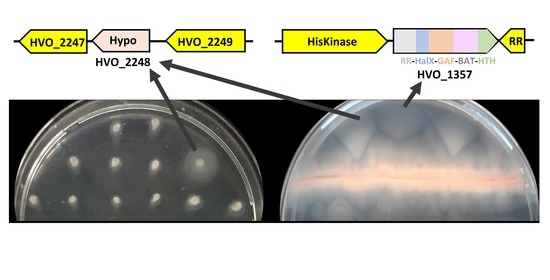
Mutations Affecting HVO_1357 or HVO_2248 Cause Hypermotility in Haloferax volcanii, Suggesting Roles in Motility Regulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Chemicals
2.2. Screens for Hfx. volcanii Hypermotility Mutants by Stabbing Individual Tn-Mutants
2.3. Hypermotility Mutant Selection by Streaking Cells of the Tn-Library across the Center of a Motility Agar Plate and Isolation of Cells That Migrated the Farthest
2.4. Genome Sequencing
2.5. Detection of Primary and Secondary Genome Alterations
2.6. Bioinformatic Analyses
3. Results and Discussion
3.1. Screening for Hypermotility by Stabbing Individual Strains
3.2. Selection for Hypermotility by Picking Cells That Moved Farthest from a Central Streak
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jung, J.; Kim, J.-S.; Taffner, J.; Berg, G.; Ryu, C.-M. Archaea, Tiny Helpers of Land Plants. Comput. Struct. Biotechnol. J. 2020, 18, 2494–2500. [Google Scholar] [CrossRef]
- Offre, P.; Spang, A.; Schleper, C. Archaea in Biogeochemical Cycles. Annu. Rev. Microbiol. 2013, 67, 437–457. [Google Scholar] [CrossRef] [Green Version]
- Spang, A.; Caceres, E.F.; Ettema, T.J.G. Genomic Exploration of the Diversity, Ecology, and Evolution of the Archaeal Domain of Life. Science 2017, 357, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Borrel, G.; Brugère, J.-F.; Gribaldo, S.; Schmitz, R.A.; Moissl-Eichinger, C. The Host-Associated Archaeome. Nat. Rev. Microbiol. 2020, 18, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Whon, T.W.; Lim, M.Y.; Kim, Y.B.; Kim, N.; Kwon, M.-S.; Kim, J.; Lee, S.H.; Choi, H.-J.; Nam, I.-H.; et al. The Human Gut Archaeome: Identification of Diverse Haloarchaea in Korean Subjects. Microbiome 2020, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.; Oesterhelt, D. Purification, Reconstitution and Polymorphic Transition of Halobacterial Flagella. J. Mol. Biol. 1987, 194, 495–499. [Google Scholar] [CrossRef]
- Albers, S.-V.; Jarrell, K.F. The Archaellum: An Update on the Unique Archaeal Motility Structure. Trends Microbiol. 2018, 26, 351–362. [Google Scholar] [CrossRef]
- Beeby, M.; Ferreira, J.L.; Tripp, P.; Albers, S.-V.; Mitchell, D.R. Propulsive Nanomachines: The Convergent Evolution of Archaella, Flagella and Cilia. FEMS Microbiol. Rev. 2020, 44, 253–304. [Google Scholar] [CrossRef]
- Meister, M.; Lowe, G.; Berg, H.C. The Proton Flux through the Bacterial Flagellar Motor. Cell 1987, 49, 643–650. [Google Scholar] [CrossRef]
- Streif, S.; Staudinger, W.F.; Marwan, W.; Oesterhelt, D. Flagellar Rotation in the Archaeon Halobacterium salinarum Depends on ATP. J. Mol. Biol. 2008, 384, 1–8. [Google Scholar] [CrossRef]
