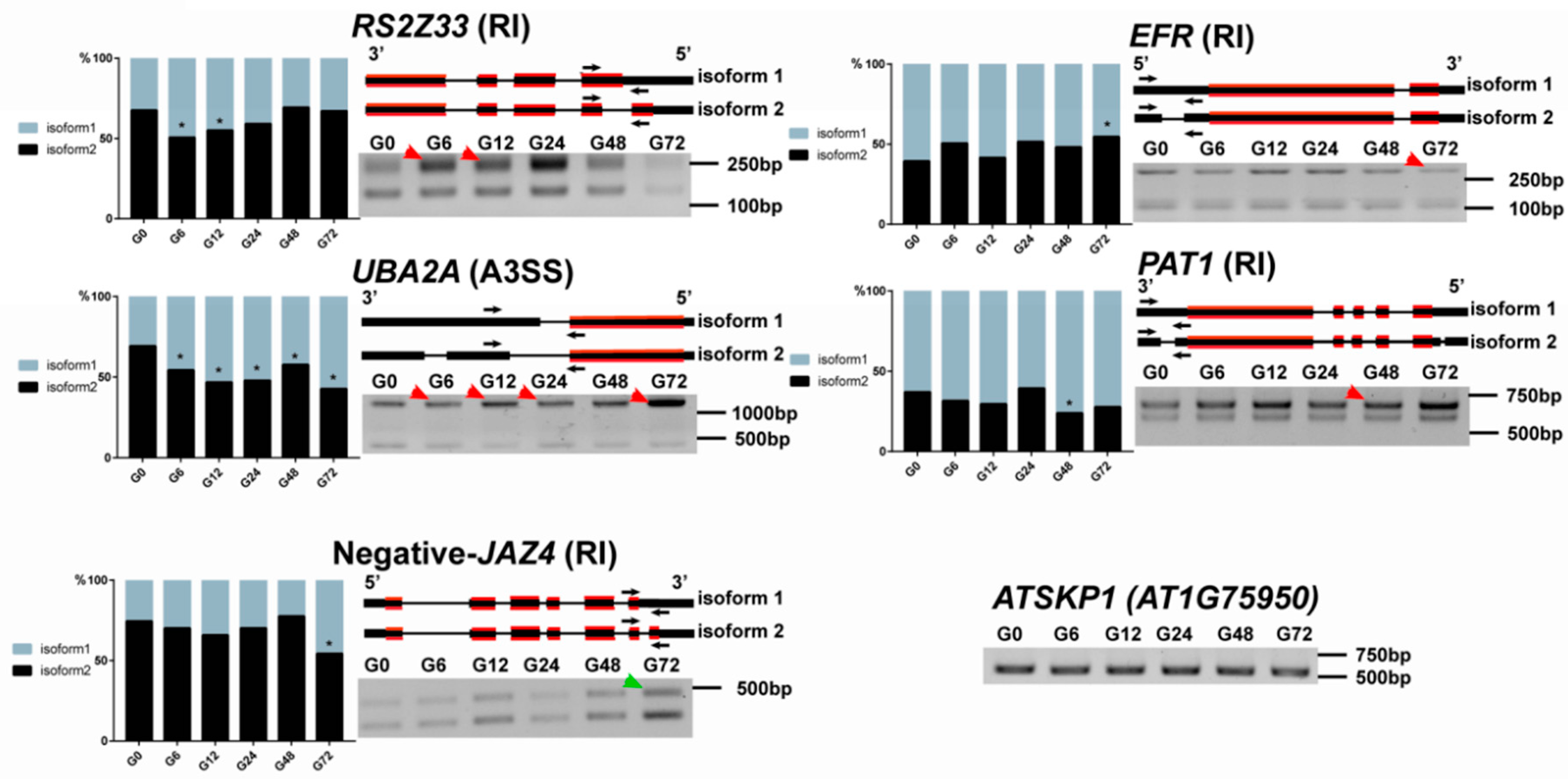
2.7. RT-PCR Validation
3. Results
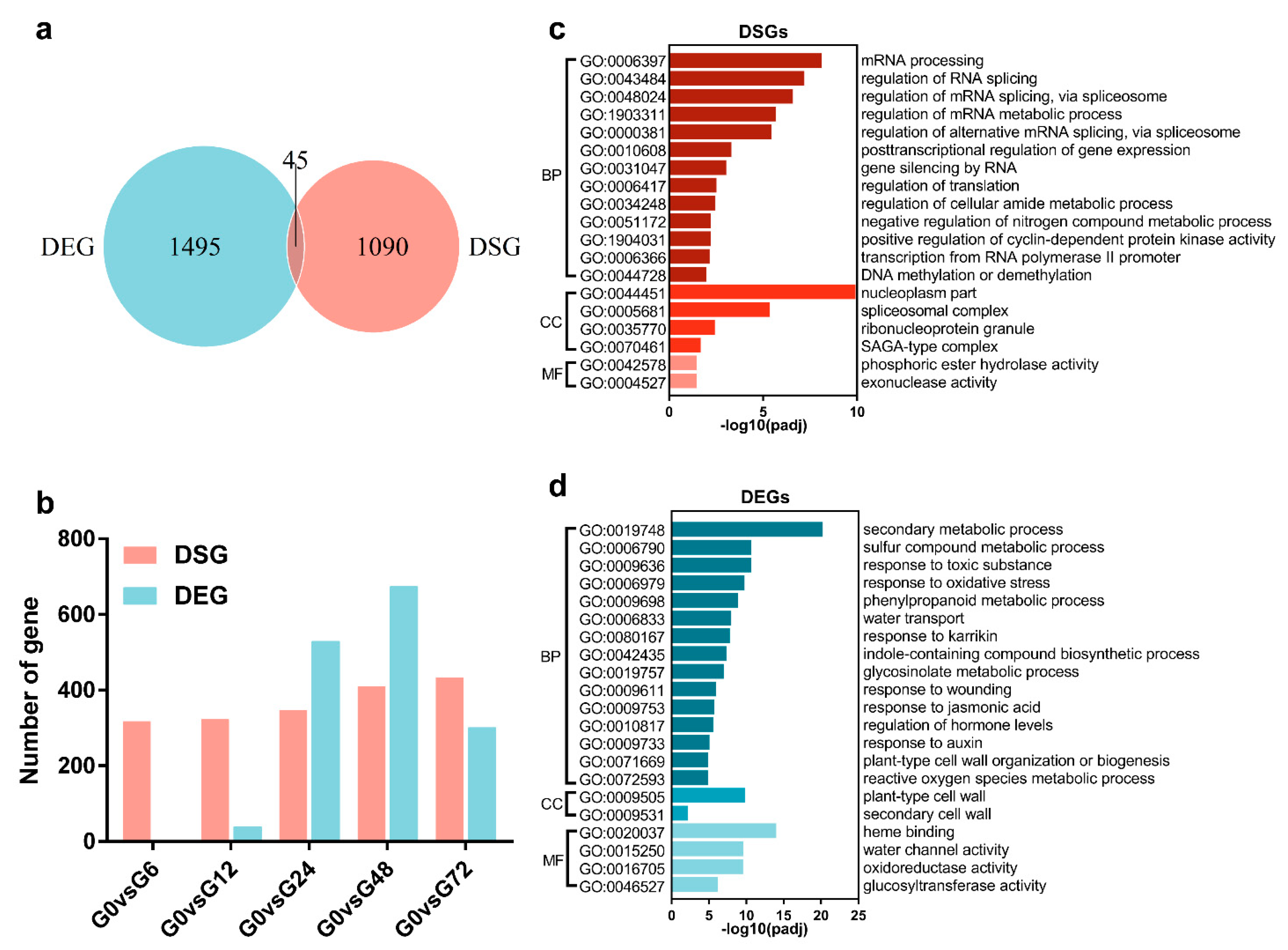
3.1. Identification of DSEs in Arabidopsis Root after GMI1000 Infection
3.2. Comparison of AS and Transcriptional Regulation in Response to R. solanacearum Infection
3.3. Analysis of DSEs in Arabidopsis Root during GMI1000 Infection
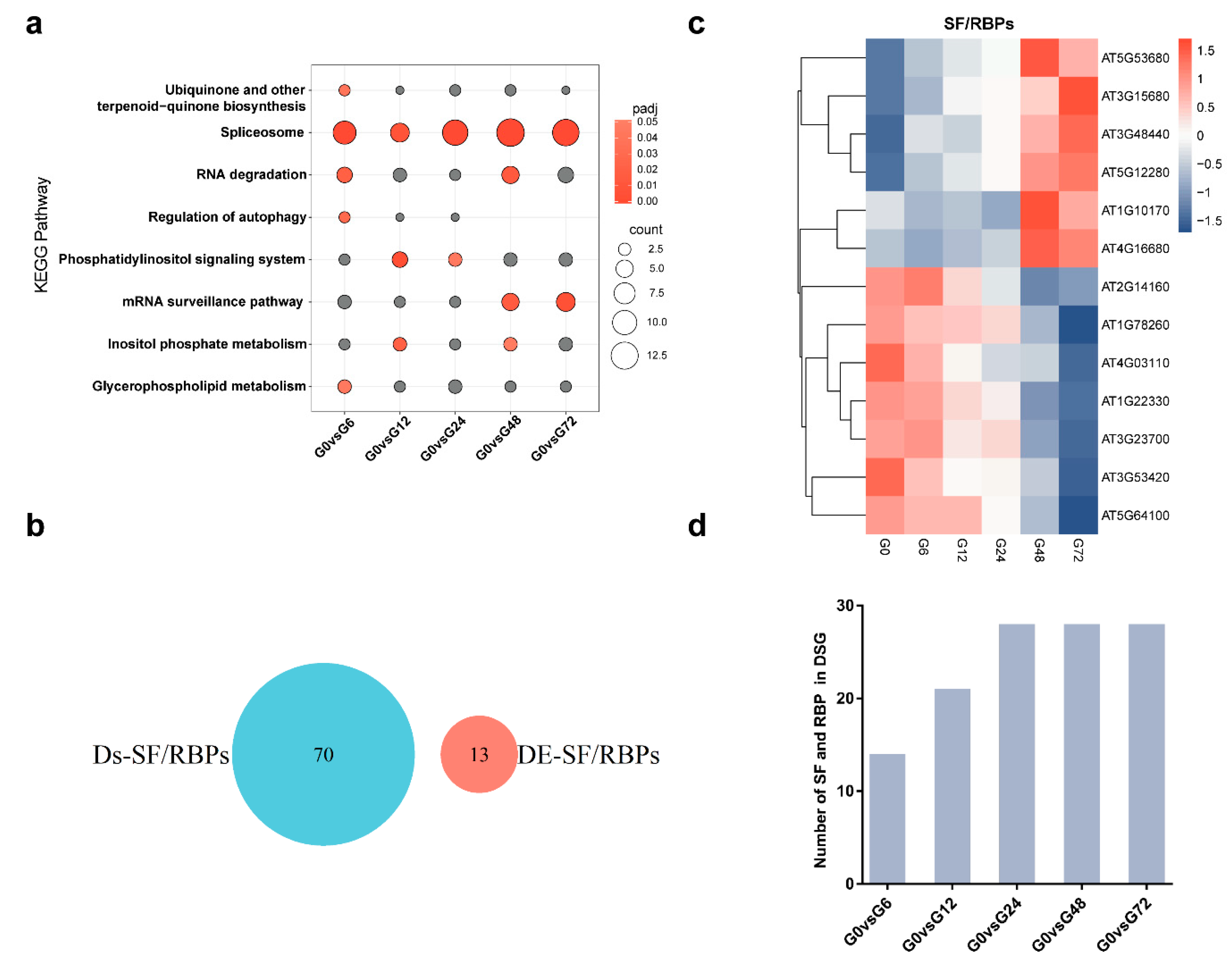
3.4. Splicing Factors and RNA-Binding Proteins are Regulated at AS and Transcriptional Levels Following R. solanacearum Infection
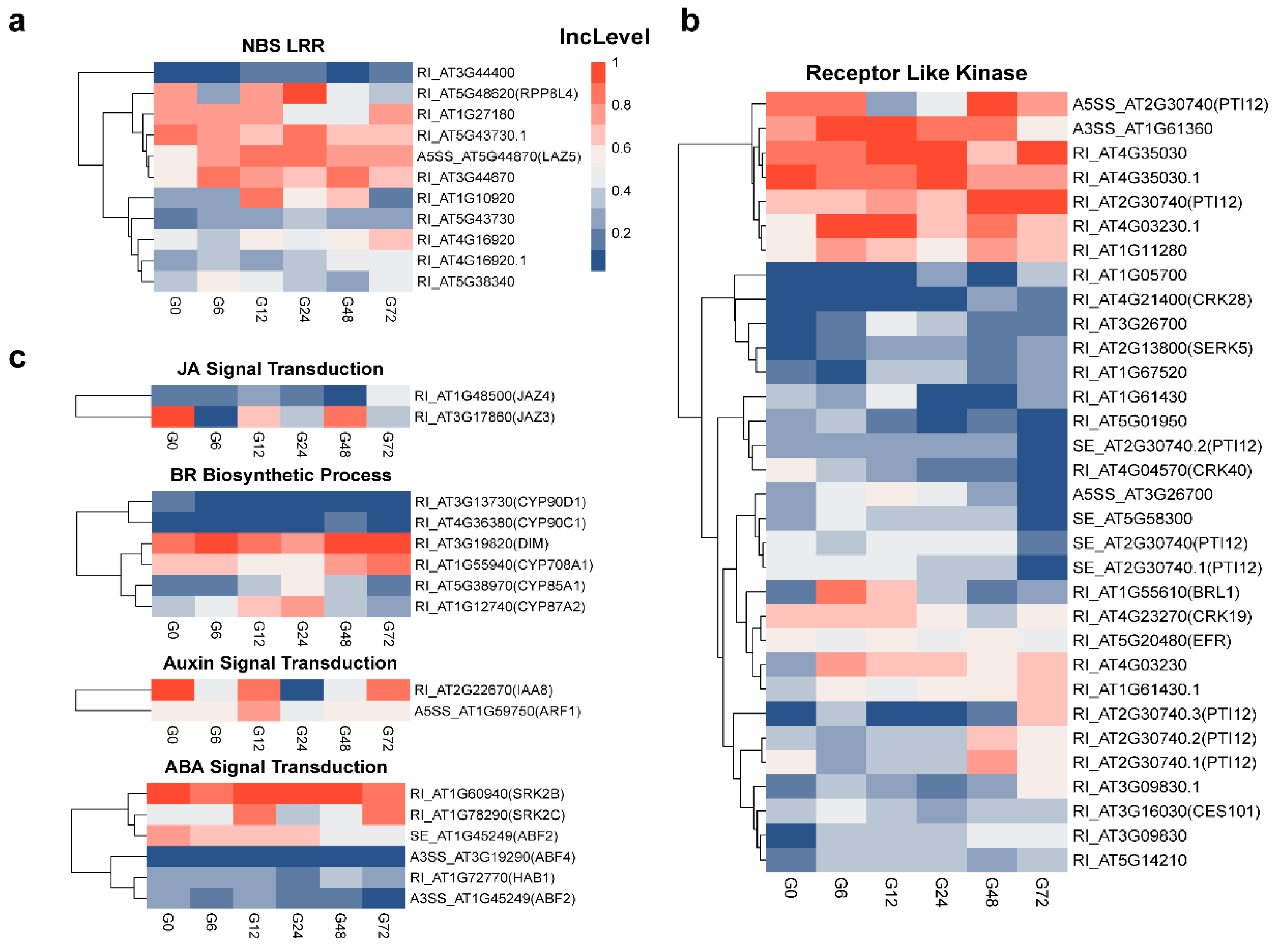
3.5. Identification of Various Defense-Related Genes Subjected to AS Modulation Following R. solanacearum Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genin, S.; Denny, T.P. Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 2012, 50, 67–89. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deslandes, L.; Pileur, F.; Liaubet, L.; Camut, S.; Can, C.; Williams, K.; Holub, E.; Beynon, J.; Arlat, M.; Marco, Y. Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum. Mol. Plant Microbe Interact. 1998, 11, 659–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef] [Green Version]
- Denance, N.; Ranocha, P.; Oria, N.; Barlet, X.; Riviere, M.P.; Yadeta, K.A.; Hoffmann, L.; Perreau, F.; Clement, G.; Maia-Grondard, A.; et al. Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 2013, 73, 225–239. [Google Scholar] [CrossRef]
