Abstract
Among other attributes, the Betaproteobacterial genus Azoarcus has biotechnological importance for plant growth-promotion and remediation of petroleum waste-polluted water and soils. It comprises at least two phylogenetically distinct groups. The “plant-associated” group includes strains that are isolated from the rhizosphere or root interior of the C4 plant Kallar Grass, but also strains from soil and/or water; all are considered to be obligate aerobes and all are diazotrophic. The other group (now partly incorporated into the new genus Aromatoleum) comprises a diverse range of species and strains that live in water or soil that is contaminated with petroleum and/or aromatic compounds; all are facultative or obligate anaerobes. Some are diazotrophs. A comparative genome analysis of 32 genomes from 30 Azoarcus-Aromatoleum strains was performed in order to delineate generic boundaries more precisely than the single gene, 16S rRNA, that has been commonly used in bacterial taxonomy. The origin of diazotrophy in Azoarcus-Aromatoleum was also investigated by comparing full-length sequences of nif genes, and by physiological measurements of nitrogenase activity using the acetylene reduction assay. Based on average nucleotide identity (ANI) and whole genome analyses, three major groups could be discerned: (i) Azoarcus comprising Az. communis, Az. indigens and Az. olearius, and two unnamed species complexes, (ii) Aromatoleum Group 1 comprising Ar. anaerobium, Ar. aromaticum, Ar. bremense, and Ar. buckelii, and (iii) Aromatoleum Group 2 comprising Ar. diolicum, Ar. evansii, Ar. petrolei, Ar. toluclasticum, Ar. tolulyticum, Ar. toluolicum, and Ar. toluvorans. Single strain lineages such as Azoarcus sp. KH32C, Az. pumilus, and Az. taiwanensis were also revealed. Full length sequences of nif-cluster genes revealed two groups of diazotrophs in Azoarcus-Aromatoleum with nif being derived from Dechloromonas in Azoarcus sensu stricto (and two Thauera strains) and from Azospira in Aromatoleum Group 2. Diazotrophy was confirmed in several strains, and for the first time in Az. communis LMG5514, Azoarcus sp. TTM-91 and Ar. toluolicum TT. In terms of ecology, with the exception of a few plant-associated strains in Azoarcus (s.s.), across the group, most strains/species are found in soil and water (often contaminated with petroleum or related aromatic compounds), sewage sludge, and seawater. The possession of nar, nap, nir, nor, and nos genes by most Azoarcus-Aromatoleum strains suggests that they have the potential to derive energy through anaerobic nitrate respiration, so this ability cannot be usefully used as a phenotypic marker to distinguish genera. However, the possession of bzd genes indicating the ability to degrade benzoate anaerobically plus the type of diazotrophy (aerobic vs. anaerobic) could, after confirmation of their functionality, be considered as distinguishing phenotypes in any new generic delineations. The taxonomy of the Azoarcus-Aromatoleum group should be revisited; retaining the generic name Azoarcus for its entirety, or creating additional genera are both possible outcomes.
1. Introduction
The genus Azoarcus (Az.) was first discovered after several strains were isolated from surface sterilised roots of the halo-tolerant C4 Poaceae species Leptochloa fusca L. (Kallar Grass) in Pakistan [1,2]. The new species Az. communis and Az. indigens were subsequently described [3], and the model endophytic Azoarcus strain, BH72, which was also isolated from Pakistani Kallar Grass, was recently incorporated into the new species, Az. olearius [4] on the basis of its high genomic similarity to the type strain, DQS-4T (which was isolated from oil-contaminated soil; [5]). Subsequent to the discovery of the plant-associated Azoarcus spp., several new species were isolated from non-plant sources, particularly from soil and/or water contaminated with aromatic compounds such as Az. evansii, Az. taiwanensis, Az. tolulyticus, Az. toluvorans, Az. toluclasticus, and Az. buckelii [6,7,8,9,10,11] as well as several strains that have not yet been classified/described to species level [12,13,14,15,16]. These are taxonomically separate from the plant-associated species [4,5,12,14,17,18] due to their ability to metabolize aromatic compounds, many of them under strict anaerobic conditions, and hence may have biotechnological potential as agents for the remediation of oil-polluted soils [19,20]. In addition, unlike the plant-associated species that are considered to be obligate aerobes, they are facultative anaerobes that can use nitrate as an alternative electron acceptor under anoxic conditions [21]. Both Azoarcus groups contain diazotrophs, but diazotrophy is not as common in the non-plant-associated group, and the nif genes of the two groups are unrelated [5,14,20,22].
So different are the two groups in terms of their molecular phylogeny and their lifestyles that there have been proposals to separate them into two genera [5,12,17,20]. Accordingly, Rabus et al. [21] have recently re-organised Azoarcus and described a new genus, Aromatoleum, comprising all species outside the “plant-associated” group including several newly-described species (Ar. aromaticum, Ar. bremense, Ar. diolicum, Ar. petrolei, and Ar. toluolicum). Azoarcus now consists only of the species Az. communis, Az. indigens, and Az. olearius. However, the molecular taxonomy in the Rabus et al. [21] study was based solely on 16S rRNA sequences and did not include several Azoarcus strains that were either not currently allocated to a species, or have been done so invalidly (e.g., Az. taiwanensis). Shortly after publication of Rabus et al. [21], one of these strains, SY39, which was isolated from seawater, was described as the type strain of a new species, Az. pumilus, that clusters loosely with Az. taiwanensis, and which is intermediate between Azoarcus and Aromatoleum [23], thus increasing uncertainty about strict taxonomic divisions in Azoarcus sensu lato. Rapid technical advances combined with modern computational solutions have greatly improved the speed and quality of the analysis and sequencing of bacterial genomes, concomitantly with a huge reduction in costs. Such genome sequences can be used to more accurately discern the phylogeny/taxonomy of groups of bacteria as the full sequences of several core genes can be used to construct robust phylogenies. Recent examples are Burkholderia-Paraburkholderia-Caballeronia [24], Trinickia and Mycetohabitans [25], and Bradyrhizobium [26,27].
The aim of the present study was to construct a robust phylogeny, utilising whole genome sequences of all described Azoarcus and Aromatoleum species together with genomes of strains that have not yet been allocated species status. A further aim was to use the genomes to investigate the lifestyle and ecology of the strains, with particular emphasis on the origin of the different types of nif genes in Azoarcus-Aromatoleum.
2. Materials and Methods
2.1. Bacterial Strains and Genomes
Fourteen Azoarcus-Aromatoleum genomes were sequenced as part of this study (Table S1a,b). These, together with previously published genome sequences and with others available in the National Center for Biotechnology Information (NCBI) database, totalling 32 Azoarcus-Aromatoleum genomes (Table 1) allowed robust reanalysis of the Azoarcus-Aromatoleum group and its separation into subgroups by different methods such as average nucleotide identity (ANI) and core genome analysis. A further 35 genomes from related genera (Table S1a) were also included in the analysis in order to place the Azoarcus-Aromatoleum group into a wider taxonomic/phylogenetic context.

Table 1.
Original sources of the strains used in phylogenies and evidence for diazotrophy.
For the purpose of genome sequencing, the following strains were obtained from various culture collections (in parentheses): Az. communis Swub3T (DSM), Az. communis LMG 5514 (LMG), Az. indigens VB32T (LMG), Az. taiwanensis NSC3 (LMG), Ar. anaerobium LuFRes1T (DSM), Ar. aromaticum pCyN1 (DSM), Ar. bremense PbN1T (DSM), Ar. buckelii U120T (DSM), Ar. diolicum 22LinT (DSM), Ar. evansii KB740T (DSM), Ar. petrolei ToN1T (DSM), Ar. toluolicum TT (DSM), and Ar. toluvorans Td21T (DSM). In addition, the Azoarcus sp. strain TTM-91 was described for the first time, and its genome was also sequenced as part of this study. Strain TTM-91 was isolated on R2A agar from uncontaminated water sampled from the Caohu River (24°04′24″ N, 120°46′04″ E) in the vicinity of Taichung City, Taiwan.
The genomes of the 14 strains in Table S1b were sequenced by MicrobesNG (UK) with the genomic DNA library prepared using the Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer’s protocol with the following modifications: 2 ng of DNA was used as input, and polymerase chain reaction (PCR) elongation time was increased to 1 min. DNA quantification and library preparation were carried out on a Microlab STAR automated liquid handling system (Hamilton, Birmingham, UK). Pooled libraries were quantified using the Kapa Biosystems Library Quantification Kit for Illumina on a Roche light cycler 96-qPCR machine (Roche, Basel, Switzerland). Libraries were sequenced on the Illumina HiSeq (Illumina) using a 250 bp paired end protocol. Read quality was verified with FastQC v0.11.5 [28] and reads were adapter trimmed and quality filtered using Trimmomatic v0.39 (http://www.usadellab.org/cms/index.php?page=trimmomatic) with a sliding window quality cut-off of Q15 [29]. Possible contaminations were removed with the bbsplit.sh script from BBMAP v37.87 [30] by removal of reads mapping to the contaminant’s genomes (e.g., human genome). De novo genome assembly was carried out with SPAdes v3.11.1 [31] and genes were annotated using Prokka v1.11 [32]. The assembly (contig) sizes were determined with QUAST v5.0.2 [33].
