Transcriptomic Analysis Revealed an Emerging Role of Alternative Splicing in Embryonal Tumor with Multilayered Rosettes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Publicly Available RNA-Seq Data
2.2. RNA-Seq Data Analysis
2.3. Functional Enrichment and PPI Network Analysis
3. Results
3.1. Identification of Differentially Expressed Genes (DEGs) between ETMR and Fetal Normal Control
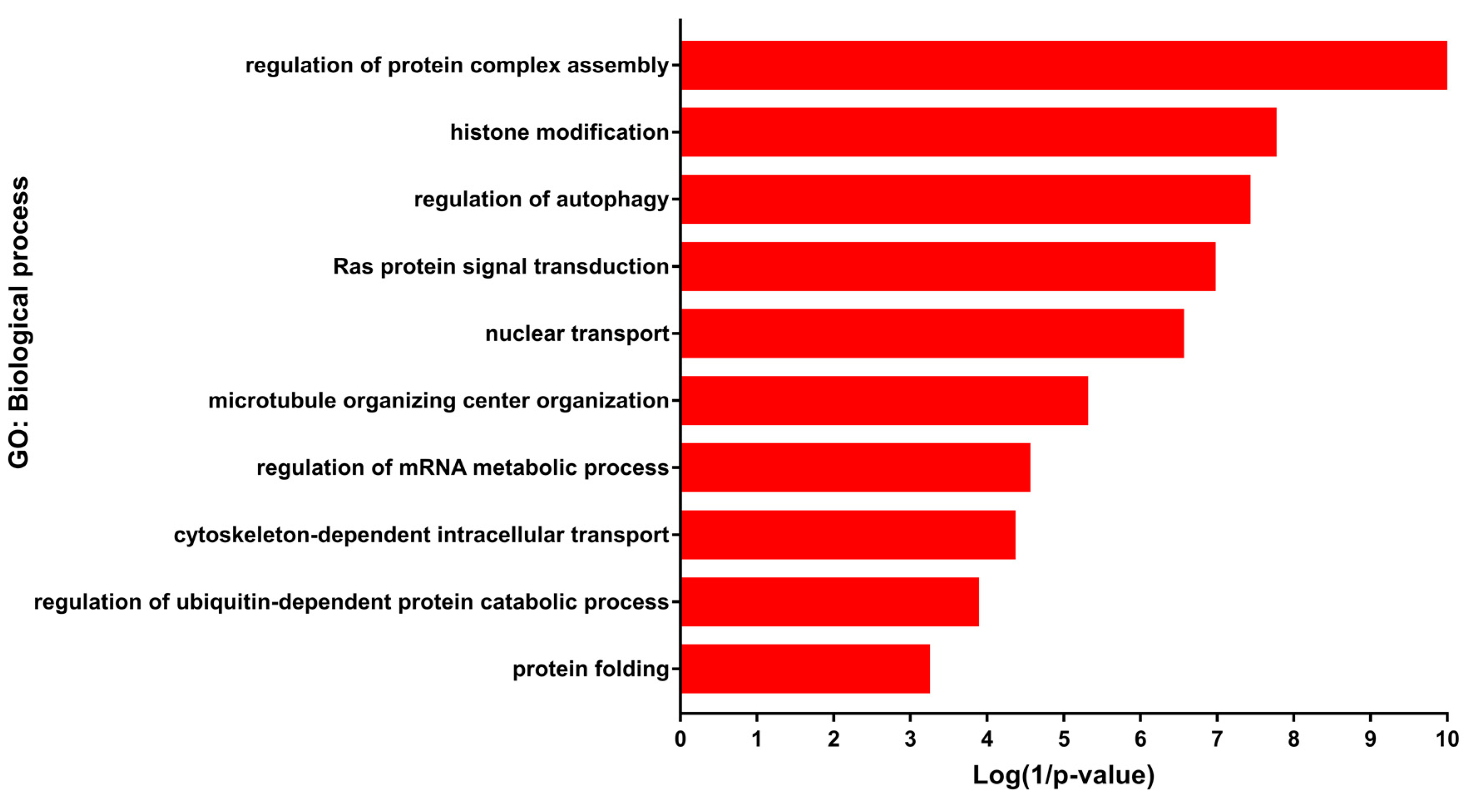
3.2. Enrichment Analysis of the DEGs
3.3. PPI Network Analysis of Up-Regulated DEGs
3.4. Alternative Splicing Events in ETMR
3.5. Up-Regulated DEGs with AS Events
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [Green Version]
- Pickles, J.C.; Hawkins, C.; Pietsch, T.; Jacques, T.S. CNS embryonal tumours: WHO 2016 and beyond. Neuropathol. Appl. Neurobiol. 2018, 44, 151–162. [Google Scholar] [CrossRef]
- Korshunov, A.; Sturm, D.; Ryzhova, M.; Hovestadt, V.; Gessi, M.; Jones, D.T.W.; Remke, M.; Northcott, P.; Perry, A.; Picard, D.; et al. Embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, and medulloepithelioma share molecular similarity and comprise a single clinicopathological entity. Acta Neuropathol. 2014, 128, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Spence, T.; Sin-Chan, P.; Picard, D.; Barszczyk, M.; Hoss, K.; Lu, M.; Kim, S.K.; Ra, Y.S.; Nakamura, H.; Fangusaro, J.; et al. CNS-PNETs with C19MC amplification and/or LIN28 expression comprise a distinct histogenetic diagnostic and therapeutic entity. Acta Neuropathol. 2014, 128, 291–303. [Google Scholar] [CrossRef] [Green Version]
- Geyer, J.R.; Sposto, R.; Jennings, M.; Boyett, J.M.; Axtell, R.A.; Breiger, D.; Broxson, E.; Donahue, B.; Finlay, J.L.; Goldwein, J.W.; et al. Multiagent chemotherapy and deferred radiotherapy in infants with malignant brain tumors: A Report from the Children’s Cancer Group. J. Clin. Oncol. 2005, 23, 7621–7631. [Google Scholar] [CrossRef]
- Choi, S.H.; Kim, S.H.; Shim, K.W.; Han, J.W.; Choi, J.; Kim, D.S.; Lyu, C.J.; Kim, J.W.; Suh, C.O.; Cho, J. Treatment outcome and prognostic molecular markers of supratentorial primitive neuroectodermal tumors. PLoS ONE 2016, 11, e0153443. [Google Scholar] [CrossRef] [Green Version]
- Picard, D.; Miller, S.; Hawkins, C.E.; Bouffet, E.; Rogers, H.A.; Chan, T.S.Y.; Kim, S.K.; Ra, Y.S.; Fangusaro, J.; Korshunov, A.; et al. Markers of survival and metastatic potential in childhood CNS primitive neuro-ectodermal brain tumours: An integrative genomic analysis. Lancet Oncol. 2012, 13, 838–848. [Google Scholar] [CrossRef] [Green Version]