- Hernandez-Blanco, C.; Feng, D.X.; Hu, J.; Sanchez-Vallet, A.; Deslandes, L.; Llorente, F.; Berrocal-Lobo, M.; Keller, H.; Barlet, X.; Sanchez-Rodriguez, C.; et al. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 2007, 19, 890–903. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, J.; Deslandes, L.; Feng, D.X.; Balague, C.; Marco, Y. Delayed Symptom Development in ein2-1, an Arabidopsis Ethylene-Insensitive Mutant, in Response to Bacterial Wilt Caused by Ralstonia solanacearum. Phytopathology 2002, 92, 1142–1148. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Wang, H.; Lu, Y.; Hu, J.; Qu, L.; Li, Z.; Wang, D.; He, Y.; Valls, M.; Coll, N.S.; et al. Deep Sequencing Reveals Early Reprogramming of Arabidopsis Root Transcriptomes Upon Ralstonia solanacearum Infection. Mol. Plant Microbe Interact. 2019, 32, 813–827. [Google Scholar] [CrossRef] [Green Version]
- Reddy, A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef] [Green Version]
- Lareau, L.F.; Inada, M.; Green, R.E.; Wengrod, J.C.; Brenner, S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 2007, 446, 926–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Kornblihtt, A.R.; Schor, I.E.; Allo, M.; Dujardin, G.; Petrillo, E.; Munoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Li, W.; Wang, S.; Zhang, X.; Coules, A.; Ding, G.; Xu, F.; Ren, J.; Lu, C.; Shi, L. Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus. Genes 2019, 10, 224. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, K.; Yabuta, Y.; Ishikawa, T.; Shigeoka, S. Identification of a cis element for tissue-specific alternative splicing of chloroplast ascorbate peroxidase pre-mRNA in higher plants. J. Biol. Chem. 2002, 277, 40623–40632. [Google Scholar] [CrossRef] [Green Version]