2.2. NCBI Sequence Datasets
All 67 (Azoarcus-Aromatoleum and related genera) genomes and nucleotide and amino acid coding sequences were downloaded from the NCBI GenBank in FASTA and GenBank formats and are presented in Table S1a.
2.3. Average Nucleotide Identity (ANI)
In order to relate our newly generated genomic resources to the pre-existing Azoarcus-Aromatoleum resources in the NCBI database, we applied whole genome comparisons by average nucleotide identity (ANI) using the alignment-free tool FastANI (version 1.31) [43] (available at: https://github.com/ParBLiSS/FastANI). We conducted two independent analyses: first, the pairwise calculation of the ANI values was performed with the 32 genomes listed in Table S2a; second, these calculations were performed with 52 genomes after the inclusion of 20 Thauera genomes (Table S2b). As a tool input requirement, the nucleotide coding sequences (CDS) were concatenated into a single multiFASTA. We performed the ANI pairwise values calculation for genomes using the FastANI default parameters. The FastANI output files (with a search summary and a lower triangular matrix containing the identity values in phylip format) were used to determine dissimilarity matrices and to construct a hierarchical neighbour-joining (NJ) tree cluster to reorder the lines and columns of the ANI matrix and construct a heat map. These analyses were performed using a Linux operating system (Ubuntu 20.4.1 LTS) and MATLAB® support programming.
2.4. Protein Clusters
The complete set of proteins translated from the CDS of each of the 67 genomes were joined in a FASTA file and clustered using the RAFTS3G tool [44] (available at: https://sourceforge.net/projects/rafts-g/) applying a minimal self-score of 0.7 as a threshold parameter for a specific protein to be included in a cluster. Two groupings were performed separately, RAFTS3G-32 for the Azoarcus-Aromatoleum group and RAFTS3g-67 for the 67 genomes. For the Azoarcus-Aromatoleum group, from an input multiFASTA of 145,695 protein sequences, RAFTS3G generated 13,959 clusters with two or more proteins (89.68% of the sequences) and 15,043 clusters with only one protein (10.32% of the sequences). For the 67 genomes, from a multiFASTA with 271,578 protein sequences, RAFTS3G generated 19,666 clusters with two or more proteins (89.39% of the sequences) and 28,811 clusters with only one protein (10.61% of the sequences).
The protein clusters were the basis for several steps in the subsequent phylogenetic/taxonomic analyses including core genome determination, nif gene cluster analysis, partial SWeeP genome representations, and specific common genes analyses.
2.5. Core Genome Analysis Based on Core Protein Groups
The core genome in the Azoarcus-Aromatoleum group was extracted and 1044 gene clusters were found in common in all 32 genomes (25.08% of the sequences). The analysis of core genome for all 67 genomes revealed only 231 common gene clusters (6.91% of the sequences).
2.6. Analysis of Nif and Other Functional Genes
Functional analyses of the nif clusters and nifH gene were performed for all 67 genomes. Analyses of the denitrification pathway and of aromatic compound degradation were performed only for the Azoarcus-Aromatoleum group.
The RAFTS3G-67 clusters were used to extract nif genes and genes for denitrification and benzoate metabolism. In this study, nif cluster analyses were performed using a manually curated “in-house” database containing 1546 genes present in the nif gene clusters of 144 diazotrophic organisms (Table S1c). This is an expanded version based on the study by Dos Santos et al. [45]. After database searches and manual validation, all nifH genes were found in the same RAFTS3G-67 grouping.
Analysis of functional genes was performed for the nitrate reduction and the aromatic compound degradation pathways. For nitrate reduction pathway analysis, we searched the RAFTS3G-32 clusters to identify the presence of genes encoding nitrate reductase (nar/nap), nitrite reductase (nir), nitric oxide reductase (nor), and nitrous oxide reductase (nos) using Aromatoleum sp. CIB corresponding marker genes (RAFTS3, [44,46]) and curated them manually. Aromatic compound degradation pathway genes including those for the anaerobic benzoate degradation pathway gene cluster (bzd), aerobic benzoate degradation pathway gene cluster (box), and genes from the “lower pathway” (LP) as marker genes were similarly searched in the RAFTS3G-32 groups.
2.7. SWeeP Phylogenies
Phylogenetic analyses were conducted using SWeeP [47] for protein set representations (available at: https://sourceforge.net/projects/spacedwordsprojection/). The purpose of this method is to transform any set of amino acid sequences into a single vector that can, for example, represent all proteins in an organism. In practice, the input file to generate the SWeeP vectors consists of a multiFASTA file containing amino acid sequences, in which the protein sequences in a particular organism are concatenated, each protein flanked by delimiters, to get a single sequence [47].
SWeeP vectors based on the complete set of proteins for each of the 32 Azoarcus-Aromatoleum genomes and for the wider group of 67 genomes were obtained to represent complete genomes. The parameters used were for spaced words, the “11011” mask, and for the projection into a size of 1369 for visualisation (37 × 37, a perfect square number, is desired for some visualisation tasks). These parameters were the same as those used for complete bacterial genomes in the study by De Pierri et al. [47] (see Figure S1).
The NJ model based on Euclidean distance matrices of SWeeP vectors was used for four analyses: phylogenetic analyses of complete genomes, phylogenetic trees and heatmaps for the analysis of the core genome, nif cluster, and nifH gene analyses. All phylogenetic trees were built using standard MATLAB functions and scripts (for an excerpt of script, see File S1, visualised with Dendroscope (version 3.7.2) [48] (available at: https://software-ab.informatik.uni-tuebingen.de/download/dendroscope3/welcome.html) and arranged with Adobe Illustrator CC 2017 (version 21.0.0, Adobe Corporation, San Jose, CA, USA).
2.8. Heat Maps
The heatmap graphs were constructed using “in-house” MATLAB® scripts and arranged using Adobe Illustrator CC 2017 (version 21.0.0). For the FastANI heatmap, the pairwise ANI values matrix was normalised by the mean values: First, ANI values were converted into dissimilarity matrices, then, the lower and upper matrices were divided by their corresponding mean values. It was decided to normalise the mean to guarantee a better correspondence between the matrices in the visualisation.
The results of FastANI were used to determine dissimilarities matrices to construct a hierarchical NJ tree cluster in which the order of lines and columns were established according to these clusters.
SWeeP vector heatmaps were generated from the distance matrices normalised by the average values, and the order of the rows and columns were determined by corresponding NJ-based dendrograms (see File S1). This technique allowed us to compare different distance/dissimilarity matrices in the same heat map.
2.9. 16S rRNA Analysis
For the 16S rRNA-based phylogeny, the genes were extracted automatically from 54 genomes from the NCBI GenBank using an “in-house” MATLAB® script and are listed in Table S1a. From the SILVA database [49], 13 16S rRNA genes from Dechloromonas sp. strains H13, Dech2017 and CZR5, D. hortensis MA-1, D. agitata is5, Thauera sp. UPWRP, T. butanivorans NBRC 103042, T. linaloolentis 47Lol_1 and 47Lol_2, T. hydrothermalis GD-2, T. phenolivorans ZV1C, Rhodocyclus tenuis DSM109T, and Rubrivivax gelatinosus CBS were collected. The 16S rRNA gene phylogenetic analyses were performed using Clustal Ômega [50] with default parameters. The phylogenetic trees were arranged with Adobe Illustrator CC 2017 (version 21.0.0).
2.10. Nitrogenase Activity
The acetylene reduction assay was used to determine nitrogenase activity on free-living cultures growing on semi-solid or liquid media [51,52]. Growth conditions for all strains are detailed in the Supplementary Materials (File S2). Ethylene (C2H4) formation was determined after incubation of the cultures in acetylene by using a Perkin Elmer Clarus 480 gas chromatograph equipped with a HayeSep® N (80–100 MESH) column (Merck Life Science Ltd., Dorset, UK). The injector and oven temperatures were kept at 100 °C, while the flame ionisation detector was set at 150 °C. The carrier gas (nitrogen) flow was set at 8–10 mL min−1. Nitrogenase activity is reported as nmol of C2H4 min−1 mg protein−1. The ethylene calibration curve was prepared from the chemical decomposition of ethephon (Merck Life Science Ltd., Dorset, UK) as described by Zhang and Wen [53]. Whole cell protein concentration was determined by the Bradford method [54] after lysis in 0.1 mM NaOH.