- Gessi, M.; Giangaspero, F.; Lauriola, L.; Gardiman, M.; Scheithauer, B.W.; Halliday, W.; Hawkins, C.; Rosenblum, M.K.; Burger, P.C.; Eberhart, C.G. Embryonal tumors with abundant neuropil and true Rosettes: A distinctive CNS primitive neuroectodermal tumor. Am. J. Surg. Pathol. 2009, 33, 211. [Google Scholar] [CrossRef] [Green Version]
- Eberhart, C.G.; Brat, D.J.; Cohen, K.J.; Burger, P.C. Pediatric neuroblastic brain tumors containing abundant neuropil and true rosettes. Pediatr. Dev. Pathol. 2000, 3, 346–352. [Google Scholar] [CrossRef]
- Li, M.; Lee, K.F.; Lu, Y.; Clarke, I.; Shih, D.; Eberhart, C.; Collins, V.P.; Van Meter, T.; Picard, D.; Zhou, L.; et al. Frequent Amplification of a chr19q13.41 MicroRNA Polycistron in Aggressive Primitive Neuroectodermal Brain Tumors. Cancer Cell 2009, 16, 533–546. [Google Scholar] [CrossRef] [Green Version]
- Kleinman, C.L.; Gerges, N.; Papillon-Cavanagh, S.; Sin-Chan, P.; Pramatarova, A.; Quang, D.A.K.; Adoue, V.; Busche, S.; Caron, M.; Djambazian, H.; et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat. Genet. 2014, 46, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.R.; Powers, J.T.; Einhorn, W.; Hoshida, Y.; Ng, T.L.; Toffanin, S.; O’Sullivan, M.; Lu, J.; Phillips, L.A.; Lockhart, V.L.; et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009, 41, 843. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Bennett, B.D.; Luo, S.; Inoue, K.; Grimm, S.A.; Schroth, G.P.; Bushel, P.R.; Kinyamu, H.K.; Archer, T.K. LIN28A modulates splicing and gene expression programs in breast cancer cells. Mol. Cell. Biol. 2015, 35, 3225–3243. [Google Scholar] [CrossRef] [Green Version]
- Wilbert, M.L.; Huelga, S.C.; Kapeli, K.; Stark, T.J.; Liang, T.Y.; Chen, S.X.; Yan, B.Y.; Nathanson, J.L.; Hutt, K.R.; Lovci, M.T.; et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell 2012, 48, 195–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Sun, N.; Lu, Z.; Sun, S.; Huang, J.; Chen, Z.; He, J. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017, 393, 40–51. [Google Scholar] [CrossRef]
- Ladomery, M. Aberrant alternative splicing is another hallmark of cancer. Int. J. Cell Biol. 2013, 2013, 463786. [Google Scholar] [CrossRef]
- David, C.J.; Manley, J.L. Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev. 2010, 24, 2343–2364. [Google Scholar] [CrossRef] [Green Version]