- Calixto, C.P.G.; Guo, W.; James, A.B.; Tzioutziou, N.A.; Entizne, J.C.; Panter, P.E.; Knight, H.; Nimmo, H.G.; Zhang, R.; Brown, J.W.S. Rapid and Dynamic Alternative Splicing Impacts the Arabidopsis Cold Response Transcriptome. Plant Cell 2018, 30, 1424–1444. [Google Scholar] [CrossRef] [Green Version]
- Howard, B.E.; Hu, Q.; Babaoglu, A.C.; Chandra, M.; Borghi, M.; Tan, X.; He, L.; Winter-Sederoff, H.; Gassmann, W.; Veronese, P.; et al. High-throughput RNA sequencing of pseudomonas-infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants. PLoS ONE 2013, 8, e74183. [Google Scholar] [CrossRef]
- Shang, X.; Cao, Y.; Ma, L. Alternative Splicing in Plant Genes: A Means of Regulating the Environmental Fitness of Plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef]
- Yang, S.; Tang, F.; Zhu, H. Alternative splicing in plant immunity. Int. J. Mol. Sci. 2014, 15, 10424–10445. [Google Scholar] [CrossRef] [Green Version]
- Rigo, R.; Bazin, J.R.M.; Crespi, M.; Charon, C.L. Alternative Splicing in the Regulation of Plant-Microbe Interactions. Plant Cell Physiol. 2019, 60, 1906–1916. [Google Scholar] [CrossRef]
- Chen, M.; Manley, J.L. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009, 10, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.D.; Ares, M., Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Barlet, X.; Deslandes, L.; Hirsch, J.; Feng, D.X.; Somssich, I.; Marco, Y. Transcriptional responses of Arabidopsis thaliana during wilt disease caused by the soil-borne phytopathogenic bacterium, Ralstonia solanacearum. PLoS ONE 2008, 3, e2589. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Naftelberg, S.; Schor, I.E.; Ast, G.; Kornblihtt, A.R. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 2015, 84, 165–198. [Google Scholar] [CrossRef]
