MicroRNAs Modulate the Pathogenesis of Alzheimer’s Disease: An In Silico Analysis in the Human Brain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. miRNAs Selection
3. Results
RNA-Seq Analysis between Healthy Subjects and AD Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef] [PubMed]
- Juzwik, C.A.; Drake, S.S.; Zhang, Y.; Paradis-Isler, N.; Sylvester, A.; Amar-Zifkin, A.; Douglas, C.; Morquette, B.; Moore, C.S.; Fournier, A.E. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog. Neurobiol. 2019, 182, 101664. [Google Scholar] [CrossRef] [PubMed]
- Sadigh-Eteghad, S.; Sabermarouf, B.; Majdi, A.; Talebi, M.; Farhoudi, M.; Mahmoudi, J. Amyloid-beta: A crucial factor in Alzheimer’s disease. Med. Princ. Pract. Int. J. Kuwait Univ. Health Sci. Cent. 2015, 24, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Swomley, A.M.; Sultana, R. Amyloid beta-peptide (1-42)-induced oxidative stress in Alzheimer disease: Importance in disease pathogenesis and progression. Antioxid. Redox Signal. 2013, 19, 823–835. [Google Scholar] [CrossRef] [Green Version]
- Hebert, S.S.; Horre, K.; Nicolai, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef] [Green Version]
- Banzhaf-Strathmann, J.; Benito, E.; May, S.; Arzberger, T.; Tahirovic, S.; Kretzschmar, H.; Fischer, A.; Edbauer, D. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. Embo J. 2014, 33, 1667–1680. [Google Scholar] [CrossRef] [Green Version]
- Santa-Maria, I.; Alaniz, M.E.; Renwick, N.; Cela, C.; Fulga, T.A.; Van Vactor, D.; Tuschl, T.; Clark, L.N.; Shelanski, M.L.; McCabe, B.D.; et al. Dysregulation of microRNA-219 promotes neurodegeneration through post-transcriptional regulation of tau. J. Clin. Investig. 2015, 125, 681–686. [Google Scholar] [CrossRef]
- Nelson, P.T.; Wang, W.-X. MiR-107 is reduced in Alzheimer’s disease brain neocortex: Validation study. J. Alzheimer’s Dis. 2010, 21, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Sethi, P.; Lukiw, W.J. Micro-RNA abundance and stability in human brain: Specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci. Lett. 2009, 459, 100–104. [Google Scholar] [CrossRef]