- Albers, S.-V.; Szabó, Z.; Driessen, A.J.M. Archaeal Homolog of Bacterial Type IV Prepilin Signal Peptidases with Broad Substrate Specificity. J. Bacteriol. 2003, 185, 3918–3925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardy, S.L.; Jarrell, K.F. Cleavage of Preflagellins by an Aspartic Acid Signal Peptidase Is Essential for Flagellation in the Archaeon Methanococcus voltae. Mol. Microbiol. 2003, 50, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Peabody, C.R.; Chung, Y.J.; Yen, M.-R.; Vidal-Ingigliardi, D.; Pugsley, A.P.; Saier, M.H. Type II Protein Secretion and Its Relationship to Bacterial Type IV Pili and Archaeal Flagella. Microbiol. Read. Engl. 2003, 149, 3051–3072. [Google Scholar] [CrossRef] [PubMed]
- Shahapure, R.; Driessen, R.P.; Haurat, M.F.; Albers, S.V.; Dame, R.T. The Archaellum: A Rotating Type IV Pilus. Mol. Microbiol. 2014, 91, 716–723. [Google Scholar] [CrossRef]
- Szabó, Z.; Stahl, A.O.; Albers, S.-V.; Kissinger, J.C.; Driessen, A.J.M.; Pohlschröder, M. Identification of Diverse Archaeal Proteins with Class III Signal Peptides Cleaved by Distinct Archaeal Prepilin Peptidases. J. Bacteriol. 2007, 189, 772–778. [Google Scholar] [CrossRef] [Green Version]
- Makarova, K.S.; Koonin, E.V.; Albers, S.-V. Diversity and Evolution of Type IV Pili Systems in Archaea. Front. Microbiol. 2016, 7, 667. [Google Scholar] [CrossRef] [Green Version]
- Patenge, N.; Berendes, A.; Engelhardt, H.; Schuster, S.C.; Oesterhelt, D. The Fla Gene Cluster Is Involved in the Biogenesis of Flagella in Halobacterium salinarum. Mol. Microbiol. 2001, 41, 653–663. [Google Scholar] [CrossRef]
- Thomas, N.A.; Mueller, S.; Klein, A.; Jarrell, K.F. Mutants in FlaI and FlaJ of the Archaeon Methanococcus voltae Are Deficient in Flagellum Assembly. Mol. Microbiol. 2002, 46, 879–887. [Google Scholar] [CrossRef]
- Chaudhury, P.; Neiner, T.; D’Imprima, E.; Banerjee, A.; Reindl, S.; Ghosh, A.; Arvai, A.S.; Mills, D.J.; Does, C.; Tainer, J.A.; et al. The Nucleotide-dependent Interaction of FlaH and FlaI Is Essential for Assembly and Function of the Archaellum Motor. Mol. Microbiol. 2016, 99, 674–685. [Google Scholar] [CrossRef]
- Reindl, S.; Ghosh, A.; Williams, G.J.; Lassak, K.; Neiner, T.; Henche, A.-L.; Albers, S.-V.; Tainer, J.A. Insights into FlaI Functions in Archaeal Motor Assembly and Motility from Structures, Conformations, and Genetics. Mol. Cell 2013, 49, 1069–1082. [Google Scholar] [CrossRef] [Green Version]
- Nutsch, T.; Oesterhelt, D.; Gilles, E.D.; Marwan, W. A Quantitative Model of the Switch Cycle of an Archaeal Flagellar Motor and Its Sensory Control. Biophys. J. 2005, 89, 2307–2323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nutsch, T.; Marwan, W.; Oesterhelt, D.; Gilles, E.D. Signal Processing and Flagellar Motor Switching During Phototaxis of Halobacterium salinarum. Genome Res. 2003, 13, 2406–2412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinosita, Y.; Uchida, N.; Nakane, D.; Nishizaka, T. Direct Observation of Rotation and Steps of the Archaellum in the Swimming Halophilic Archaeon Halobacterium salinarum. Nat. Microbiol. 2016, 1, 16148. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.K.; Staudinger, W.F.; Siedler, F.; Oesterhelt, D. Physiological Sites of Deamidation and Methyl Esterification in Sensory Transducers of Halobacterium salinarum. J. Mol. Biol. 2008, 380, 285–302. [Google Scholar] [CrossRef] [PubMed]