- Wang, X.; Hu, L.; Wang, X.; Li, N.; Xu, C.; Gong, L.; Liu, B. DNA Methylation Affects Gene Alternative Splicing in Plants: An Example from Rice. Mol. Plant 2016, 9, 305–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bove, J.; Kim, C.Y.; Gibson, C.A.; Assmann, S.M. Characterization of wound-responsive RNA-binding proteins and their splice variants in Arabidopsis. Plant Mol. Biol. 2008, 67, 71–88. [Google Scholar] [CrossRef]
- Roux, M.E.; Rasmussen, M.W.; Palma, K.; Lolle, S.; Regue, A.M.; Bethke, G.; Glazebrook, J.; Zhang, W.; Sieburth, L.; Larsen, M.R.; et al. The mRNA decay factor PAT1 functions in a pathway including MAP kinase 4 and immune receptor SUMM2. EMBO J. 2015, 34, 593–608. [Google Scholar] [CrossRef] [Green Version]
- Sweat, T.A.; Lorang, J.M.; Bakker, E.G.; Wolpert, T.J. Characterization of natural and induced variation in the LOV1 gene, a CC-NB-LRR gene conferring victorin sensitivity and disease susceptibility in Arabidopsis. Mol. Plant Microbe Interact. 2008, 21, 7–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palma, K.; Thorgrimsen, S.; Malinovsky, F.G.; Fiil, B.K.; Nielsen, H.B.; Brodersen, P.; Hofius, D.; Petersen, M.; Mundy, J. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 2010, 6, e1001137. [Google Scholar] [CrossRef]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef]