3. Results and Discussion
3.1. Whole-Genome Sequences and Identity of Azoarcus sp. Strain TTM-91
Features about the genomes of the 14 Azoarcus-Aromatoleum strains that were sequenced for this study are presented in Table 1 and Table S1a,b. The genomes of an additional 18 strains were obtained from the NCBI, and these are listed in Table 1 together with their original sources. Azoarcus sp. TTM-91 was the only strain that was isolated specifically for this study. The 16S rRNA sequence of Azoarcus sp. strain TTM-91 suggests that it is closely-related to Az. indigens (>99% similarity), and it also has a very similar genome size (5,393,782 bp) and G + C content (67.70%) to Az. indigens VB32T (Table 1 and Table S1b). The genomes of the remaining strains sequenced for this study were relatively standard in their size (ranging from 4,227,546 for Ar. buckelii to 6,025,652 for Ar. toluiolicum) and G + C content (ranging from 62.36% for Az. communis Swub3T to 67.70% for Azoarcus sp. TTM-91) in the wider context of the Azoarcus-Aromatoleum group (Table 1 and Table S1b).
3.2. Average Nucleotide Identity (ANI) and Core Genome Analysis
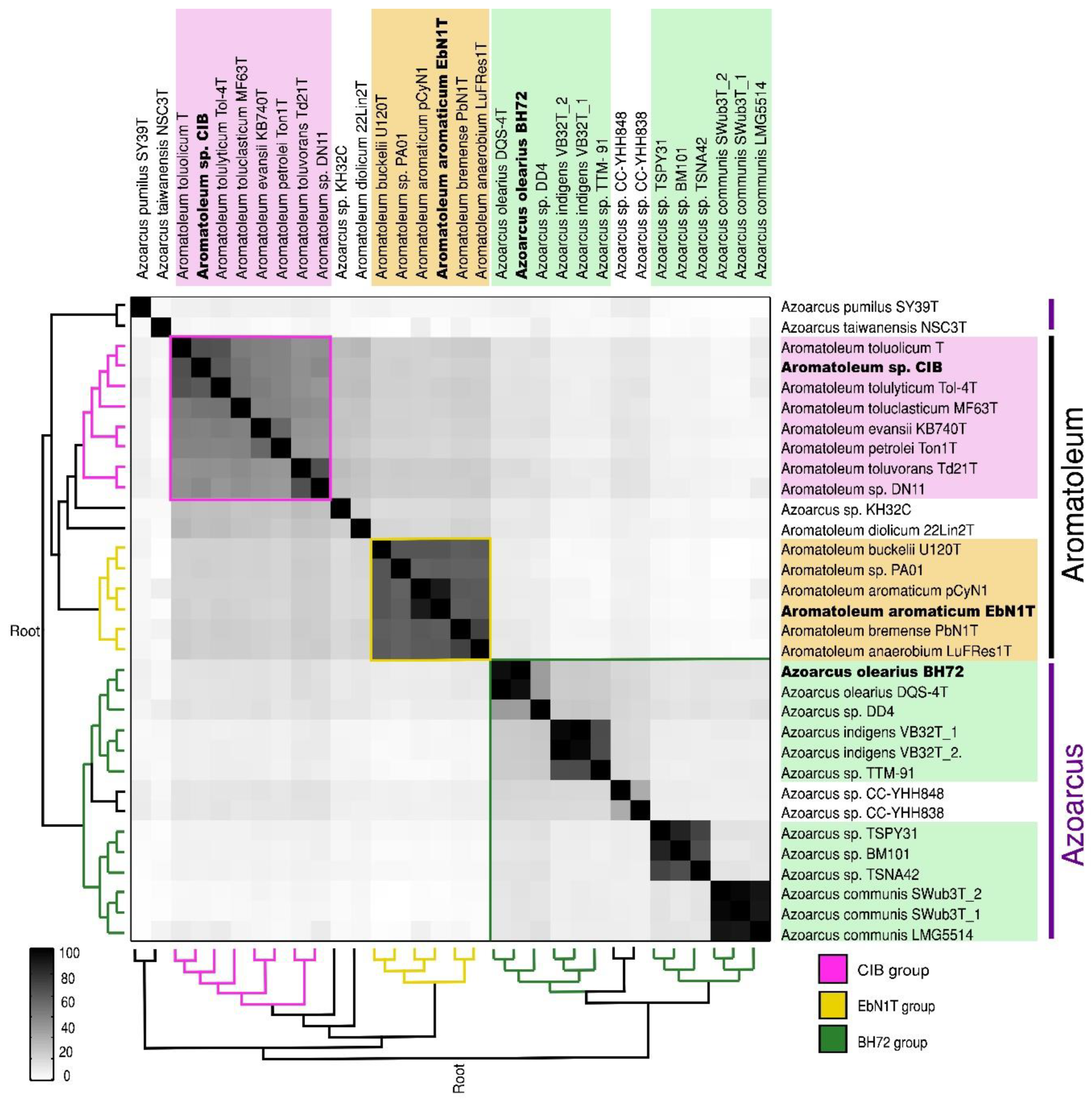
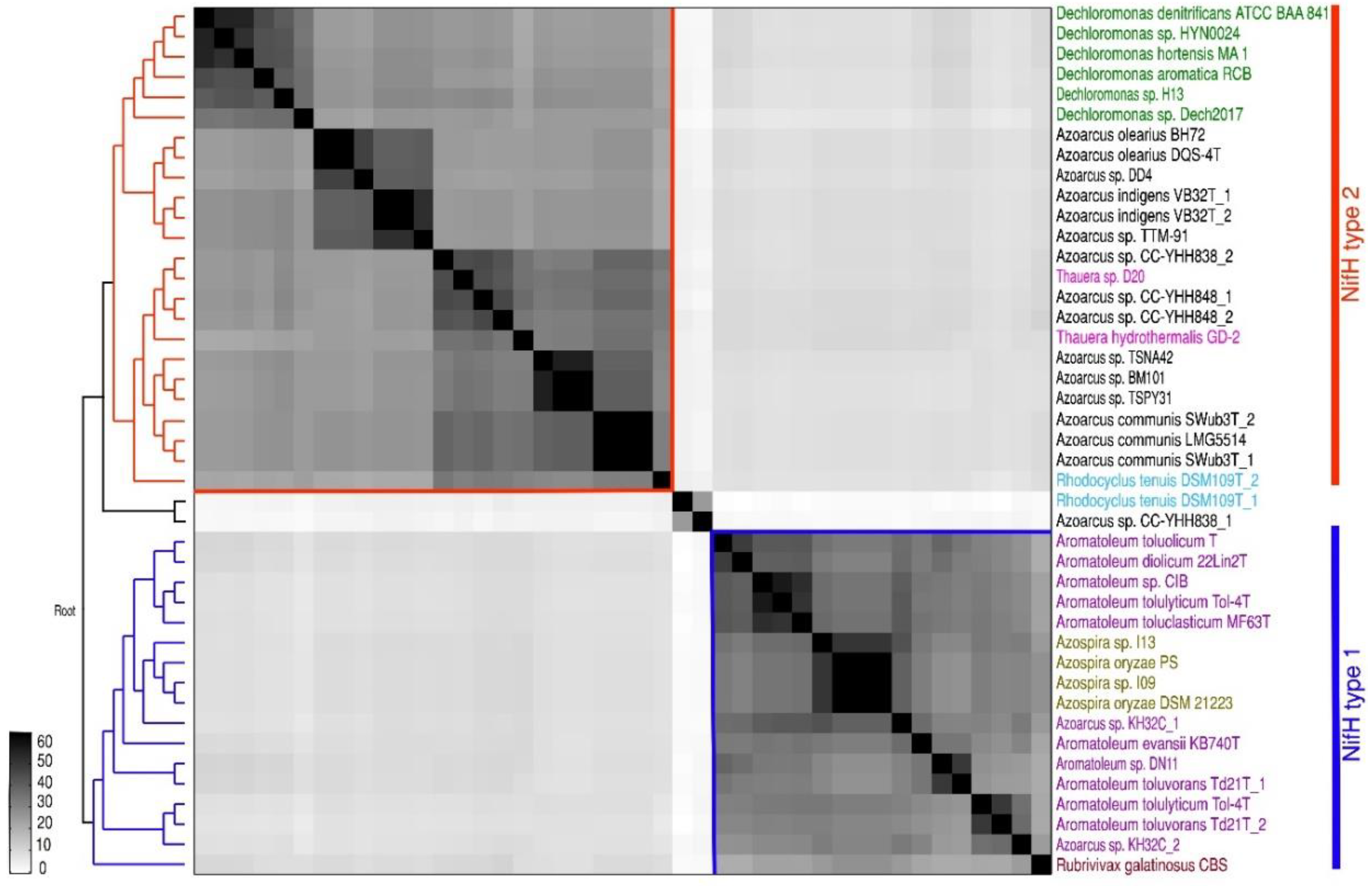
A matrix comparing the ANI of all 30 strains comprising 32 genomes (as two sequences were available for Az. communis Swub3T and Az. indigens VB32T) is presented in Figure 1, and the ANI percentage values are given in Table S2a. The genomes of the 30 Azoarcus-Aromatoleum strains are broadly divided into the three main groups representing the genera Azoarcus and two large clades in Aromatoleum, but other groupings are also discernible. Aromatoleum contains two clearly separate groups: Group 1 (“EbN1 group”) comprising the species Ar. anaerobium, Ar. aromaticum, Ar buckelii, and Ar. bremense, together with the strain PAO1; and Group 2 (“CIB group”) comprising the species Ar. diolicum, Ar. evansii, Ar. petrolei, Ar. toluclasticum, Ar. tolulyticum, Ar. toluolicum, and Ar. toluvorans, together with the “Azoarcus” strains CIB and DN11 [20,37]. Aromatoleum diolicum sits just outside Group 2, and the strain KH32C appears not to belong to either group, but is slightly closer to the Aromatoleum Group 2 than to Group 1. In the ANI matrix, the genus Azoarcus is divided into “species complexes” represented by the type strains (plus any related genome-sequenced strains) of the three formally described species, Az. communis, Az. indigens (possibly including strain TTM-91 from the present study), and Az. olearius, plus a fourth, undescribed species related to Az. communis comprising two strains from South Korea (TSPY31, TSPN42) and the metagenome-derived strain BM101 from the USA. Outside these four well-defined, and related Azoarcus species complexes, are two Taiwanese strains “Az. nasutitermitis” CC-YHH838 and “Az. rhizosphaerae” CC-YHH848 [42], and the type strains of the single strain species Az. pumilus and “Az. taiwanensis”, which are distantly related to each other.