- Cui, J.G.; Li, Y.Y.; Zhao, Y.; Bhattacharjee, S.; Lukiw, W.J. Differential regulation of Interleukin-1 Receptor-associated Kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-κB in stressed human astroglial cells and in Alzheimer disease. J. Biol. Chem. 2010, 285, 38951–38960. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Zhao, J.; Lu, G. miR-106b inhibits tau phosphorylation at Tyr18 by targeting Fyn in a model of Alzheimer’s disease. Biochem. Biophys. Res. Commun. 2016, 478, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Moncini, S.; Lunghi, M.; Valmadre, A.; Grasso, M.; Del Vescovo, V.; Riva, P.; Denti, M.A.; Venturin, M. The miR-15/107 family of microRNA genes regulates CDK5R1/p35 with implications for Alzheimer’s disease pathogenesis. Mol. Neurobiol. 2017, 54, 4329–4342. [Google Scholar] [CrossRef]
- Shioya, M.; Obayashi, S.; Tabunoki, H.; Arima, K.; Saito, Y.; Ishida, T.; Satoh, J.-I. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol. Appl. Neurobiol. 2010, 36, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Vijayan, M.; Reddy, P.H. MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer’s disease. Hum. Mol. Genet. 2017, 26, 3808–3822. [Google Scholar] [CrossRef] [PubMed]
- Long, J.M.; Maloney, B.; Rogers, J.T.; Lahiri, D.K. Novel upregulation of amyloid-β precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5′-untranslated region: Implications in Alzheimer’s disease. Mol. Psychiatr. 2019, 24, 345–363. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Reddy, P.H. MicroRNA-455-3p as a Potential Biomarker for Alzheimer’s Disease: An Update. Front. Aging Neurosci. 2018, 10, 41. [Google Scholar] [CrossRef] [Green Version]
- Madadi, S.; Schwarzenbach, H.; Saidijam, M.; Mahjub, R.; Soleimani, M. Potential microRNA-related targets in clearance pathways of amyloid-beta: Novel therapeutic approach for the treatment of Alzheimer’s disease. Cell Biosci. 2019, 9, 91. [Google Scholar] [CrossRef] [Green Version]
- Bazrgar, M.; Khodabakhsh, P.; Mohagheghi, F.; Prudencio, M.; Ahmadiani, A. Brain microRNAs dysregulation: Implication for missplicing and abnormal post-translational modifications of tau protein in Alzheimer’s disease and related tauopathies. Pharmacol. Res. 2020, 155, 104729. [Google Scholar] [CrossRef]
- Amakiri, N.; Kubosumi, A.; Tran, J.; Reddy, P.H. Amyloid Beta and MicroRNAs in Alzheimer’s Disease. Front. Neurosci. 2019, 13, 430. [Google Scholar] [CrossRef] [Green Version]