- Oltean, S.; Bates, D.O. Hallmarks of alternative splicing in cancer. Oncogene 2014, 33, 5311–5318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venables, J.P. Unbalanced alternative splicing and its significance in cancer. BioEssays 2006, 28, 378–386. [Google Scholar] [CrossRef]
- Ghigna, C.; Valacca, C.; Biamonti, G. Alternative Splicing and Tumor Progression. Curr. Genom. 2008, 9, 556–570. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.D.; Lee, N.H. Aberrant RNA splicing in cancer and drug resistance. Cancers 2018, 10, 458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Hu, Z.; Zhao, Y.; Huang, S.; He, X. Transcriptome-Wide Analysis Reveals the Landscape of Aberrant Alternative Splicing Events in Liver Cancer. Hepatology 2019, 69, 359–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutertre, M.; Vagner, S.; Auboeuf, D. Alternative splicing and breast cancer. RNA Biol. 2010, 7, 403–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonomi, S.; Gallo, S.; Catillo, M.; Pignataro, D.; Biamonti, G.; Ghigna, C. Oncogenic alternative splicing switches: Role in cancer progression and prospects for therapy. Int. J. Cell Biol. 2013, 2013, 962038. [Google Scholar] [CrossRef] [PubMed]
- Omenn, G.S.; Yocum, A.K.; Menon, R. Alternative splice variants, a new class of protein cancer biomarker candidates: Findings in pancreatic cancer and breast cancer with systems biology implications. Dis. Markers 2010, 28, 241–251. [Google Scholar] [CrossRef]
- Samur, A.A.; Samur, M.K.; Lopez, M.A.; Derebail, S.; Anderson, K.C.; Avet-Loiseau, H.; Parmigiani, G.; Fulciniti, M.; Munshi, N.C. Genome wide transcriptomic analysis identifies dysregulated splicing factor profile with molecular and functional role in multiple myeloma. Blood 2019, 134, 361. [Google Scholar] [CrossRef]
- Lindsay, S.J.; Xu, Y.; Lisgo, S.N.; Harkin, L.F.; Copp, A.J.; Gerrelli, D.; Clowry, G.J.; Talbot, A.; Keogh, M.J.; Coxhead, J.; et al. HDBR Expression: A Unique Resource for Global and Individual Gene Expression Studies during Early Human Brain Development. Front. Neuroanat. 2016, 10, 86. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Pimentel, H.; Bray, N.L.; Puente, S.; Melsted, P.; Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 2017, 14, 687. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2018. R Package Version 1.3.5. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 4 August 2020).