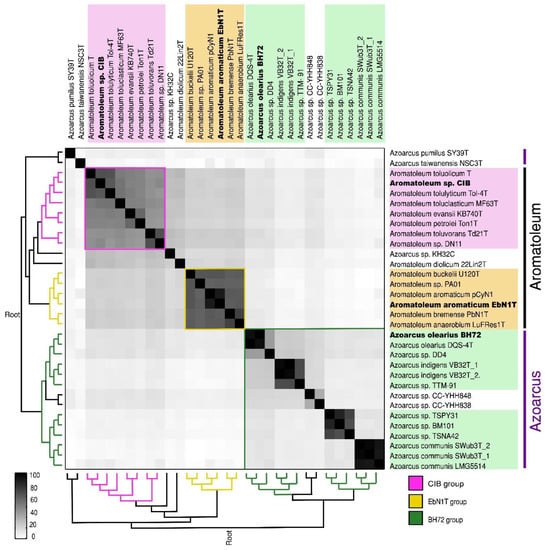
Figure 1.
Average nucleotide identity (ANI) of whole genome comparison of 32 (18 database and 14 newly sequenced) Azoarcus and Aromatoleum genomes from 30 strains showing subgroups of the known genera Azoarcus and Aromatoleum. The depicted heatmap is a matrix generated by FastANI, ordered according to the dendrograms (left/below) obtained with the neighbour joining clustering method in MATLAB, considering upper and lower triangular complementary ANI dissimilarity matrices as input. All genome accession numbers are listed in Table 1 and Table S1a.
A two principal components projection for k-means clusters (k = 4) generated with SWeeP vectors demonstrated that the wider group of 67 genome-sequenced strains was clustered into four groups (Figure S2a), and these were linked with a core genome phylogeny (Figure S2b). This analysis allowed us to take a wider look at the Azoarcus-Aromatoleum group, particularly in the context of its neighbouring genus Thauera. It suggests that Thauera (or at least those 20 strains within it whose genomes have been fully sequenced) comprises two groups: a larger one in k3 consisting of 18 of the sequenced Thauera strains, and a much smaller subgroup containing only T. hydrothermalis GD-2T and Thauera sp. D20 that is placed in k2 (which also contains most of the Azoarcus strains). Both Thauera groups were more closely related to Azoarcus than to Aromatoleum, with T. hydrothermalis GD-2T and Thauera sp. D20 obviously nested within Azoarcus.
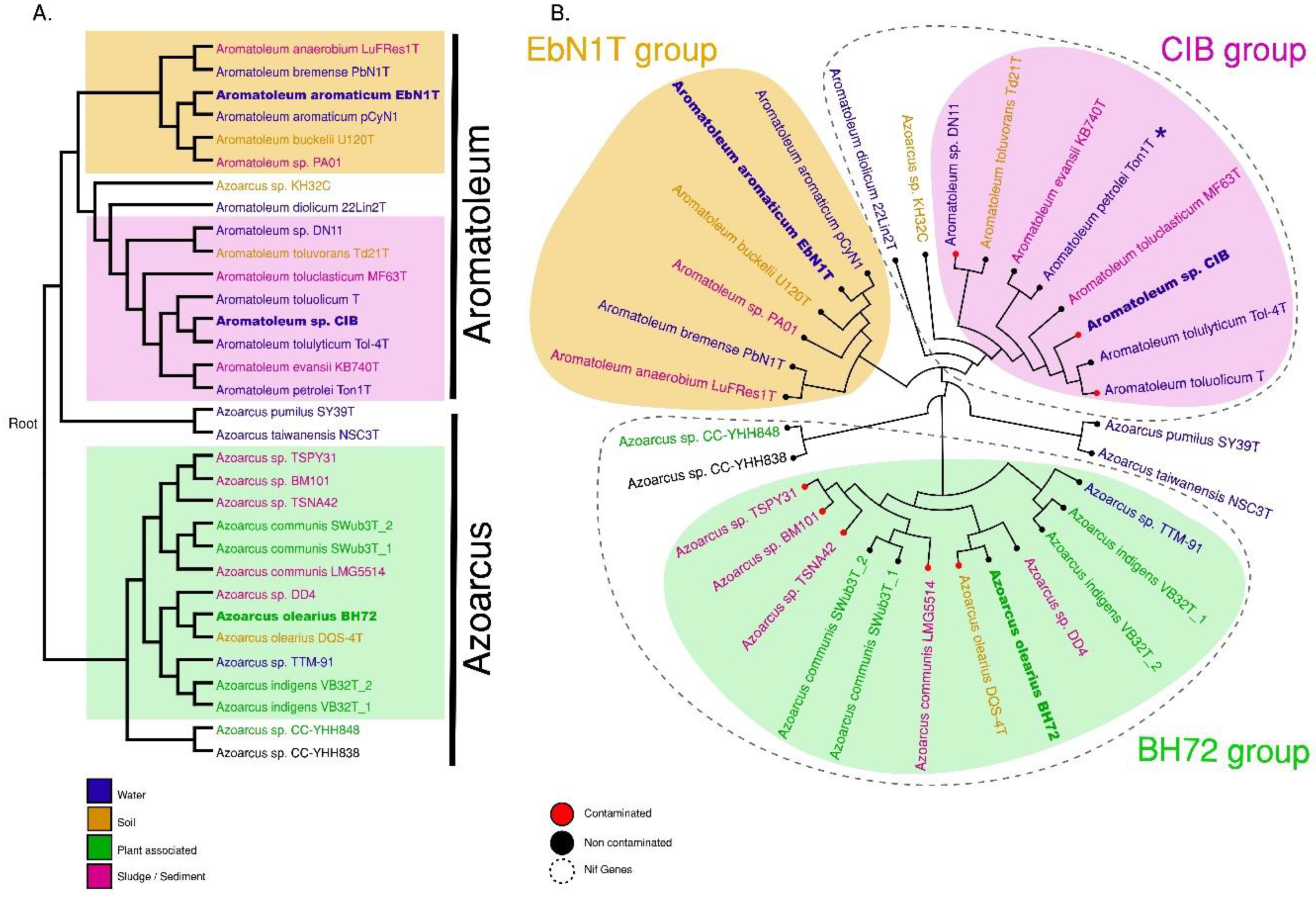
A phylogenetic analysis was performed based on the whole genome and the core genome (Figure 2A,B). Three major groups can be discerned comprising (1) Azoarcus consisting of 14 strains including all currently accepted Azoarcus species plus the informally named Az. nasutitermitis (CC-YHH838), Az. rhizosphaerae (CC-YHH848) and Az. taiwanensis; (2) The “EbN1-Group” of six Aromatoleum strains including Ar. aromaticum EbN1T; and (3) the “CIB-Group” of 10 Aromatoleum strains plus “Azoarcus” sp. KH32C. Three small sub-groups are apparent in Azoarcus; the one closest to Azoarcus sensu stricto (s.s.) consists of two Taiwanese strains (CC-YHH838, CC-YHH848), and the other groups, which are more distant from Azoarcus (s.s.), consist of Az. pumilus and Az. taiwanensis from China and Taiwan, respectively. The EbN1-Group in Aromatoleum is monophyletic and could thus be termed Aromatoleum sensu stricto (s.s.), but the CIB-Group is looser and could be considered a polytomy (Figure 2A).
