TATA Binding Protein (TBP) Promoter Drives Ubiquitous Expression of Marker Transgene in the Adult Sea Anemone Nematostella vectensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sea Anemone Culture
2.2. Cloning
2.3. Transgenesis
2.4. Cell Dissociation
2.5. Flow Cytometry
2.6. Fluorescence Microscopy
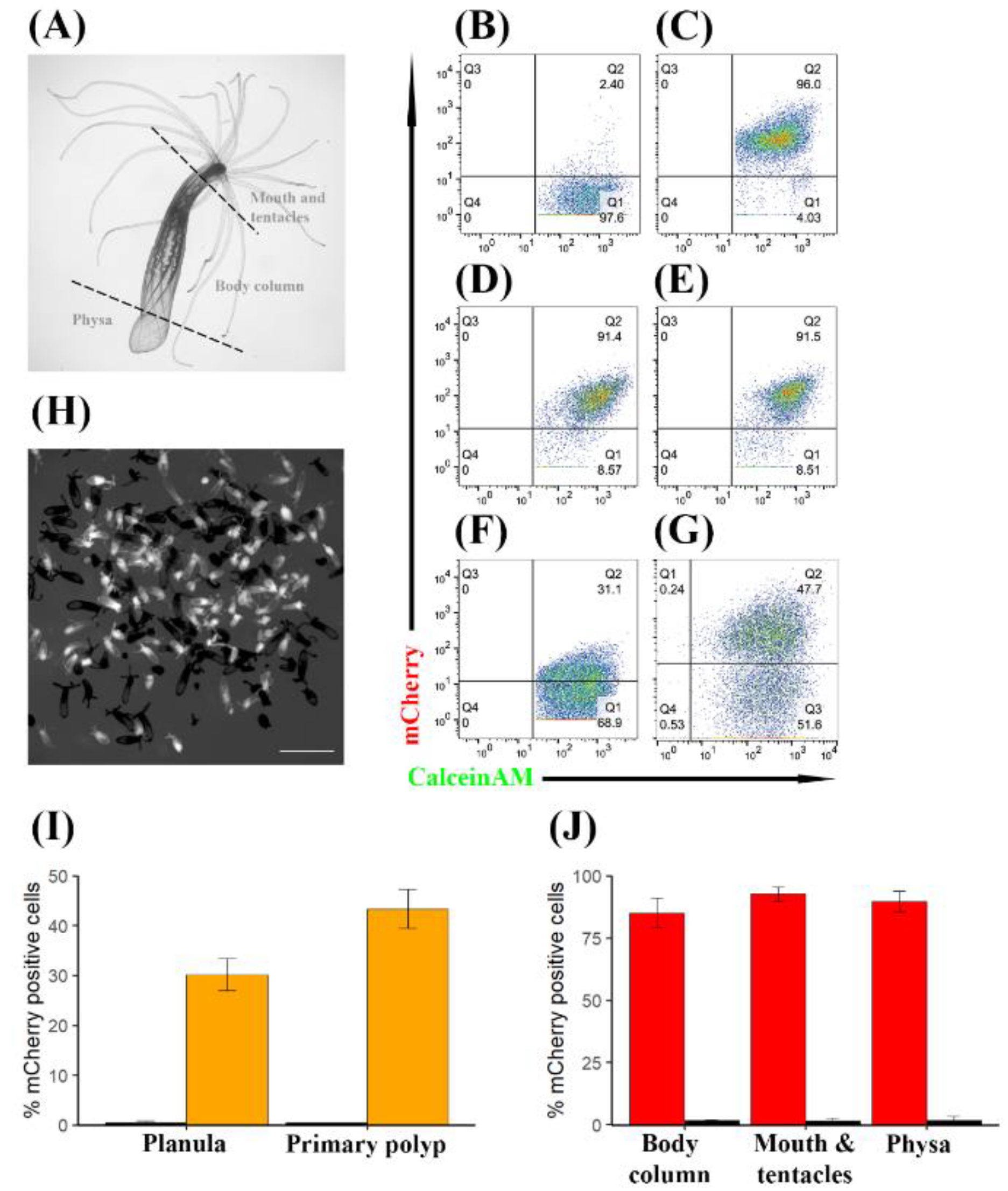
3. Results
3.1. Generation of the TBP::mCherry Nematostella vectensis Transgenic Line
3.2. TBP Promoter Drives the Expression of mCherry in Diverse Cell Types
3.3. TBP::mCherry Is Expressed Stably at Different Life Stages and Throughout Generations in a High Percentage of Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Layden, M.J.; Rentzsch, F.; Röttinger, E. The rise of the starlet sea anemone Nematostella vectensis as a model system to investigate development and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 408–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Technau, U.; Steele, R.E. Evolutionary crossroads in developmental biology: Cnidaria. Development 2011, 138, 1447–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hand, C.; Uhlinger, K.R. The culture, sexual and asexual reproduction, and growth of the sea Anemone Nematostella vectensis. Biol. Bull. 1992, 182, 169–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefanik, D.J.; Friedman, L.E.; Finnerty, J.R. Collecting, rearing, spawning and inducing regeneration of the starlet sea anemone, Nematostella vectensis. Nat. Protoc. 2013, 8, 916–923. [Google Scholar] [CrossRef] [PubMed]
- Fritzenwanker, J.H.; Technau, U. Induction of gametogenesis in the basal cnidarian Nematostella vectensis (Anthozoa). Dev. Genes Evol. 2002, 212, 99–103. [Google Scholar] [CrossRef]
- Columbus-Shenkar, Y.Y.; Sachkova, M.Y.; Macrander, J.; Fridrich, A.; Modepalli, V.; Reitzel, A.M.; Sunagar, K.; Moran, Y. Dynamics of venom composition across a complex life cycle. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Putnam, N.H.; Srivastava, M.; Hellsten, U.; Dirks, B.; Chapman, J.; Salamov, A.; Terry, A.; Shapiro, H.; Lindquist, E.; Kapitonov, V.V.; et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317, 86–94. [Google Scholar] [CrossRef] [Green Version]
- Renfer, E.; Amon-Hassenzahl, A.; Steinmetz, P.R.H.; Technau, U. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc. Natl. Acad. Sci. USA 2010, 107, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Nakanishi, N.; Renfer, E.; Technau, U.; Rentzsch, F. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development 2012, 139, 347–357. [Google Scholar] [CrossRef] [Green Version]
- Layden, M.J.; Johnston, H.; Amiel, A.R.; Havrilak, J.; Steinworth, B.; Chock, T.; Röttinger, E.; Martindale, M.Q. MAPK signaling is necessary for neurogenesis in Nematostella vectensis. BMC Biol. 2016, 14. [Google Scholar] [CrossRef] [Green Version]
- Richards, G.S.; Rentzsch, F. Transgenic analysis of a SoxB gene reveals neural progenitor cells in the cnidarian Nematostella vectensis. Development 2014, 4681–4689. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Del Viso, F.; Chen, C.Y.; Ikmi, A.; Kroesen, A.E.; Gibson, M.C. An axial Hox code controls tissue segmentation and body patterning in Nematostella vectensis. Science 2018, 361, 1377–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinmetz, P.R.H.; Aman, A.; Kraus, J.E.M.; Technau, U. Gut-Like ectodermal tissue in a sea anemone challenges germ layer homology. Nat. Ecol. Evol. 2017, 1, 1535–1542. [Google Scholar] [CrossRef] [PubMed]