- Quax, T.E.F.; Albers, S.-V.; Pfeiffer, F. Taxis in Archaea. Emerg. Top. Life Sci. 2018, 2, 535–546. [Google Scholar] [CrossRef] [Green Version]
- Rudolph, J.; Oesterhelt, D. Chemotaxis and Phototaxis Require a CheA Histidine Kinase in the Archaeon Halobacterium salinarium. EMBO J. 1995, 14, 667–673. [Google Scholar] [CrossRef]
- Rudolph, J.; Oesterhelt, D. Deletion Analysis of the Che Operon in the Archaeon Halobacterium salinarium. J. Mol. Biol. 1996, 258, 548–554. [Google Scholar] [CrossRef]
- Schlesner, M.; Miller, A.; Streif, S.; Staudinger, W.F.; Müller, J.; Scheffer, B.; Siedler, F.; Oesterhelt, D. Identification of Archaea-Specific Chemotaxis Proteins Which Interact with the Flagellar Apparatus. BMC Microbiol. 2009, 9, 56. [Google Scholar] [CrossRef] [Green Version]
- Schlesner, M.; Miller, A.; Besir, H.; Aivaliotis, M.; Streif, J.; Scheffer, B.; Siedler, F.; Oesterhelt, D. The Protein Interaction Network of a Taxis Signal Transduction System in a Halophilic Archaeon. BMC Microbiol. 2012, 12, 272. [Google Scholar] [CrossRef] [Green Version]
- Wong-Ng, J.; Celani, A.; Vergassola, M. Exploring the Function of Bacterial Chemotaxis. Curr. Opin. Microbiol. 2018, 45, 16–21. [Google Scholar] [CrossRef]
- Yang, W.; Briegel, A. Diversity of Bacterial Chemosensory Arrays. Trends Microbiol. 2020, 28, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Quax, T.E.F.; Altegoer, F.; Rossi, F.; Li, Z.; Rodriguez-Franco, M.; Kraus, F.; Bange, G.; Albers, S.-V. Structure and Function of the Archaeal Response Regulator CheY. Proc. Natl. Acad. Sci. USA 2018, 115, E1259–E1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Kinosita, Y.; Rodriguez-Franco, M.; Nußbaum, P.; Braun, F.; Delpech, F.; Quax, T.E.F.; Albers, S.-V. Positioning of the Motility Machinery in Halophilic Archaea. mBio 2019, 10, e00377-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Rodriguez-Franco, M.; Albers, S.; Quax, T.E.F. The Switch Complex ArlCDE Connects the Chemotaxis System and the Archaellum. Mol. Microbiol. 2020, 114, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Duggin, I.G.; Aylett, C.H.S.; Walsh, J.C.; Michie, K.A.; Wang, Q.; Turnbull, L.; Dawson, E.M.; Harry, E.J.; Whitchurch, C.B.; Amos, L.A.; et al. CetZ Tubulin-like Proteins Control Archaeal Cell Shape. Nature 2015, 519, 362–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiljunen, S.; Pajunen, M.I.; Dilks, K.; Storf, S.; Pohlschroder, M.; Savilahti, H. Generation of Comprehensive Transposon Insertion Mutant Library for the Model Archaeon, Haloferax volcanii, and Its Use for Gene Discovery. BMC Biol. 2014, 12, 103. [Google Scholar] [CrossRef]
- Legerme, G.; Yang, E.; Esquivel, R.; Kiljunen, S.; Savilahti, H.; Pohlschroder, M. Screening of a Haloferax volcanii Transposon Library Reveals Novel Motility and Adhesion Mutants. Life 2016, 6, 41. [Google Scholar] [CrossRef] [Green Version]