- Long, J.M.; Ray, B.; Lahiri, D.K. MicroRNA-339-5p down-regulates protein expression of beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J. Biol. Chem. 2014, 289, 5184–5198. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Song, Y.; Zhou, X.; Deng, Y.; Liu, T.; Weng, G.; Yu, D.; Pan, S. MicroRNA-29c targets beta-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol. Med. Rep. 2015, 12, 3081–3088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, P.Y.; Hernandez-Rapp, J.; Jolivette, F.; Lecours, C.; Bisht, K.; Goupil, C.; Dorval, V.; Parsi, S.; Morin, F.; Planel, E.; et al. miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet. 2015, 24, 6721–6735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, A.A.; Ganepola, G.A.P.; Rutledge, J.R.; Chang, D.H. The Potential Role of Dysregulated miRNAs in Alzheimer’s Disease Pathogenesis and Progression. J. Alzheimer’s Dis. 2019, 67, 1123–1145. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Chen, D.; Chen, N. The Regulation of microRNAs in Alzheimer’s Disease. Front. Neurol. 2020, 11, 288. [Google Scholar] [CrossRef] [PubMed]
- Angelucci, F.; Cechova, K.; Valis, M.; Kuca, K.; Zhang, B.; Hort, J. MicroRNAs in Alzheimer’s Disease: Diagnostic Markers or Therapeutic Agents? Front. Pharmacol. 2019, 10, 665. [Google Scholar] [CrossRef]
- Kaplan, J.T.; Gimbel, S.I.; Harris, S. Neural correlates of maintaining one’s political beliefs in the face of counterevidence. Sci. Rep. 2016, 6, 39589. [Google Scholar] [CrossRef] [Green Version]
- Scheff, S.W.; Price, D.A. Synapse loss in the temporal lobe in Alzheimer’s disease. Ann. Neurol. 1993, 33, 190–199. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; The International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [Green Version]
- Xiao, F.; Zuo, Z.; Cai, G.; Kang, S.; Gao, X.; Li, T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009, 37, D105–D110. [Google Scholar] [CrossRef]
- Hsu, S.D.; Lin, F.M.; Wu, W.Y.; Liang, C.; Huang, W.C.; Chan, W.L.; Tsai, W.T.; Chen, G.Z.; Lee, C.J.; Chiu, C.M.; et al. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011, 39, D163–D169. [Google Scholar] [CrossRef] [Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebert, S.S.; Wang, W.X.; Zhu, Q.; Nelson, P.T. A study of small RNAs from cerebral neocortex of pathology-verified Alzheimer’s disease, dementia with lewy bodies, hippocampal sclerosis, frontotemporal lobar dementia, and non-demented human controls. J. Alzheimer’s Dis. 2013, 35, 335–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [Green Version]