- Fritz, A.E.; Ikmi, A.; Seidel, C.; Paulson, A.; Gibson, M.C. Mechanisms of tentacle morphogenesis in the sea anemone Nematostella vectensis. Development 2013, 140, 2212–2223. [Google Scholar] [CrossRef] [Green Version]
- Casola, S. Mouse models for miRNA expression: The ROSA26 locus. Methods Mol. Biol. 2010, 667, 145–163. [Google Scholar] [CrossRef]
- Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999, 21, 70–71. [Google Scholar] [CrossRef]
- Kisseberth, W.C.; Brettingen, N.T.; Lohse, J.K.; Sandgren, E.P. Ubiquitous expression of marker transgenes in mice and rats. Dev. Biol. 1999, 214, 128–138. [Google Scholar] [CrossRef] [Green Version]
- Srinivas, S.; Watanabe, T.; Lin, C.S.; William, C.M.; Tanabe, Y.; Jessell, T.M.; Costantini, F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001, 1, 4. [Google Scholar] [CrossRef] [Green Version]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef] [Green Version]
- Mosimann, C.; Kaufman, C.K.; Li, P.; Pugach, E.K.; Tamplin, O.J.; Zon, L.I. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 2011, 138, 169–177. [Google Scholar] [CrossRef] [Green Version]
- Wittlieb, J.; Khalturin, K.; Lohmann, J.U.; Anton-Erxleben, F.; Bosch, T.C.G. Transgenic Hydra allow In Vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 6208–6211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brockschnieder, D.; Pechmann, Y.; Sonnenberg-Riethmacher, E.; Riethmacher, D. An improved mouse line for Cre-induced cell ablation due to diphtheria toxin A, expressed from the Rosa26 locus. Genesis 2006, 44, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.G.; Ono, N.; Kalajzic, I. Methods in lineage tracing. In Principles of Bone Biology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1887–1898. ISBN 9780128148419. [Google Scholar]
- Lee, T.I.; Young, R.A. Transcription of Eukaryotic Protein-Coding Genes. Annu. Rev. Genet. 2000, 34, 77–137. [Google Scholar] [CrossRef]
- Hoffmann, A.; Sinn, E.; Yamamoto, T.; Wang, J.; Roy, A.; Horikoshi, M.; Roeder, R.G. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature 1990, 346, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Peterson, M.G.; Tanese, N.; Pugh, B.F.; Tjian, R. Functional domains and upstream activation properties of cloned human TATA binding protein. Science 1990, 248, 1625–1630. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yan, C.; Fang, J.; Inouye, C.; Tjian, R.; Ivanov, I.; Nogales, E. Near-Atomic resolution visualization of human transcription promoter opening. Nature 2016, 533, 359–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwaiger, M.; Schönauer, A.; Rendeiro, A.F.; Pribitzer, C.; Schauer, A.; Gilles, A.F.; Schinko, J.B.; Renfer, E.; Fredman, D.; Technau, U. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res. 2014, 24, 639–650. [Google Scholar] [CrossRef] [Green Version]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.G.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef]