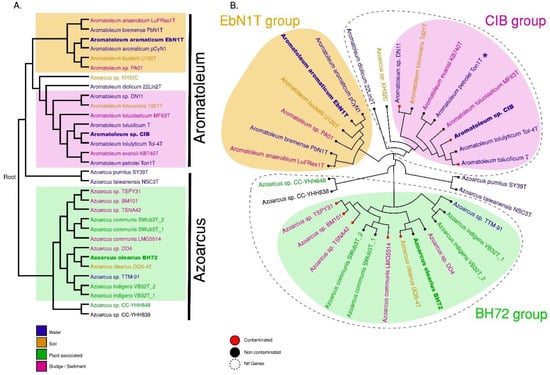
Figure 2.
Phylogenetic trees of whole genomes (A) and clustering of core genomes (B). These analyses confirm the grouping of 30 Azoarcus and Aromatoleum strains (represented by 32 genomes) into three major groups named the EBN1, CIB, and BH72 groups. Both trees were constructed based on distance matrices of SWeeP vectors. The colours in the names indicate the source of the initial isolation of the corresponding strain: water (blue), soil (brown), sludge/sediment (pink) or plant-associated (green). Whether the substrate was contaminated (red) by organic solvents or non-contaminated (black) is indicated by the tip-point colour in the circular tree. The presence of nif genes in a group is indicated with dashed outlines. Asterisk (*) indicates the absence of nif genes in Ar. petrolei Ton1T.
In terms of ecology/habitat, no obvious pattern could be discerned in the genotaxonomy of Azoarcus-Aromatoleum (Figure 2B) (i.e., the 30 taxa come from a wide range of habitats, and no habitat is particular to any of the three major groups). These include water from widely different sources (oceans, rivers, hot springs, aquifers, and wastewater treatment plants) as the primary habitat for the genome-sequenced Azoarcus-Aromatoleum strains, comprising 13 of the strains, sludge/sediment as the secondary habitat with eight strains, and soil as the tertiary habitat with four strains. Of the remaining five strains, all of which are in Azoarcus, one came from a termite nest in Taiwan (Az. nasutitermitis CC-YHH838), one from the rhizosphere of Ficus religiosa in Taiwan (Az. rhizosphaerae CC-YHH848), and three from roots of Kallar Grass including the model endophyte Az. olearius BH72, and the type strains of Az. communis (SwuB3T) and Az. indigens (VB32T). The genome-sequenced strains were also scored for habitats that were indicated as being contaminated by oil/petroleum; this showed that all the major groups in Azoarcus-Aromatoleum contained strains (seven of the genome-sequenced strains) that had an association with contaminated water or soils, but this association is by no means ubiquitous (Figure 2B).
In summary, although it should be stressed that the 30 strains presented in Figure 2B are only a “snapshot” of the habitats of Azoarcus-Aromatoleum strains, and their selection for our analysis is inevitably biased by the requirement for fully genome-sequenced strains, it is, nevertheless, clear from the present study and from perusing the databases for deposited gene sequences that water, sludge, and soil (contaminated and uncontaminated) are the principal habitats of all three Azoarcus-Aromatoleum groups. Plants are not a common habitat; only three strains in our analysis were actually isolated from plants (i.e., Kallar Grass), and all of these were from a single field in Pakistan [1,3]. Moreover, in each case, these plant-associated strains (Az. olearius BH72, Az. communis Swub3T, and Az. indigens VB32T) have sister strains in the same (or closely-related) species that were not isolated from plants such as Az. olearius DQS-4T (oil-contaminated soil; [5]), Az. communis LMG 5514 (refinery oil sludge; [3]), and Azoarcus sp. TTM-91 (river water; this study). Other examples are Azoarcus sp. DD4, which is quite closely related to Az. olearius and may belong to this species (Figure 2A,B); this was isolated from activated sludge from a wastewater treatment plant [16]. Moreover, the other genome-sequenced strains in the Az. communis species complex (BM101, TSPY31, TSNA42), were isolated from sediments. Therefore, even though Azoarcus was the only group in the present study that contained plant-associated strains/species, these were few in number, and the most intensively studied among them, Az. olearius BH72, did not appear to possess genes that make it any more specialised in terms of associating with plants than its non-plant-associated sister Az. olearius DQS-4T [5]. Therefore, widely used terms in the literature like “the plant-associated Azoarcus genus/group” (see Introduction) should henceforth be avoided as these give a misleading impression about the much wider habitat preferences of Azoarcus.
Phylogenetic whole genome analyses including Thauera (Figure S2a,b) also indicated that T. hydrothermalis GD-2T and Thauera sp. D20 were actually nested within Azoarcus, and are apparently most closely related to the Azoarcus strains CC-YHH838 and CC-YHH848. Thauera hydrothermalis GD-2T was isolated from the sediment of a hot spring in Tibet [55], and is a facultative anaerobe. Nothing has been published as yet about Thauera sp. D20, except that it was isolated from a saline lake in Inner Mongolia [56].
The analyses of whole genomes and of the core genomes of the 30 Azoarcus-Aromatoleum strains (Figure 2) has closely supported the groupings revealed by the ANI analysis. In addition, the taxonomy revealed by these genome-based analyses does not conflict with that shown by 16S rRNA sequences extracted from the genomes (Figure S3), which in itself is largely concordant with that of Rabus et al. [21], who used fewer (and mainly type) strains to construct their 16S rRNA phylogeny. The main differences between the whole genome analysis (Figures S2b and S4) and that based on 16S rRNA (Figure S3) is that the former places T. hydrothermalis GD-2T and Thauera sp. D20 in Azoarcus (see above), but the latter places them in Thauera together with the Azoarcus strains CC-YHH838 and CC-YHH848 (Figure S3). Further studies including a polyphasic analysis should help determine if T. hydrothermalis GD-2T and Thauera sp. D20 actually belong to Azoarcus or if they belong to Thauera together with CC-YHH838 and CC-YHH848.
At present, the delineation of bacterial species and, to some extent, genera is still based upon the 16S rRNA gene with suggested cut-offs of <97% similarity between related taxa for species, and a more arbitrary value of 94–95% for genera [57,58,59,60]. In addition, for generic delineations, it is recommended that phenotypic and lifestyle attributes be taken into account in addition to genetics. Accordingly, Rabus et al. [21] described the new genus Aromatoleum on the basis that they are facultative aerobes that can use nitrate as an alternative electron acceptor under anoxia, and that they have the ability to degrade aromatic compounds under anaerobic conditions. Although they also recognised that 16S rRNA sequences showed that Aromatoleum contained two clades corresponding to the Groups 1 (EbN1T) and 2 (CIB) described in the present study, Rabus et al. [21] recommended that in spite of this genetic separation that Aromatoleum should not be further divided as suggested by Martin-Moldes et al. [20], as all members of the new genus as delineated by them had the overarching and prominent property of anaerobic biodegradation of aromatic compounds. However, they did not include several strains that are pertinent to this debate such as Azoarcus sp. KH32C, Az. pumilus, and Az. taiwanensis, which fit into neither Aromatoleum nor Azoarcus as delineated by Rabus et al. [21]. Reinhold-Hurek et al. [17] described three new genera within Azoarcus sensu lato as it stood at that time (viz. Azonexus, Azopira, and Azovibrio), at least partly on the basis that their 16S rRNA sequences were only 93–94% similar to Azoarcus (s.s.). If 16S rRNA sequences were to be used in this manner to discern genera within the Azoarcus-Aromatoleum strains in the present study, it would suggest that Azoarcus-Aromatoleum constitutes a single genus with the exception of Az. pumilus and Az. taiwanensis, which would be placed into separate genera (Figure S3, Table S3a,b). However, we are rapidly moving into an era of genome-based taxonomies, and so 16S rRNA sequences are likely to be used less and less frequently as the sole gene sequence-based criterion for describing new taxa.
Although it has recently been demonstrated that ANI cannot be used for genus definition [61], it can still give indications of where generic boundaries might genuinely lie. In the present study, together with whole genome and core phylogenetics analysis from large numbers of genes, it is suggested that in terms of their core genomes, the two clades in Aromatoleum could constitute separate genera, but also that separate genera for Az. pumilus and Az. taiwanensis could be created, and possibly another for Azoarcus strain KH32C (Figures S2b and S4).