- Boschi, F.; Schvartzman, C.; Murchio, S.; Ferreira, V.; Siri, M.I.; Galvan, G.A.; Smoker, M.; Stransfeld, L.; Zipfel, C.; Vilaro, F.L.; et al. Enhanced Bacterial Wilt Resistance in Potato Through Expression of Arabidopsis EFR and Introgression of Quantitative Resistance from Solanum commersonii. Front. Plant Sci. 2017, 8, 1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacombe, S.; Rougon-Cardoso, A.; Sherwood, E.; Peeters, N.; Dahlbeck, D.; van Esse, H.P.; Smoker, M.; Rallapalli, G.; Thomma, B.P.; Staskawicz, B.; et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 2010, 28, 365–369. [Google Scholar] [CrossRef]
- Oblessuc, P.R.; Obulareddy, N.; DeMott, L.; Matiolli, C.C.; Thompson, B.K.; Melotto, M. JAZ4 is involved in plant defense, growth, and development in Arabidopsis. Plant J. 2020, 101, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Zhou, W.; Baldwin, I.T.; Xu, S. Insect herbivory elicits genome-wide alternative splicing responses in Nicotiana attenuata. Plant J. 2015, 84, 228–243. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, R.; Wang, Y.; Zhang, L.; Wang, C.; Lv, S.; Liu, X.; Wang, Y.; Ji, W. Transcriptome-wide alternative splicing modulation during plant-pathogen interactions in wheat. Plant Sci. 2019, 288, 110160. [Google Scholar] [CrossRef]
- Mauger, O.; Lemoine, F.; Scheiffele, P. Targeted Intron Retention and Excision for Rapid Gene Regulation in Response to Neuronal Activity. Neuron 2016, 92, 1266–1278. [Google Scholar] [CrossRef] [Green Version]