- Genikhovieh, G.; Technau, U. Induction of spawning in the starlet sea anemone nematostella vectensis, In Vitro fertilization of gametes, and dejellying of zygotes. Cold Spring Harb. Protoc. 2009, 4. [Google Scholar] [CrossRef]
- Renfer, E.; Technau, U. Meganuclease-Assisted generation of stable transgenics in the sea anemone Nematostella vectensis. Nat. Protoc. 2017, 12, 1844–1854. [Google Scholar] [CrossRef] [PubMed]
- Sebé-Pedrós, A.; Saudemont, B.; Chomsky, E.; Plessier, F.; Mailhé, M.P.; Renno, J.; Loe-Mie, Y.; Lifshitz, A.; Mukamel, Z.; Schmutz, S.; et al. Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell 2018, 173, 1520–1534.e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imbalzano, A.N.; Kwon, H.; Green, M.R.; Kingston, R.E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 1994, 370, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Antonova, S.V.; Boeren, J.; Timmers, H.T.M.; Snel, B. Epigenetics and transcription regulation during eukaryotic diversification: The saga of TFIID. Genes Dev. 2019, 33, 888–902. [Google Scholar] [CrossRef] [Green Version]
- Ikmi, A.; Gibson, M.C. Identification and In Vivo characterization of NvFP-7R, a developmentally regulated red fluorescent protein of Nematostella vectensis. PLoS ONE 2010, 5, e11807. [Google Scholar] [CrossRef]
- Warner, J.F.; Guerlais, V.; Amiel, A.R.; Johnston, H.; Nedoncelle, K.; Röttinger, E. NvERTx: A gene expression database to compare embryogenesis and regeneration in the sea anemone Nematostella vectensis. Development 2018, 145. [Google Scholar] [CrossRef] [Green Version]
- Helm, R.R.; Siebert, S.; Tulin, S.; Smith, J.; Dunn, C.W. Characterization of differential transcript abundance through time during Nematostella vectensis development. BMC Genom. 2013, 14, 266. [Google Scholar] [CrossRef] [Green Version]
- Fischer, A.H.L.; Mozzherin, D.; Eren, A.M.; Lans, K.D.; Wilson, N.; Cosentino, C.; Smith, J. SeaBase: A multispecies transcriptomic resource and platform for gene network inference. Integr. Comp. Biol. 2014, 54, 250–263. [Google Scholar] [CrossRef] [Green Version]
- Babonis, L.S.; Ryan, J.F.; Enjolras, C.; Martindale, M.Q. Genomic analysis of the tryptome reveals molecular mechanisms of gland cell evolution. EvoDevo 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Frank, P.; Bleakney, J.S. Histology and sexual reproduction of the anemone Nematostella vectensis stephenson 1935. J. Nat. Hist. 1976, 10, 441–449. [Google Scholar] [CrossRef]
- Jahnel, S.M.; Walzl, M.; Technau, U. Development and epithelial organisation of muscle cells in the sea anemone Nematostella vectensis. Front. Zool. 2014, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zenkert, C.; Takahashi, T.; Diesner, M.O.; Özbek, S. Morphological and molecular analysis of the Nematostella vectensis cnidom. PLoS ONE 2011, 6, e22725. [Google Scholar] [CrossRef] [PubMed]