- Silvestro, S.; Bramanti, P.; Mazzon, E. Role of miRNAs in Alzheimer’s Disease and Possible Fields of Application. Int. J. Mol. Sci. 2019, 20, 3979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konovalova, J.; Gerasymchuk, D.; Parkkinen, I.; Chmielarz, P.; Domanskyi, A. Interplay between MicroRNAs and Oxidative Stress in Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 6055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadlon, A.; Takousis, P.; Alexopoulos, P.; Evangelou, E.; Prokopenko, I.; Perneczky, R. miRNAs Identify Shared Pathways in Alzheimer’s and Parkinson’s Diseases. Trends Mol. Med. 2019, 25, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Villa, C.; Ridolfi, E.; Fenoglio, C.; Ghezzi, L.; Vimercati, R.; Clerici, F.; Marcone, A.; Gallone, S.; Serpente, M.; Cantoni, C.; et al. Expression of the transcription factor Sp1 and its regulatory hsa-miR-29b in peripheral blood mononuclear cells from patients with Alzheimer’s disease. J. Alzheimer’s Dis. 2013, 35, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.A.; Tomas, J.F.; Queiroz, J.A.; Figueiras, A.R.; Sousa, F. Recombinant pre-miR-29b for Alzheimer s disease therapeutics. Sci. Rep. 2016, 6, 19946. [Google Scholar] [CrossRef]
- Jahangard, Y.; Monfared, H.; Moradi, A.; Zare, M.; Mirnajafi-Zadeh, J.; Mowla, S.J. Therapeutic Effects of Transplanted Exosomes Containing miR-29b to a Rat Model of Alzheimer’s Disease. Front. Neurosci. 2020, 14, 564. [Google Scholar] [CrossRef]
- Maldonado-Lasuncion, I.; Atienza, M.; Sanchez-Espinosa, M.P.; Cantero, J.L. Aging-Related Changes in Cognition and Cortical Integrity are Associated With Serum Expression of Candidate MicroRNAs for Alzheimer Disease. Cereb. Cortex 2019, 29, 4426–4437. [Google Scholar] [CrossRef]
- Roshan, R.; Shridhar, S.; Sarangdhar, M.A.; Banik, A.; Chawla, M.; Garg, M.; Singh, V.P.; Pillai, B. Brain-specific knockdown of miR-29 results in neuronal cell death and ataxia in mice. RNA 2014, 20, 1287–1297. [Google Scholar] [CrossRef] [Green Version]