- Medina, M.W.; Gao, F.; Naidoo, D.; Rudel, L.L.; Temel, R.E.; McDaniel, A.L.; Marshall, S.M.; Krauss, R.M. Coordinately regulated alternative splicing of genes involved in cholesterol biosynthesis and uptake. PLoS ONE 2011, 6, e19420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazin, J.; Mariappan, K.; Jiang, Y.; Blein, T.; Voelz, R.; Crespi, M.; Hirt, H. Role of MPK4 in pathogen-associated molecular pattern-triggered alternative splicing in Arabidopsis. PLoS Pathog. 2020, 16, e1008401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, Y.; Ding, P.; Li, Y.; Kong, Q.; Zhang, Y. Splicing of receptor-like kinase-encoding SNC4 and CERK1 is regulated by two conserved splicing factors that are required for plant immunity. Mol. Plant 2014, 7, 1766–1775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.N.; Shi, Y.; Powers, J.J.; Gowda, N.B.; Zhang, C.; Ibrahim, H.M.M.; Ball, H.B.; Chen, S.L.; Lu, H.; Mount, S.M. Transcriptome analyses reveal SR45 to be a neutral splicing regulator and a suppressor of innate immunity in Arabidopsis thaliana. BMC Genomics 2017, 18, 772. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, V.; Joe, A.; Jeong, B.R.; Korneli, C.; Boutrot, F.; Westedt, I.; Staiger, D.; Alfano, J.R.; Zipfel, C. Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 2013, 32, 701–712. [Google Scholar] [CrossRef] [Green Version]
- Sanabria, N.M.; Dubery, I.A. Alternative splicing of the receptor-like kinase Nt-Sd-RLK in tobacco cells responding to lipopolysaccharides: Suggestive of a role in pathogen surveillance and perception? FEBS Lett. 2016, 590, 3628–3638. [Google Scholar] [CrossRef]