- Legerme, G.; Pohlschroder, M. Limited Cross-Complementation Between Haloferax volcanii PilB1-C1 and PilB3-C3 Paralogs. Front. Microbiol. 2019, 10, 700. [Google Scholar] [CrossRef]
- Abdul Halim, M.F.; Pfeiffer, F.; Zou, J.; Frisch, A.; Haft, D.; Wu, S.; Tolić, N.; Brewer, H.; Payne, S.H.; Paša-Tolić, L.; et al. Haloferax volcanii Archaeosortase Is Required for Motility, Mating, and C-Terminal Processing of the S-Layer Glycoprotein. Mol. Microbiol. 2013, 88, 1164–1175. [Google Scholar] [CrossRef]
- Hoffmann, L.; Schummer, A.; Reimann, J.; Haurat, M.F.; Wilson, A.J.; Beeby, M.; Warscheid, B.; Albers, S.-V. Expanding the Archaellum Regulatory Network-The Eukaryotic Protein Kinases ArnC and ArnD Influence Motility of Sulfolobus acidocaldarius. MicrobiologyOpen 2017, 6, e00414. [Google Scholar] [CrossRef] [Green Version]
- Maier, L.-K.; Benz, J.; Fischer, S.; Alstetter, M.; Jaschinski, K.; Hilker, R.; Becker, A.; Allers, T.; Soppa, J.; Marchfelder, A. Deletion of the Sm1 Encoding Motif in the Lsm Gene Results in Distinct Changes in the Transcriptome and Enhanced Swarming Activity of Haloferax Cells. Biochimie 2015, 117, 129–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripepi, M.; Esquivel, R.N.; Wirth, R.; Pohlschröder, M. Haloferax volcanii Cells Lacking the Flagellin FlgA2 Are Hypermotile. Microbiology 2013, 159, 2249–2258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allers, T.; Ngo, H.-P.; Mevarech, M.; Lloyd, R.G. Development of Additional Selectable Markers for the Halophilic Archaeon Haloferax volcanii Based on the LeuB and TrpA Genes. Appl. Environ. Microbiol. 2004, 70, 943–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Silva, R.T.; Abdul-Halim, M.F.; Pittrich, D.A.; Brown, H.J.; Pohlschroder, M.; Duggin, I.G. Improved Growth and Morphological Plasticity of Haloferax volcanii. bioRxiv 2020, 1–29. [Google Scholar] [CrossRef]
- Hartman, A.L.; Norais, C.; Badger, J.H.; Delmas, S.; Haldenby, S.; Madupu, R.; Robinson, J.; Khouri, H.; Ren, Q.; Lowe, T.M.; et al. The Complete Genome Sequence of Haloferax volcanii DS2, a Model Archaeon. PLoS ONE 2010, 5, e9605. [Google Scholar] [CrossRef] [Green Version]
- Wendoloski, D.; Ferrer, C.; Dyall-Smith, M.L. A New Simvastatin (Mevinolin)-Resistance Marker from Haloarcula hispanica and a New Haloferax volcanii Strain Cured of Plasmid PHV2. Microbiol. Read. Engl. 2001, 147, 959–964. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, M.; Malla, S.; Blythe, M.J.; Nieduszynski, C.A.; Allers, T. Accelerated Growth in the Absence of DNA Replication Origins. Nature 2013, 503, 544–547. [Google Scholar] [CrossRef]
- Delmas, S.; Shunburne, L.; Ngo, H.-P.; Allers, T. Mre11-Rad50 Promotes Rapid Repair of DNA Damage in the Polyploid Archaeon Haloferax volcanii by Restraining Homologous Recombination. PLoS Genet. 2009, 5, e1000552. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://sourceforge.net/projects/bbmap/ (accessed on 19 October 2020).