- Patrick, E.; Rajagopal, S.; Wong, H.A.; McCabe, C.; Xu, J.; Tang, A.; Imboywa, S.H.; Schneider, J.A.; Pochet, N.; Krichevsky, A.M.; et al. Dissecting the role of non-coding RNAs in the accumulation of amyloid and tau neuropathologies in Alzheimer’s disease. Mol. Neurodegener. 2017, 12, 51. [Google Scholar] [CrossRef]
- Zeng, Z.; Liu, Y.; Zheng, W.; Liu, L.; Yin, H.; Zhang, S.; Bai, H.; Hua, L.; Wang, S.; Wang, Z.; et al. MicroRNA-129-5p alleviates nerve injury and inflammatory response of Alzheimer’s disease via downregulating SOX6. Cell Cycle 2019, 18, 3095–3110. [Google Scholar] [CrossRef]
- Wang, T.; Cai, Q.; Yang, W.J.; Fan, H.H.; Yi, J.F.; Xu, F. MicroRNA-219 alleviates glutamate-induced neurotoxicity in cultured hippocampal neurons by targeting calmodulin-dependent protein kinase II gamma. Neural Regen Res. 2018, 13, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, W.; Yi, Y.; Tong, Q. miR-219-5p inhibits tau phosphorylation by targeting TTBK1 and GSK-3beta in Alzheimer’s disease. J. Cell. Biochem. 2019, 120, 9936–9946. [Google Scholar] [CrossRef] [PubMed]
- Delay, C.; Dorval, V.; Fok, A.; Grenier-Boley, B.; Lambert, J.C.; Hsiung, G.Y.; Hebert, S.S. MicroRNAs targeting Nicastrin regulate Abeta production and are affected by target site polymorphisms. Front. Mol. Neurosci. 2014, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Di Gregoli, K.; Jenkins, N.; Salter, R.; White, S.; Newby, A.C.; Johnson, J.L. MicroRNA-24 regulates macrophage behavior and retards atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1990–2000. [Google Scholar] [CrossRef] [Green Version]
- Lugli, G.; Cohen, A.M.; Bennett, D.A.; Shah, R.C.; Fields, C.J.; Hernandez, A.G.; Smalheiser, N.R. Plasma Exosomal miRNAs in Persons with and without Alzheimer Disease: Altered Expression and Prospects for Biomarkers. PLoS ONE 2015, 10, e0139233. [Google Scholar] [CrossRef] [Green Version]
- Lusardi, T.A.; Phillips, J.I.; Wiedrick, J.T.; Harrington, C.A.; Lind, B.; Lapidus, J.A.; Quinn, J.F.; Saugstad, J.A. MicroRNAs in Human Cerebrospinal Fluid as Biomarkers for Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 55, 1223–1233. [Google Scholar] [CrossRef] [Green Version]