- Zhang, X.C.; Gassmann, W. RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 2003, 15, 2333–2342. [Google Scholar] [CrossRef]
- Feng, D.X.; Tasset, C.; Hanemian, M.; Barlet, X.; Hu, J.; Tremousaygue, D.; Deslandes, L.; Marco, Y. Biological control of bacterial wilt in Arabidopsis thaliana involves abscissic acid signalling. New Phytol. 2012, 194, 1035–1045. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, N.; Zhang, R.; Zhang, M.; Niu, Y.; Fu, S.; Wang, Y.; Wang, D.; Chen, Y.; Zhao, C.; Chen, Q.; et al. Global Profiling of Dynamic Alternative Splicing Modulation in Arabidopsis Root upon Ralstonia solanacearum Infection. Genes 2020, 11, 1078. https://doi.org/10.3390/genes11091078
Qin N, Zhang R, Zhang M, Niu Y, Fu S, Wang Y, Wang D, Chen Y, Zhao C, Chen Q, et al. Global Profiling of Dynamic Alternative Splicing Modulation in Arabidopsis Root upon Ralstonia solanacearum Infection. Genes. 2020; 11(9):1078. https://doi.org/10.3390/genes11091078
Chicago/Turabian StyleQin, Ning, Ruize Zhang, Mancang Zhang, Yang Niu, Shouyang Fu, Yisa Wang, Dongdong Wang, Yue Chen, Cuizhu Zhao, Qin Chen, and et al. 2020. "Global Profiling of Dynamic Alternative Splicing Modulation in Arabidopsis Root upon Ralstonia solanacearum Infection" Genes 11, no. 9: 1078. https://doi.org/10.3390/genes11091078
APA StyleQin, N., Zhang, R., Zhang, M., Niu, Y., Fu, S., Wang, Y., Wang, D., Chen, Y., Zhao, C., Chen, Q., & Lu, H. (2020). Global Profiling of Dynamic Alternative Splicing Modulation in Arabidopsis Root upon Ralstonia solanacearum Infection. Genes, 11(9), 1078. https://doi.org/10.3390/genes11091078