- Becker, E.A.; Seitzer, P.M.; Tritt, A.; Larsen, D.; Krusor, M.; Yao, A.I.; Wu, D.; Madern, D.; Eisen, J.A.; Darling, A.E.; et al. Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-Response. PLoS Genet. 2014, 10, e1004784. [Google Scholar] [CrossRef]
- Zdobnov, E.M.; Tegenfeldt, F.; Kuznetsov, D.; Waterhouse, R.M.; Simão, F.A.; Ioannidis, P.; Seppey, M.; Loetscher, A.; Kriventseva, E.V. OrthoDB v9.1: Cataloging Evolutionary and Functional Annotations for Animal, Fungal, Plant, Archaeal, Bacterial and Viral Orthologs. Nucleic Acids Res. 2017, 45, D744–D749. [Google Scholar] [CrossRef]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB V10: Sampling the Diversity of Animal, Plant, Fungal, Protist, Bacterial and Viral Genomes for Evolutionary and Functional Annotations of Orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberto, J. SyntTax: A Web Server Linking Synteny to Prokaryotic Taxonomy. BMC Bioinform. 2013, 14, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R.; et al. IMG/M v.5.0: An Integrated Data Management and Comparative Analysis System for Microbial Genomes and Microbiomes. Nucleic Acids Res. 2019, 47, D666–D677. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Stamatis, D.; Bertsch, J.; Ovchinnikova, G.; Katta, H.Y.; Mojica, A.; Chen, I.-M.A.; Kyrpides, N.C.; Reddy, T. Genomes OnLine Database (GOLD) v.7: Updates and New Features. Nucleic Acids Res. 2019, 47, D649–D659. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Phyletic Distribution and Lineage-Specific Domain Architectures of Archaeal Two-Component Signal Transduction Systems. J. Bacteriol. 2018, 200, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ruepp, A.; Wanner, G.; Soppa, J. A 71-KDa Protein from Halobacterium salinarium Belongs to a Ubiquitous P-Loop ATPase Superfamily with Head-Rod-Tail Structure. Arch. Microbiol. 1998, 169, 1–9. [Google Scholar] [CrossRef]
- Miranda, H.V.; Antelmann, H.; Hepowit, N.; Chavarria, N.E.; Krause, D.J.; Pritz, J.R.; Bäsell, K.; Becher, D.; Humbard, M.A.; Brocchieri, L.; et al. Archaeal Ubiquitin-like SAMP3 Is Isopeptide-Linked to Proteins via a UbaA-Dependent Mechanism. Mol. Cell. Proteom. 2014, 13, 220–239. [Google Scholar] [CrossRef] [Green Version]
- Schulze, S.; Adams, Z.; Cerletti, M.; De Castro, R.; Ferreira-Cerca, S.; Fufezan, C.; Giménez, M.I.; Hippler, M.; Jevtic, Z.; Knüppel, R.; et al. The Archaeal Proteome Project Advances Knowledge about Archaeal Cell Biology through Comprehensive Proteomics. Nat. Commun. 2020, 11, 3145. [Google Scholar] [CrossRef]
- Gropp, F.; Betlach, M.C. The Bat Gene of Halobacterium halobium Encodes a Trans-Acting Oxygen Inducibility Factor. Proc. Natl. Acad. Sci. USA 1994, 91, 5475–5479. [Google Scholar] [CrossRef] [Green Version]