- Song, D.; Li, G.; Hong, Y.; Zhang, P.; Zhu, J.; Yang, L.; Huang, J. miR199a decreases Neuritin expression involved in the development of Alzheimer’s disease in APP/PS1 mice. Int. J. Mol. Med. 2020, 46, 384–396. [Google Scholar] [CrossRef]
- Mellios, N.; Feldman, D.A.; Sheridan, S.D.; Ip, J.P.K.; Kwok, S.; Amoah, S.K.; Rosen, B.; Rodriguez, B.A.; Crawford, B.; Swaminathan, R.; et al. MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol. Psychiatr. 2018, 23, 1051–1065. [Google Scholar] [CrossRef] [Green Version]
- Denk, J.; Boelmans, K.; Siegismund, C.; Lassner, D.; Arlt, S.; Jahn, H. MicroRNA Profiling of CSF Reveals Potential Biomarkers to Detect Alzheimer’s Disease. PLoS ONE 2015, 10, e0126423. [Google Scholar] [CrossRef] [Green Version]
- Ryan, B.; Williams, J.M.; Curtis, M.A. Plasma MicroRNAs Are Altered Early and Consistently in a Mouse Model of Tauopathy. Neuroscience 2019, 411, 164–176. [Google Scholar] [CrossRef]
- Singh, B.K.; Vatsa, N.; Kumar, V.; Shekhar, S.; Sharma, A.; Jana, N.R. Ube3a deficiency inhibits amyloid plaque formation in APPswe/PS1deltaE9 mouse model of Alzheimer’s disease. Hum. Mol. Genet. 2017, 26, 4042–4054. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.M.; Tang, Z.Y.; Sun, X.L. miR-411 suppresses acute spinal cord injury via downregulation of Fas ligand in rats. Biochem. Biophys. Res. Commun. 2018, 501, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Vetere, G.; Barbato, C.; Pezzola, S.; Frisone, P.; Aceti, M.; Ciotti, M.; Cogoni, C.; Ammassari-Teule, M.; Ruberti, F. Selective inhibition of miR-92 in hippocampal neurons alters contextual fear memory. Hippocampus 2014, 24, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.P.; Baker, K.E.; Fisher, A.; Hoff, L.; Pak, E.S.; Murashov, A.K. miRNA-431 Prevents Amyloid-beta-Induced Synapse Loss in Neuronal Cell Culture Model of Alzheimer’s Disease by Silencing Kremen1. Front. Cell. Neurosci. 2018, 12, 87. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Murashov, A.K. MicroRNA-431 regulates axon regeneration in mature sensory neurons by targeting the Wnt antagonist Kremen1. Front. Mol. Neurosci. 2013, 6, 35. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.S.; Liu, F.F.; Liu, D.; Huang, H.Z.; Wei, N.; Tan, L.; Chen, J.G.; Man, H.Y.; Gong, C.X.; Lu, Y.; et al. Opposite effects of two estrogen receptors on tau phosphorylation through disparate effects on the miR-218/PTPA pathway. Aging Cell 2015, 14, 867–877. [Google Scholar] [CrossRef]