3.3. Genes for Nitrate Reduction (Nar/Nap, Nir, Nor, Nos) and for the Anaerobic (Bzd) or Aerobic (Box) Degradation of Benzoate
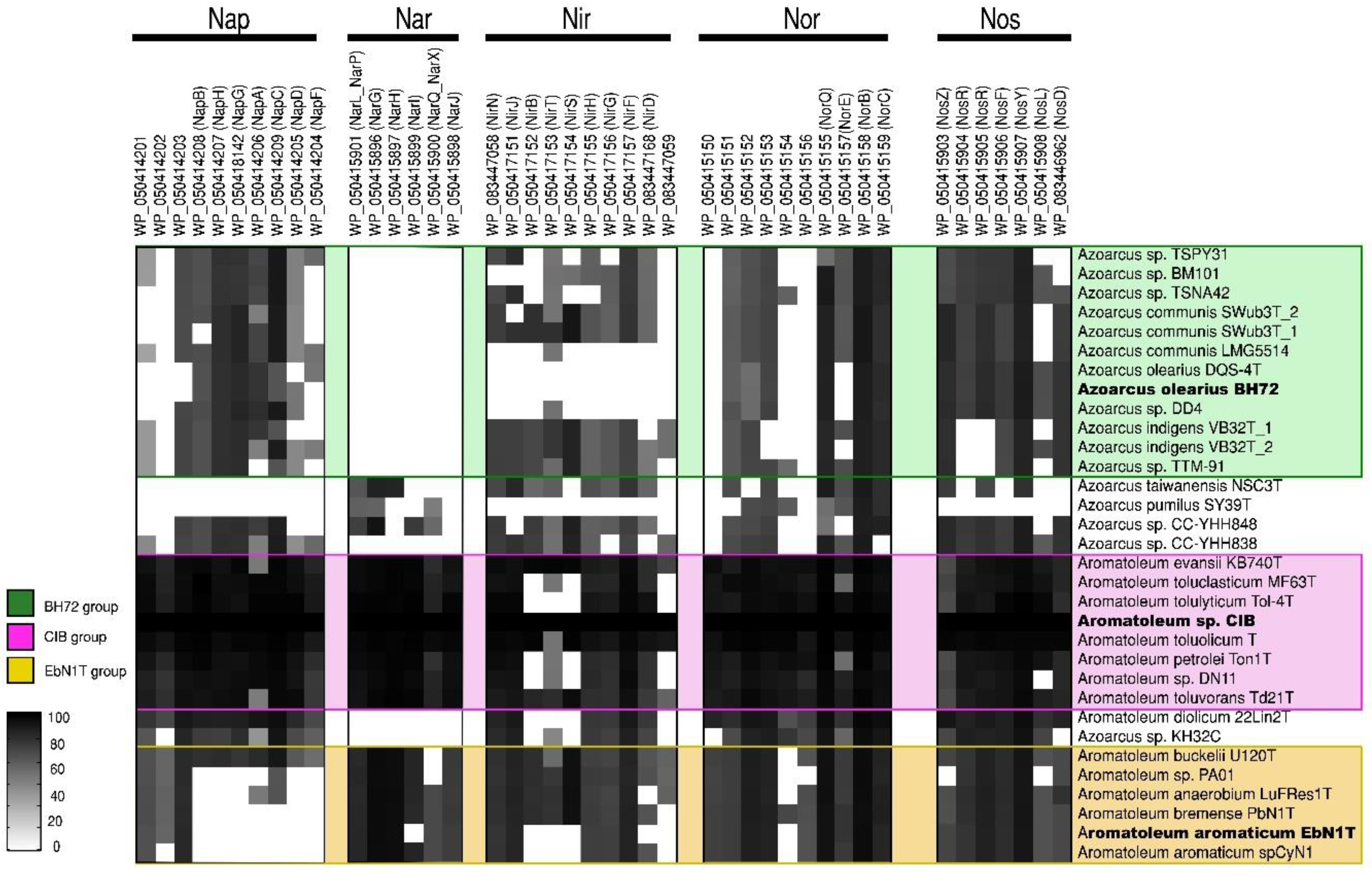
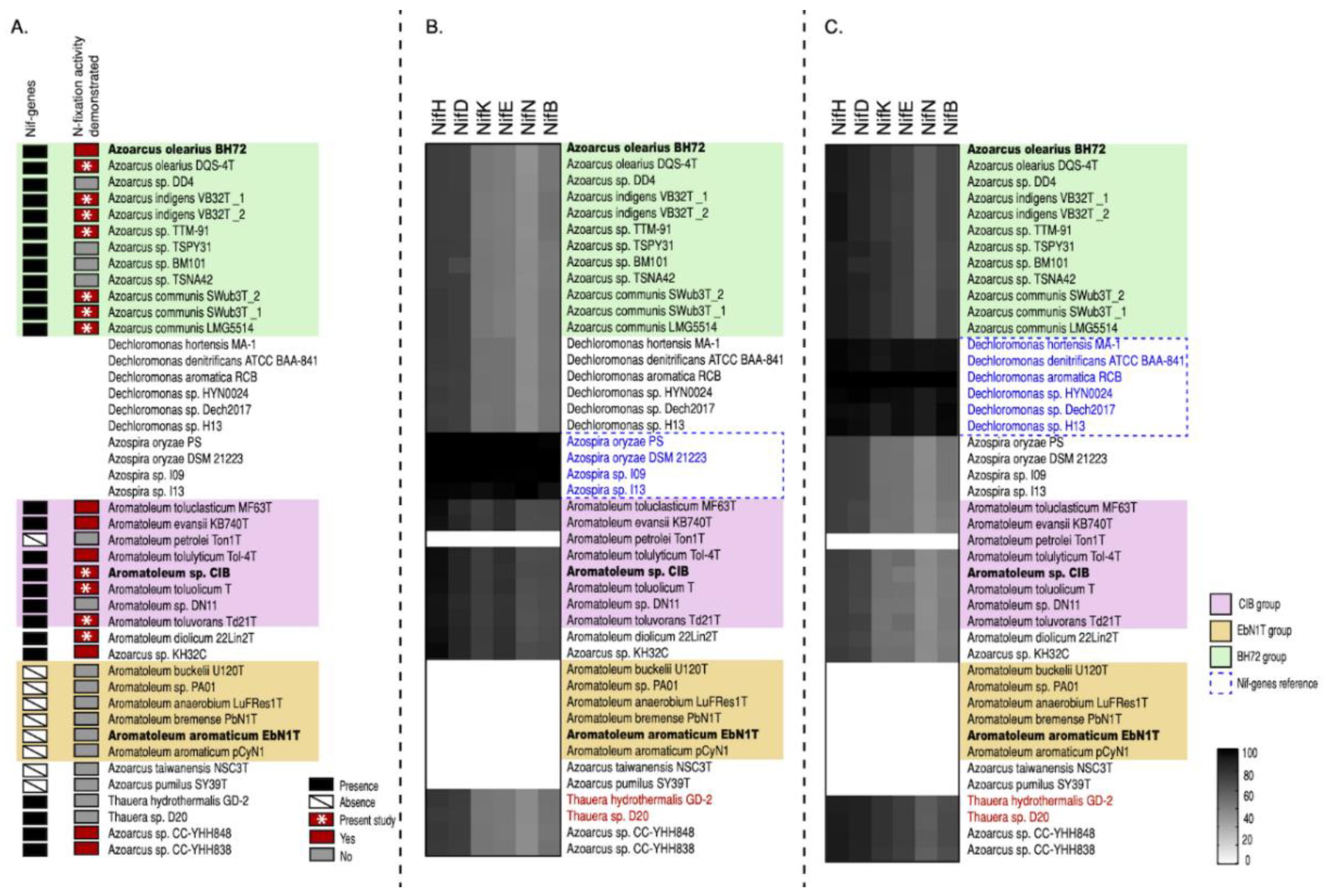
Further insights into the taxonomy of Azoarcus-Aromatoleum were gained by comparing sets of genes involved in the use of nitrate as an alternative electron acceptor (nar/nap, nir, nor, and nos; [62]), thus allowing for anaerobic respiration (Figure 3) as well as genes involved in the anaerobic and aerobic catabolism of benzoate (bzd and box, respectively; [63,64]) (Figure 4). Using the genome of Aromatoleum sp. CIB as a reference (Figure 3), it was revealed that with the exception of Az. pumilus SY39T, all the genome-sequenced Azoarcus and Aromatoleum strains possessed genes for nitrate reduction (nap and/or nar), but nar appeared not to be present in the Azoarcus genomes, except for that of the strain CC-YHH848 and that of Az. taiwanensis. Moreover, although Azoarcus possessed nap genes, the Aromatoleum strains had a fuller complement of them; interestingly, strains in the CIB group were particularly well endowed with both nap and nar genes, whereas those in the EbN1 group (except for Ar. buckelii) were apparently lacking in key nap genes, and hence may be unable to utilize this pathway for nitrate reduction. The only other strains that lacked nap genes were Az. pumilus SY39T and Az. taiwanensis. Other anomalous strains in terms of nitrate reduction were Ar. diolicum and Azoarcus sp. KH32C in that they were similar to Azoarcus s.s. in possessing nap, but not nar genes. All the examined genomes contained genes for nitrite reduction (nir), except for those of the three Az. olearius strains (DQS-4T, BH72, DD4), Az. communis LMG5514, and Az. pumilus SY39T (Figure 3). It has long been known that BH72 does not possess nitrite reductase genes [40], but the present study suggests that this appears to be a characteristic of its species, Az. olearius. Of the key nir genes, nirK and nirS [62], the former was only identified in Azoarcus sp. KH32C. Moreover, although nirS was present in most, but not all of the genomes, it is currently unclear whether some of the strains that apparently lack both nirS and nirK (e.g., Ar. aromaticum, Ar. petrolei, Ar. tolulyticum, and Ar. toluclasticum) can actually reduce nitrite. Genes for the final two steps in denitrification, nor (nitric oxide reductase) and nos (nitrous oxide reductase), were present in all the sequenced Azoarcus-Aromatoleum genomes, except that nos genes could not be detected in Az. pumilus SY39T.