- Leong, D.; Boyer, H.; Betlach, M. Transcription of Genes Involved in Bacterio-Opsin Gene Expression in Mutants of a Halophilic Archaebacterium. J. Bacteriol. 1988, 170, 4910–4915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laass, S.; Monzon, V.A.; Kliemt, J.; Hammelmann, M.; Pfeiffer, F.; Förstner, K.U.; Soppa, J. Characterization of the Transcriptome of Haloferax volcanii, Grown under Four Different Conditions, with Mixed RNA-Seq. PLoS ONE 2019, 14, e0215986. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Affected Genes 1 | |||||
|---|---|---|---|---|---|
| Tn-Insertion Position 2 | Total Count | Primary | Secondary | Count | Isolates 3 |
| 2,734,510 | 3 | HVO_2248 | HVO_1357 | 1 | MC3 |
| HVO_2248 | - | 1 | MC50 | ||
| HVO_2248 | yes | 1 | SAH4 | ||
| 2,734,434 | 9 | HVO_2248 | HVO_1357 | 1 | MC14 |
| HVO_2248 | - | 6 | SAH5, MC9, 15, 16, 17, 26 | ||
| HVO_2248 | yes | 2 | MC27, 49 | ||
| 2,734,755 | 10 | HVO_2248 (Near) | HVO_1357, + | 3 | MC1, 28, 31 |
| HVO_2248 (Near) | - | 4 | MC8, 23, 30, 35 | ||
| HVO_2248 (Near) | yes | 3 | MC4, 6, 7 | ||
| 1,147,136 | 2 | HVO_0576 | HVO_2248, + | 2 | MC36, 45 |
| 2,404,062 | 1 | HVO_1926 | HVO_2248, + | 1 | MC34 |
| 2,723,422 | 1 | HVO_2229 | HVO_2248 | 1 | MC54 |
| 2,871,511 | 1 | HVO_2377 | HVO_2248, + | 1 | MC44 |
| 621,497 | 1 | HVO_A0546 | HVO_2248, + | 1 | MC52 |
| 1,867,146 | 4 | HVO_1357 | HVO_2248, + | 1 | MC11 |
| HVO_1357 | - | 1 | MC19 | ||
| HVO_1357 | yes | 2 | MC24, 37 | ||
| 1,867,189 | 13 | HVO_1357 | - | 7 | MC5, 10, 22, 29, 40, 41, 48 |
| HVO_1357 | yes | 6 | MC2, 12, 20, 21, 25, 33 | ||
| 1,865,880 | 2 | HVO_1357 | - | 2 | MC42, 46 |
| 2,678,278 | 1 | HVO_2176 (Near) | - | 1 | SAH1 |
| 2,225,641 | 1 | HVO_1726 | - | 1 | SAH2 |
| 1,020,227 | 1 | HVO_0430 | extensive | 1 | SAH3 |
| 3,126,642 | 1 | HVO_2649 | extensive | 1 | MC47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Collins, M.; Afolayan, S.; Igiraneza, A.B.; Schiller, H.; Krespan, E.; Beiting, D.P.; Dyall-Smith, M.; Pfeiffer, F.; Pohlschroder, M. Mutations Affecting HVO_1357 or HVO_2248 Cause Hypermotility in Haloferax volcanii, Suggesting Roles in Motility Regulation. Genes 2021, 12, 58. https://doi.org/10.3390/genes12010058
Collins M, Afolayan S, Igiraneza AB, Schiller H, Krespan E, Beiting DP, Dyall-Smith M, Pfeiffer F, Pohlschroder M. Mutations Affecting HVO_1357 or HVO_2248 Cause Hypermotility in Haloferax volcanii, Suggesting Roles in Motility Regulation. Genes. 2021; 12(1):58. https://doi.org/10.3390/genes12010058
Chicago/Turabian StyleCollins, Michiyah, Simisola Afolayan, Aime B. Igiraneza, Heather Schiller, Elise Krespan, Daniel P. Beiting, Mike Dyall-Smith, Friedhelm Pfeiffer, and Mechthild Pohlschroder. 2021. "Mutations Affecting HVO_1357 or HVO_2248 Cause Hypermotility in Haloferax volcanii, Suggesting Roles in Motility Regulation" Genes 12, no. 1: 58. https://doi.org/10.3390/genes12010058
APA StyleCollins, M., Afolayan, S., Igiraneza, A. B., Schiller, H., Krespan, E., Beiting, D. P., Dyall-Smith, M., Pfeiffer, F., & Pohlschroder, M. (2021). Mutations Affecting HVO_1357 or HVO_2248 Cause Hypermotility in Haloferax volcanii, Suggesting Roles in Motility Regulation. Genes, 12(1), 58. https://doi.org/10.3390/genes12010058