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Kahles, A.; Ong, C.S.; Zhong, Y.; Rätsch, G. SplAdder: Identification, quantification and testing of alternative splicing events from RNA-Seq data. Bioinformatics 2016, 32, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [Green Version]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; Von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Pagliarini, V.; Naro, C.; Sette, C. Splicing regulation: A molecular device to enhance cancer cell adaptation. BioMed Res. Int. 2015, 2015, 543067. [Google Scholar] [CrossRef] [Green Version]
- Wan, Q.; Sang, X.; Jin, L.; Wang, Z. Alternative splicing events as indicators for the prognosis of uveal melanoma. Genes 2020, 11, 227. [Google Scholar] [CrossRef] [Green Version]
- Xue, D.; Cheng, P.; Jiang, J.; Ren, Y.; Wu, D.; Chen, W. Systemic Analysis of the Prognosis-Related RNA Alternative Splicing Signals in Melanoma. Med. Sci. Monit. 2020, 26, e921133-1. [Google Scholar] [CrossRef]
- Zhang, D.; Hu, Q.; Liu, X.; Ji, Y.; Chao, H.-P.; Liu, Y.; Tracz, A.; Kirk, J.; Buonamici, S.; Zhu, P.; et al. Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat. Commun. 2020, 11, 2089. [Google Scholar] [CrossRef]
- Suzuki, H.; Kumar, S.A.; Shuai, S.; Diaz-Navarro, A.; Gutierrez-Fernandez, A.; De Antonellis, P.; Cavalli, F.M.G.; Juraschka, K.; Farooq, H.; Shibahara, I.; et al. Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature 2019, 574, 707–711. [Google Scholar] [CrossRef]
- Huang, H.H.; Ferguson, I.D.; Thornton, A.M.; Bastola, P.; Lam, C.; Lin, Y.H.T.; Choudhry, P.; Mariano, M.C.; Marcoulis, M.D.; Teo, C.F.; et al. Proteasome inhibitor-induced modulation reveals the spliceosome as a specific therapeutic vulnerability in multiple myeloma. Nat. Commun. 2020, 11, 1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbanski, L.M.; Leclair, N.; Anczuków, O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA 2018, 9, e1476. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, A.; Ryzhova, M.; Jones, D.T.W.; Northcott, P.A.; Van Sluis, P.; Volckmann, R.; Koster, J.; Versteeg, R.; Cowdrey, C.; Perry, A.; et al. LIN28A immunoreactivity is a potent diagnostic marker of embryonal tumor with multilayered rosettes (ETMR). Acta Neuropathol. 2012, 124, 875–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Ng, S.C.; Segr, A.V.; Shinoda, G.; Shah, S.P.; Einhorn, W.S.; Takeuchi, A.; Engreitz, J.M.; Hagan, J.P.; Kharas, M.G.; et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef] [Green Version]
- Spence, T.; Perotti, C.; Sin-Chan, P.; Picard, D.; Wu, W.; Singh, A.; Anderson, C.; Blough, M.D.; Cairncross, J.G.; Lafay-Cousin, L.; et al. A novel C19MC amplified cell line links Lin28/let-7 to mTOR signaling in embryonal tumor with multilayered rosettes. Neuro. Oncol. 2014, 16, 62–71. [Google Scholar] [CrossRef] [Green Version]
- Xiong, H.; Zhao, W.; Wang, J.; Seifer, B.J.; Ye, C.; Chen, Y.; Jia, Y.; Chen, C.; Shen, J.; Wang, L.; et al. Oncogenic mechanisms of Lin28 in breast cancer: New functions and therapeutic opportunities. Oncotarget 2017, 8, 25721. [Google Scholar] [CrossRef]
- Sun, W.L. Ambra1 in autophagy and apoptosis: Implications for cell survival and chemotherapy resistance. Oncol. Lett. 2016, 12, 367–374. [Google Scholar] [CrossRef] [Green Version]
- Bozaykut, P.; Ozer, N.K.; Karademir, B. Regulation of protein turnover by heat shock proteins. Free Radic. Biol. Med. 2014, 77, 195–209. [Google Scholar] [CrossRef]
- Zoppino, F.C.M.; Guerrero-Gimenez, M.E.; Castro, G.N.; Ciocca, D.R. Comprehensive transcriptomic analysis of heat shock proteins in the molecular subtypes of human breast cancer. BMC Cancer 2018, 18, 700. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Ding, W.; Wang, Y.; Ji, T.; Sun, S.; Mo, Q.; Chen, P.; Fang, Y.; Liu, J.; Wang, B.; et al. Ubiquitin B in cervical cancer: Critical for the maintenance of cancer stem-like cell characters. PLoS ONE 2013, 8, e84457. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.; Park, S.; Lee, E.K.; Yoo, Y.J. Downregulation of ubiquitin level via knockdown of polyubiquitin gene Ubb as potential cancer therapeutic intervention. Sci. Rep. 2013, 3, 2623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, V.; Pereira, J.F.S.; Jordan, P. Signaling pathways driving aberrant splicing in cancer cells. Genes 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Marabti, E.; Younis, I. The cancer spliceome: Reprograming of alternative splicing in cancer. Front. Mol. Biosci. 2018, 5, 80. [Google Scholar] [CrossRef]