- Giel-Moloney, M.; Krause, D.S.; Chen, G.; Van Etten, R.A.; Leiter, A.B. Ubiquitous and uniform In Vivo fluorescence in ROSA26-EGFP BAC transgenic mice. Genesis 2007, 45, 83–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiki, Y.; Tao, K.; Bianchi, D.W.; Giel-Moloney, M.; Leiter, A.B.; Johnson, K.L. Quantification of green fluorescent protein by in vivo imaging, PCR, and flow cytometry: Comparison of transgenic strains and relevance for fetal cell microchimerism. Cytom. Part A 2008, 73, 11–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bestor, T.H. Gene silencing as a threat to the success of gene therapy. J. Clin. Investig. 2000, 105, 409–411. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.I.; Whitelaw, E. The vagaries of variegating transgenes. Bioessays 1996, 18, 919–923. [Google Scholar] [CrossRef]
- Kioussis, D.; Festenstein, R. Locus control regions: Overcoming heterochromatin-induced gene inactivation in mammals. Curr. Opin. Genet. Dev. 1997, 7, 614–619. [Google Scholar] [CrossRef]
- Sadelain, M.; Papapetrou, E.P.; Bushman, F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer 2011, 12, 51–58. [Google Scholar] [CrossRef]
- Ikmi, A.; McKinney, S.A.; Delventhal, K.M.; Gibson, M.C. TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Pastrana, E. Optogenetics: Controlling cell function with light. Nat. Methods 2011, 8, 24–25. [Google Scholar] [CrossRef]
- Deisseroth, K.; Feng, G.; Majewska, A.K.; Miesenböck, G.; Ting, A.; Schnitzer, M.J. Next-Generation optical technologies for illuminating genetically targeted brain circuits. J. Neurosci. 2006, 26, 10380–10386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marvin, J.S.; Shimoda, Y.; Magloire, V.; Leite, M.; Kawashima, T.; Jensen, T.P.; Kolb, I.; Knott, E.L.; Novak, O.; Podgorski, K.; et al. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat. Methods 2019, 16, 763–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portugues, R.; Severi, K.E.; Wyart, C.; Ahrens, M.B. Optogenetics in a transparent animal: Circuit function in the larval zebrafish. Curr. Opin. Neurobiol. 2013, 23, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yuste, R. In Vivo imaging of neural activity. Nat. Methods 2017, 14, 349–359. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Admoni, Y.; Kozlovski, I.; Lewandowska, M.; Moran, Y. TATA Binding Protein (TBP) Promoter Drives Ubiquitous Expression of Marker Transgene in the Adult Sea Anemone Nematostella vectensis. Genes 2020, 11, 1081. https://doi.org/10.3390/genes11091081
Admoni Y, Kozlovski I, Lewandowska M, Moran Y. TATA Binding Protein (TBP) Promoter Drives Ubiquitous Expression of Marker Transgene in the Adult Sea Anemone Nematostella vectensis. Genes. 2020; 11(9):1081. https://doi.org/10.3390/genes11091081
Chicago/Turabian StyleAdmoni, Yael, Itamar Kozlovski, Magda Lewandowska, and Yehu Moran. 2020. "TATA Binding Protein (TBP) Promoter Drives Ubiquitous Expression of Marker Transgene in the Adult Sea Anemone Nematostella vectensis" Genes 11, no. 9: 1081. https://doi.org/10.3390/genes11091081
APA StyleAdmoni, Y., Kozlovski, I., Lewandowska, M., & Moran, Y. (2020). TATA Binding Protein (TBP) Promoter Drives Ubiquitous Expression of Marker Transgene in the Adult Sea Anemone Nematostella vectensis. Genes, 11(9), 1081. https://doi.org/10.3390/genes11091081