- Venkataraman, S.; Birks, D.K.; Balakrishnan, I.; Alimova, I.; Harris, P.S.; Patel, P.R.; Handler, M.H.; Dubuc, A.; Taylor, M.D.; Foreman, N.K.; et al. MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma. J. Biol. Chem. 2013, 288, 1918–1928. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.; Sun, B.; Xu, P.; Zhu, Y.; Meng, X.H.; Teng, G.J.; Xiao, Z.D. MiR-218 Induces Neuronal Differentiation of ASCs in a Temporally Sequential Manner with Fibroblast Growth Factor by Regulation of the Wnt Signaling Pathway. Sci. Rep. 2017, 7, 39427. [Google Scholar] [CrossRef]
- Pallares-Albanell, J.; Zomeno-Abellan, M.T.; Escaramis, G.; Pantano, L.; Soriano, A.; Segura, M.F.; Marti, E. A High-Throughput Screening Identifies MicroRNA Inhibitors That Influence Neuronal Maintenance and/or Response to Oxidative Stress. Mol. Therapy. Nucleic Acids 2019, 17, 374–387. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, R.B.; Mufson, E.J.; Counts, S.E. Evidence for a neuroprotective microRNA pathway in amnestic mild cognitive impairment. Front. Neurosci. 2015, 9, 430. [Google Scholar] [CrossRef] [Green Version]
- Garza-Manero, S.; Arias, C.; Bermudez-Rattoni, F.; Vaca, L.; Zepeda, A. Identification of age- and disease-related alterations in circulating miRNAs in a mouse model of Alzheimer’s disease. Front. Cell. Neurosci. 2015, 9, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
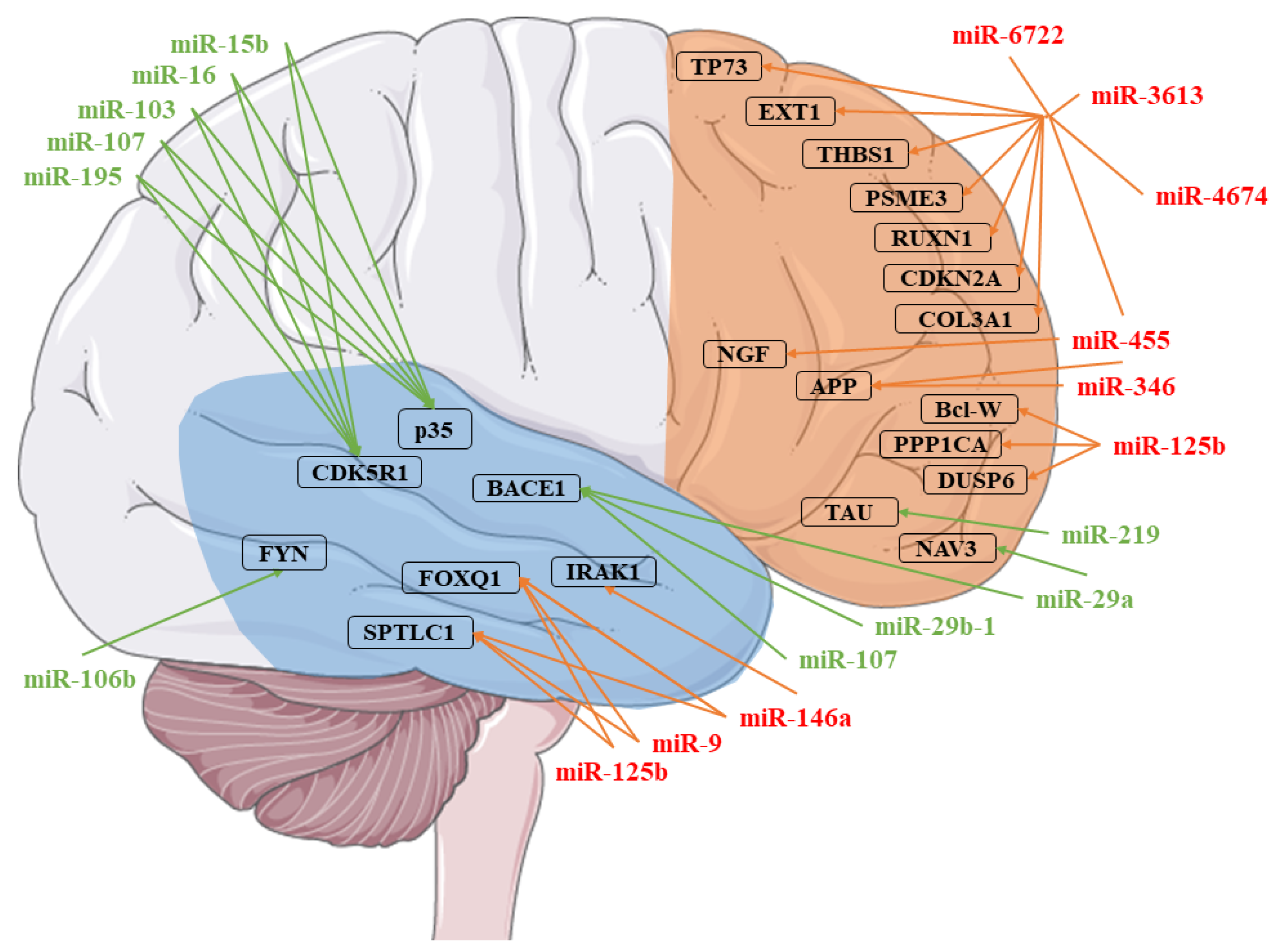

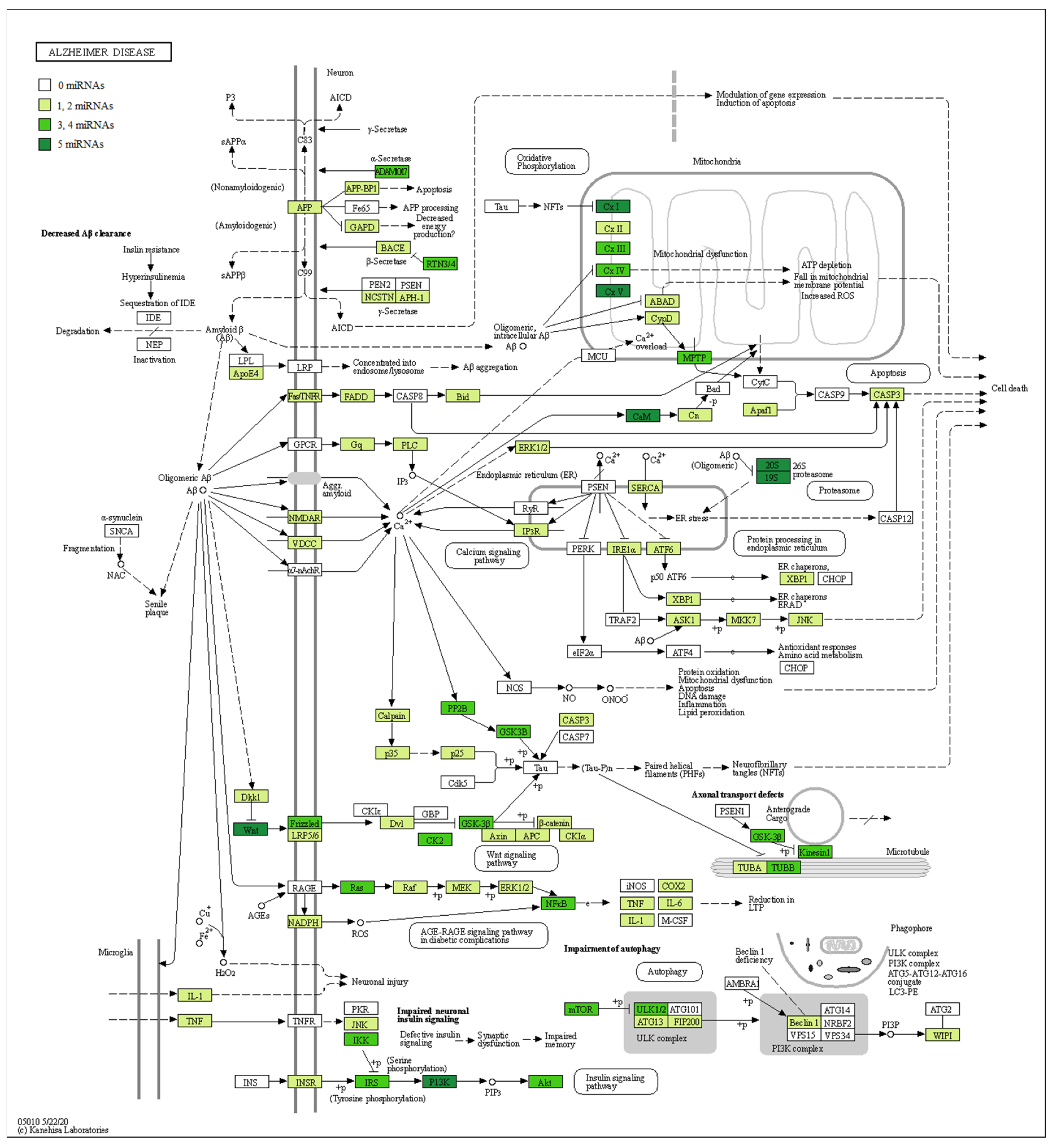
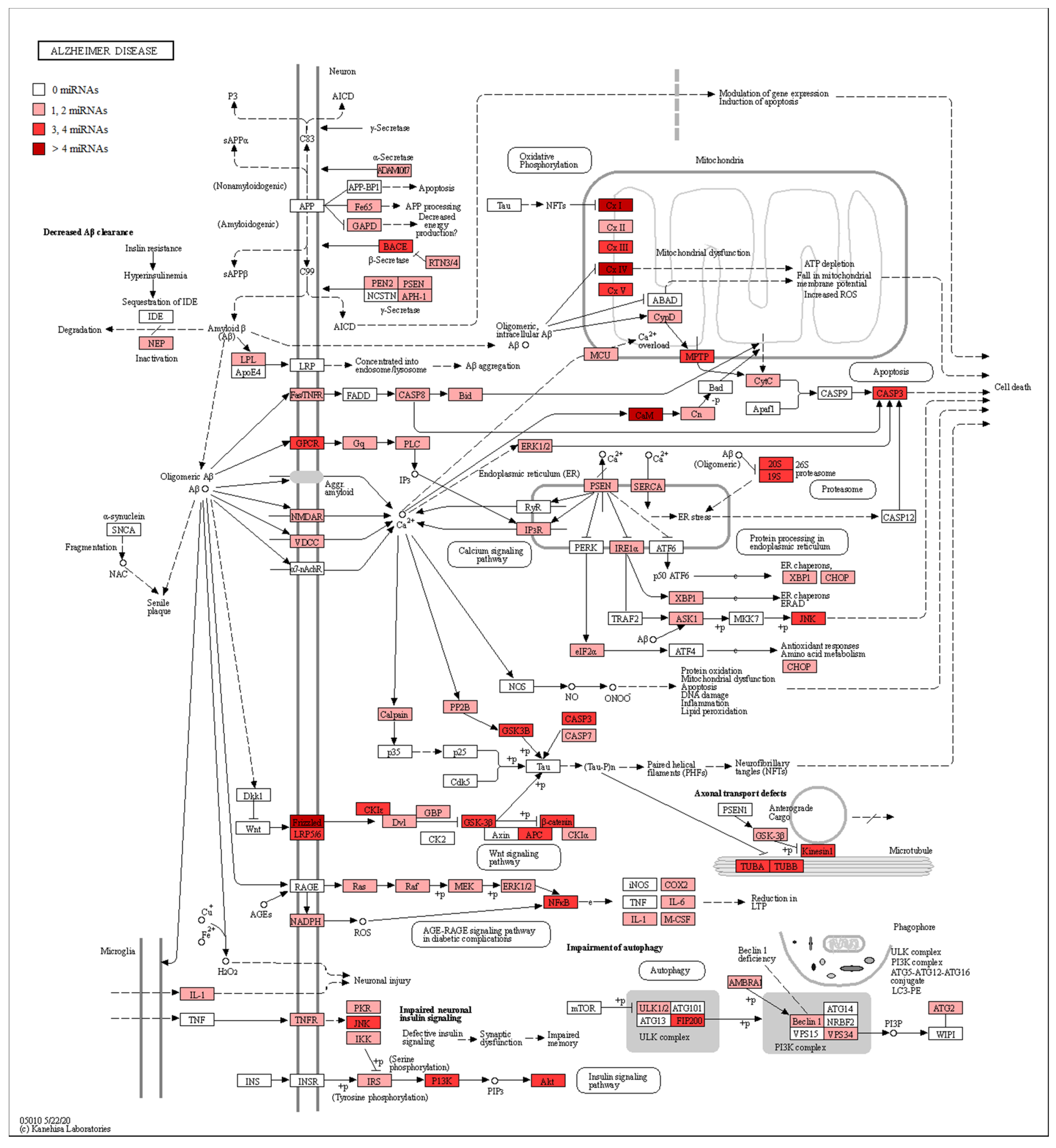
| miRNA | Healthy Subjects Expression ± SD | AD Patients Expression ± SD | Fold Change | q-Value |
|---|---|---|---|---|
| MIR129-2 | 4553.64 ± 11,498.81 | 387.41 ± 385.06 | −2.74 | 1.46 × 10−2 |
| MIR1296 | 302.98 ± 602.29 | 77.88 ± 55.42 | −2.41 | 2.43 × 10−2 |
| MIR199A2 | 1.74 ± 1.97 | 7.45 ± 10.04 | 2.38 | 1.82 × 10−2 |
| MIR218-2 | 4.51 ± 2.75 | 97.15 ± 239.28 | 3.46 | 1.82 × 10−2 |
| MIR219A1 | 5571.76 ± 14,780.51 | 3.36 ± 7.21 | −9.28 | 8.24 × 10−8 |
| MIR24-2 | 6.50 ± 3.93 | 16.23 ± 21.56 | 1.72 | 3.19 × 10−2 |
| MIR29B1 | 246.48 ± 301.94 | 17.75 ± 19.93 | −2.72 | 8.24 × 10−4 |
| MIR375 | 34.98 ± 13.42 | 12.94 ± 12.35 | −1.26 | 9.35 × 10−3 |