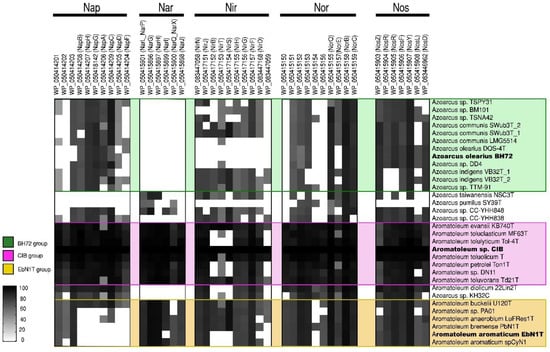
Figure 3.
Nitrate reduction pathway analysis. Heatmap revealing the presence/absence of genes from the nap, nar, nir, nor, and nos gene clusters from 32 genomes (from 30 strains) of the Azoarcus-Aromatoleum group. The indicated marker genes were identified in the RAFTS3G-32 clusters containing corresponding genes in Aromatoleum sp. CIB and verified manually. The homology of the identified genes to reference genes is indicated with a grey scale gradient from white (0%) to black (100%).
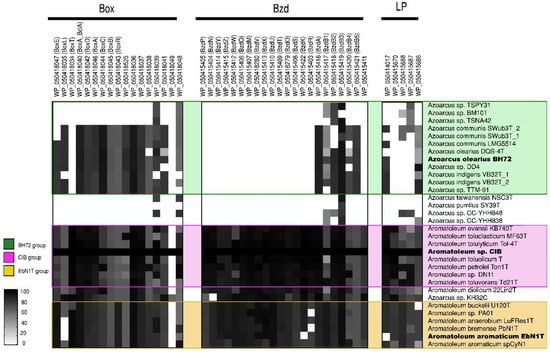
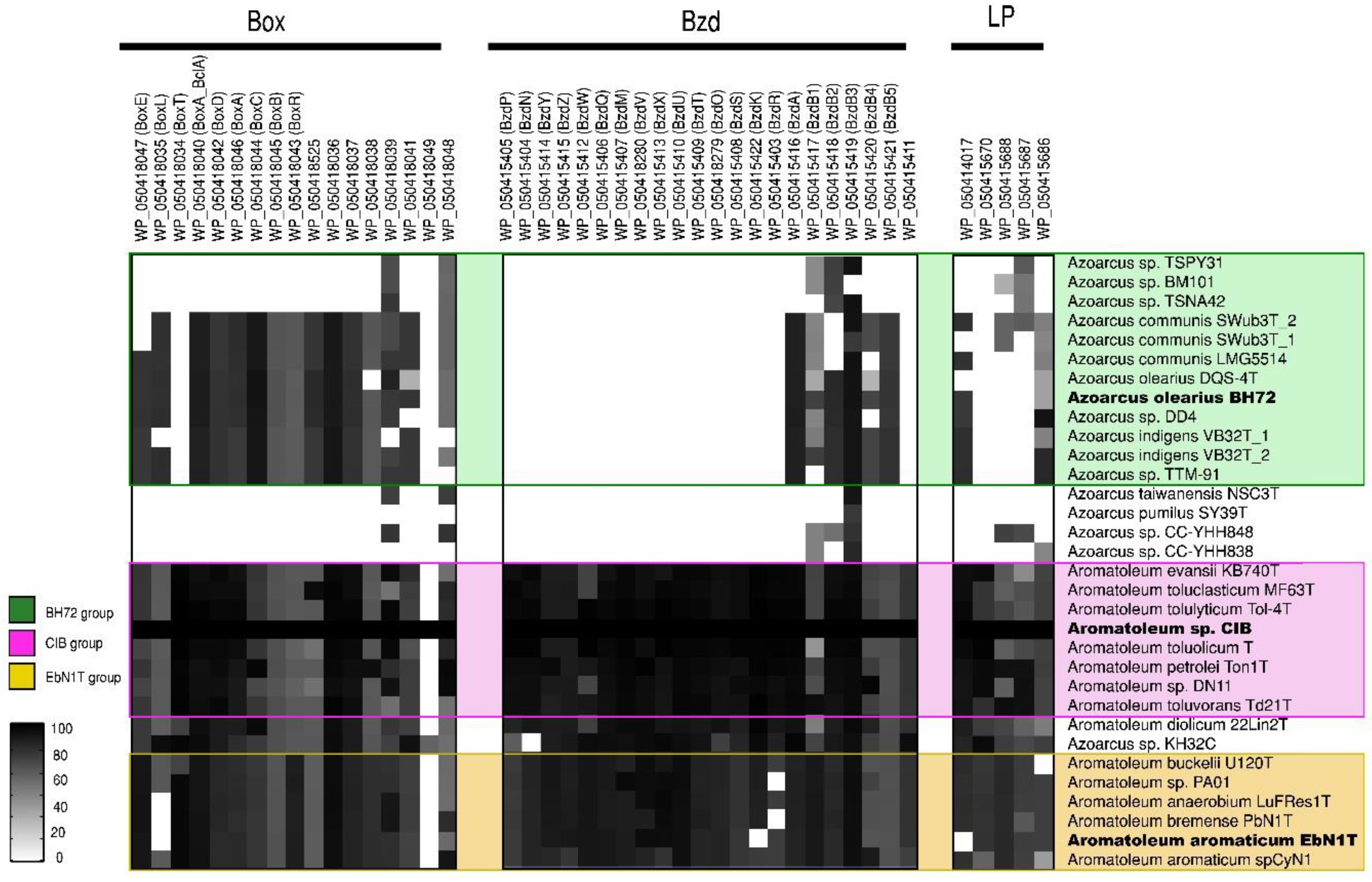
Figure 4.
Aromatic compound degradation pathway analysis. Heatmap revealing the presence/absence of different genes from the benzoate (bzd) and benzoyl-coenzyme clusters (box) as well as of genes from the “lower pathway” (LP) present in 32 genomes (from 30 strains) of the Azoarcus-Aromatoleum group. The indicated marker genes were identified in the RAFTS3G-32 clusters containing corresponding genes in Aromatoleum sp. CIB and verified manually. The homology of the identified genes to reference genes is indicated with a grey scale gradient from white (0%) to black (100%).
bzd genes were present in all of the Aromatoleum strains plus Azoarcus sp. KH32C (Figure 4), and some were also observed in Az. olearius, Az. indigens, and Az. communis, although it is unlikely that the latter has the capacity to metabolize benzoate under anaerobic conditions since the genes that encode for the key enzyme involved in the benzoyl-CoA reduction (bzdNOPQ) are absent in these strains. On the other hand, all the aforementioned strains also possess box genes, so it is probable that they might degrade benzoate aerobically [64], but the absence of bzd and box genes in the Az. communis strains TSPY31, TSPN42, and BM101 in Az. pumilus SY39T and in Az. taiwanensis suggests that they cannot metabolize benzoate at all.
In summary, if the presence of nap and/or nar genes are indicative of a capacity to derive energy from respiratory nitrate reduction [62], then it is possible that all the Azoarcus-Aromatoleum strains in the present study possess it, with the exception of Az. pumilus SY39T. Indeed, Zamarro et al. [65] experimentally demonstrated nitrate reductase-based anaerobic metabolism for a modified variant of Az. communis Swub3T with a bzd cassette inserted into its genome. In addition, it is also likely that all the strains, except for Az. olearius, Az. communis LMG5514, and Az. pumilus SY39T, have the genetic capacity to perform the complete denitrification of nitrate to N2.