- Tran, E.J.; King, M.C.; Corbett, A.H. Macromolecular transport between the nucleus and the cytoplasm: Advances in mechanism and emerging links to disease. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 2784–2795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gravina, G.L.; Senapedis, W.; McCauley, D.; Baloglu, E.; Shacham, S.; Festuccia, C. Nucleo-cytoplasmic transport as a therapeutic target of cancer. J. Hematol. Oncol. 2014, 7, 85. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.P.; Jacquemyn, M.; Lipecka, J.; Chhuon, C.; Aushev, V.N.; Meunier, B.; Singh, M.K.; Carpi, N.; Piel, M.; Codogno, P.; et al. STK 38 kinase acts as XPO 1 gatekeeper regulating the nuclear export of autophagy proteins and other cargoes. EMBO Rep. 2019, 20, e48150. [Google Scholar] [CrossRef]
- Niu, M.; Wu, S.; Mao, L.; Yang, Y. CRM1 is a cellular target of curcumin: New insights for the myriad of biological effects of an ancient spice. Traffic 2013, 14, 1042–1052. [Google Scholar] [CrossRef]
- Yashiroda, Y.; Yoshida, M. Nucleo-Cytoplasmic Transport of Proteins as a Target for Therapeutic Drugs. Curr. Med. Chem. 2005, 10, 741–748. [Google Scholar] [CrossRef]
- Parikh, K.; Cang, S.; Sekhri, A.; Liu, D. Selective inhibitors of nuclear export (SINE)-A novel class of anti-cancer agents. J. Hematol. Oncol. 2014, 7, 78. [Google Scholar] [CrossRef] [Green Version]




| Gene | log2FC | Adjusted p-Value | Description |
|---|---|---|---|
| UBB | 8.085482 | 2.67 × 10−96 | Polyubiquitin-B |
| LIN28A | 8.048782 | 8.71 × 10−55 | Protein lin-28 homolog A |
| XPO1 | 7.413878 | 1.61 × 10−22 | Exportin-1 |
| CELF2 | 7.108907 | 3.64 × 10−67 | CUGBP Elav-like family member 2 |
| MPRIP | 6.943921 | 5.21 × 10−45 | Myosin phosphatase Rho-interacting protein |
| TTC3 | 6.149392 | 1.34 × 10−16 | E3 ubiquitin-protein ligase TTC3 |
| MARCH6 | 6.124846 | 5.56 × 10−28 | E3 ubiquitin-protein ligase MARCH6 |
| HSPA4 | 6.113184 | 5.34 × 10−28 | Heat shock protein family A member 4 |
| PEX1 | 6.093165 | 1.01 × 10−28 | Peroxisome biogenesis factor 1 |
| PARD3 | 6.049788 | 1.30 × 10−20 | Partitioning defective 3 homolog |
| AKAP9 | 6.028525 | 2.06 × 10−07 | A-kinase anchor protein 9 |
| HIF1A | 6.020426 | 6.87 × 10−10 | Hypoxia-inducible factor 1-α |
| THOC2 | 6.000342 | 1.32 × 10−07 | THO complex subunit 2 |
| PUF60 | 5.996787 | 3.37 × 10−13 | Poly(U)-binding-splicing factor PUF60 |
| ATM | 5.95952 | 2.46 × 10−09 | Serine-protein kinase ATM |
| HSPA8 | 5.240461 | 0.000871 | Heat shock cognate 71 kDa protein |
| HNRNPC | 5.153226 | 3.94 × 10−26 | Heterogeneous nuclear ribonucleoproteins C1/C2 |
| AMBRA1 | 3.968858 | 0.001036 | Activating molecule in BECN1-regulated autophagy protein 1 |
| HDAC6 | 3.413792 | 2.43 × 10−06 | Histone deacetylase 6 |
| UBE3A | 3.258943 | 0.000783 | Ubiquitin-protein ligase E3A |
| HNRNPA1 | 3.215216 | 8.56 × 10−07 | Heterogeneous nuclear ribonucleoprotein A1 |
| HNRNPL | 3.004006 | 0.015223 | Heterogeneous nuclear ribonucleoprotein L |
| MBNL1 | 2.853727 | 0.000229 | Muscleblind-like protein 1 |
| DDX58 | 2.621277 | 0.011365 | Probable ATP-dependent RNA helicase DDX58 |
| ELAVL1 | 2.438159 | 3.69 × 10−05 | ELAV-like protein 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hesham, D.; El-Naggar, S. Transcriptomic Analysis Revealed an Emerging Role of Alternative Splicing in Embryonal Tumor with Multilayered Rosettes. Genes 2020, 11, 1108. https://doi.org/10.3390/genes11091108
Hesham D, El-Naggar S. Transcriptomic Analysis Revealed an Emerging Role of Alternative Splicing in Embryonal Tumor with Multilayered Rosettes. Genes. 2020; 11(9):1108. https://doi.org/10.3390/genes11091108
Chicago/Turabian StyleHesham, Dina, and Shahenda El-Naggar. 2020. "Transcriptomic Analysis Revealed an Emerging Role of Alternative Splicing in Embryonal Tumor with Multilayered Rosettes" Genes 11, no. 9: 1108. https://doi.org/10.3390/genes11091108