| MIR411 | 866.63 ± 671.94 | 442.34 ± 230.96 | −1.19 | 1.30 × 10−2 |
| MIR431 | 44.84 ± 55.42 | 19.07 ± 14.79 | −1.92 | 1.61 × 10−2 |
| MIR92A1 | 109.22 ± 114.82 | 448.56 ± 939.73 | 2.17 | 4.92 × 10−2 |
| MIR99A | 3250.67 ± 1624.41 | 5695.06 ± 2568.75 | 1.15 | 3.73 × 10−3 |
| miRNA | Biological Role Linked to AD | Pathway | Target |
|---|---|---|---|
| MIR129-2 | Nerve injury, inflammatory response, Aβ, and NFT plaques | - | - |
| MIR1296 | Neural response to oxidative stress | - | - |
| MIR199A2 | Neurogenesis, neural migration, early brain development, autophagy | - | Neuritin, ERK |
| MIR218-2 | Tau phosphorylation, mitochondrial respiratory chain | Wnt signaling pathway, non-amyloidogenic pathway | PTPα, ADAM17 |
| MIR219A1 | Tau phosphorylation, glutamate neurotoxicity | - | Tau, CAMK2G, TTBK1, GSK-3β |
| MIR24-2 | Aβ production | - | NCSTN, MMP14 |
| MIR29B1 | Neuron survival | Amyloidogenic pathway | BACE1, Sp1 |
| MIR375 | Tauopathy | - | UBE3A |
| MIR411 | Neuroprotective effects | - | FasL |
| MIR431 | Aβ protection | Wnt/β-catenin signaling pathway | Krm1 |
| MIR92A1 | Structural plasticity | - | CPEB3, MEF2D, KCC2 |
| MIR99A | NFT | - | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gugliandolo, A.; Chiricosta, L.; Boccardi, V.; Mecocci, P.; Bramanti, P.; Mazzon, E. MicroRNAs Modulate the Pathogenesis of Alzheimer’s Disease: An In Silico Analysis in the Human Brain. Genes 2020, 11, 983. https://doi.org/10.3390/genes11090983
Gugliandolo A, Chiricosta L, Boccardi V, Mecocci P, Bramanti P, Mazzon E. MicroRNAs Modulate the Pathogenesis of Alzheimer’s Disease: An In Silico Analysis in the Human Brain. Genes. 2020; 11(9):983. https://doi.org/10.3390/genes11090983
Chicago/Turabian StyleGugliandolo, Agnese, Luigi Chiricosta, Virginia Boccardi, Patrizia Mecocci, Placido Bramanti, and Emanuela Mazzon. 2020. "MicroRNAs Modulate the Pathogenesis of Alzheimer’s Disease: An In Silico Analysis in the Human Brain" Genes 11, no. 9: 983. https://doi.org/10.3390/genes11090983
APA StyleGugliandolo, A., Chiricosta, L., Boccardi, V., Mecocci, P., Bramanti, P., & Mazzon, E. (2020). MicroRNAs Modulate the Pathogenesis of Alzheimer’s Disease: An In Silico Analysis in the Human Brain. Genes, 11(9), 983. https://doi.org/10.3390/genes11090983