3.4. Nif Genes and Nitrogenase Activity
The presence of nif genes was analysed in all 30 Azoarcus-Aromatoleum genomes used in the present study as well as the related genera Azospira, Dechloromonas, and Thauera (Figure 5 and Figure 6, Figure S5; Table 1, Table S1a). The RAFTS3G-32 cluster analyses showed that nifH genes were present in 43 genomes in total. The following organisms lack the nifH gene and homology to all other nif genes (except nifUS): Ar. buckelii U120T, Aromatoleum sp. PA01, Ar. anaerobium LuFRes1, Ar. bremense PbN1T, Ar. aromaticum EbN1T, Ar. aromaticum pCyN1, Az. taiwanensis NSC3T, and Az. pumilus SY39T. In contrast, two copies of the nifH gene were found in Azoarcus sp. CC YHH838, Azoarcus sp. CC YHH848, Rhodocyclus tenuis DSM109T, Azoarcus sp. KH32C, and Ar. toluvorans Td21T. All 12 Azoarcus strains contained the minimal set of nif genes necessary for nitrogen fixation (56) suggesting that all are diazotrophs including some not yet demonstrated experimentally for nitrogenase activity such as Azoarcus sp. strain DD4 (the nitrogenase activity of Az. communis LMG5514 and Azoarcus sp. TTM-91 was confirmed in the present study; Table 2). Interestingly, all members of the Aromatoleum CIB-Group, with the exception of Ar. petrolei, are diazotrophs, albeit with a very different nif gene complement to those in Azoarcus. This includes the recently described species Ar. diolicum and Ar toluolicum; both have been demonstrated to express nitrogenase activity in the present study (Table 2), and Ar. diolicum by Rabus et al. [21]. Although we were unable to demonstrate nitrogenase activity for Ar. evansii under any of the conditions tested (Table 2), the original description of this species as Az. evansii stated that activity could be detected [6], but did not give details about how the assays were performed. None of the Aromatoleum EbN1 Group possessed nif genes, as previously reported [12,21].
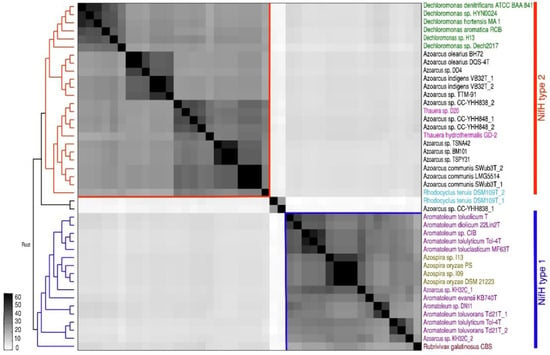
Figure 5.
Single gene nifH phylogeny and heat map confirms the presence of two types of nifH genes in Azoarcus-Aromatoleum genomes. The nifH genes all belonged to a RATFTS3G-67 cluster that was identified by RAFTS3 searches and verified manually.
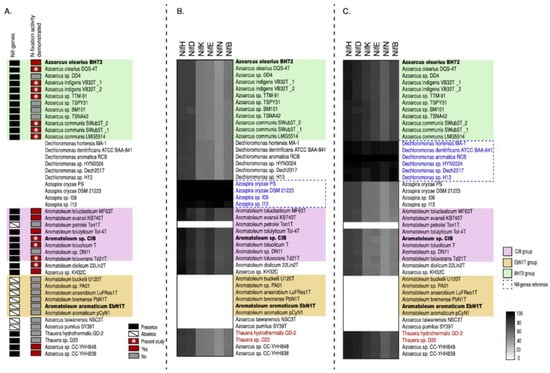
Figure 6.
Analysis of the minimum set of nif genes for diazotrophy (nifHDKENB) reveals the presence of Azospira-type nif genes in the CIB-Group and Dechloromonas-type nif genes in the BH72-Group in Azoarcus-Aromatoleum. The presence of nif genes in black and experimentally proven nitrogenase (acetylene reduction) activity (ARA) in red are shown in panel (A) with ARA data obtained during the present study indicated by *. Known nif genes from the reference strains Azospira oryzae DSM 21223 (B) or Dechloromonas aromatica RCB (C) were used to identify homologous genes in the 32 analysed Azoarcus (green) and Aromatoleum (yellow and pink) genomes as well as in 20 related Thauera (note that only the two Thauera genomes harbouring nif genes are shown). For comparison, four Azospira (A) and six Dechloromonas (B) genomes are also included. The heatmaps indicate homology with a grey scale gradient from white (0%) to black (100%).

Table 2.
Nitrogenase (acetylene reduction) activity of selected strains of Azoarcus and Aromatoleum.
The potential origin of the nif genes as suggested by their closest relatives was investigated. On the basis of partial nifD and nifH gene sequences, Faoro et al. [5] suggested that these might be Dechloromonas for Azoarcus and Azospira for Aromatoleum. Dechloromonas, Azospira, Azoarcus, Aromatoleum, and Thauera are all in the order Rhodocyclales [18], but were in different families: Dechloromonas is in the Azonexaceae, Azospira in the Rhodocyclaceae, and Azoarcus, Aromatoleum and Thauera belong to the Zoogloeaceae. Phylogenies built from nearly full-length nifH (Figure 5) and from nif cluster genes (Figure S5) again confirmed the different nif origins of the two groups of diazotrophs in Azoarcus-Aromatoleum, and the relatedness of the nif cluster in the Aromatoleum CIB-Group to Azospira and that of Azoarcus to Dechloromonas. The minimal nif gene set required for nitrogen fixation (nifH, nifD, nifK, nifE, nifN, and nifB) [45] of Azoarcus-Aromatoleum were compared to Azospira oryzae 6a3T [17] and to D. aromatica strain RCBT [66] by RAFTS3G-32 cluster analyses. The heatmaps clearly show that the nif genes of the Aromatoleum CIB-Group strains are more closely related to Azospira oryzae (Figure 6B) and those of Azoarcus to D. aromatica (Figure 6C). They also illustrate the absence of nif genes in the Aromatoleum EbN1 Group and in Thauera. Interestingly, both the analyses using the single nifH phylogenies (Figure 5) and core nif genes (Figure 6) placed two Thauera strains (T. hydrothermalis GD-2T and Thauera sp. D20) within Azoarcus, and these were the same strains that were nested within Azoarcus in the whole-genome analysis (Figures S1 and S4). Rather than suggesting that the genus Thauera possessed some diazotrophic members, it lends support to our earlier contention that these “Thauera” strains should be more appropriately included within the genus Azoarcus.
Frequent gene duplications in the genome of Azoarcus sp. KH32C, Ar. toluvorans Td21T, and Azoarcus sp. CC-YHH848 were revealed by our analysis as well as the presence of two types of nifH genes in the genomes of Azoarcus sp. CC-YHH838 and Rhodocyclus tenuis DSM109T (Figure 5).
The separate origins of the nif genes in Azoarcus (s.s.) and in Aromatoleum CIB Group (Aromatoleum Group 2) suggests that they were obtained from different sources via horizontal gene transfer (HGT). It is not clear when and why this happened. Azospira oryzae, the possible donor of nif to Aromatoleum Group 2 is a facultative anaerobe associated with Kallar Grass and rice with an ecology similar to Azoarcus spp. [17,67], whereas Dechloromonas, the possible donor of nif to Azoarcus, is also a facultative anaerobe, but is not normally found associated with plants [66] (i.e., the nif donors to the two groups of diazotrophs in Azoarcus-Aromatoleum have the opposite ecology to that which would have been intuitively expected). On the other hand, this makes more sense if the nif gene regulation in Azoarcus and Aromatoleum strains is compared (i.e., the Az. olearius BH72-related strains possess nifL and nifA linked to the rnf1 gene cluster, whereas the Aromatoleum CIB group strains do not encode NifL, and in this case nifA is linked to nifB) (Figure S6). In contrast to the Azoarcus strains, the Aromatoleum CIB group strains encode a class of NifA proteins that possesses a cysteine-containing interdomain linker that confers oxygen sensitivity [68]. The more elaborate NifL-NifA regulatory system, when linked to the Rnf1 complex, which can support electron transfer to nitrogenase under aerobic conditions [69], is the one that is more likely to be associated with diazotrophic organisms that are more dependent on aerobic respiration such as Azoarcus [70]. This again illustrates how difficult it is to ascribe a definitive habitat, ecology, and lifestyle to any of the groups in Azoarcus-Aromatoleum. Good examples are the two Az. olearius strains, BH72 and DQS-4T, that were isolated, respectively, from plants and from oil-contaminated soil, and yet they are almost identical in terms of genes, putatively allowing them to have an endophytic lifestyle [5]. In addition, the former Azoarcus sp. strain CIB, which is now placed in Aromatoleum Group 2, a group of organisms not previously known to be associated with plants, contains many “plant-associated” features in its genome, and can associate with rice endophytically and even express nitrogenase (and possibly produce indole acetic acid) in planta [14,20,71].
4. Conclusions
The aim of this study was neither to describe nor to re-circumscribe new taxa within Azoarcus-Aromatoleum, but it does provide an opportunity to suggest how these new genomic data may be used for this purpose in future studies (combined with appropriate phenotypic criteria). The paraphyletic nature of the Azoarcus-Aromatoleum group revealed by the ANI analysis, and by phylogenetic analysis of the core genomes and the 16S rRNA sequences of 30 genome-sequenced strains suggests that its taxonomy could be revised solely on the basis of genetics to either (1) retain the generic name Azoarcus for its entirety, or (2) that if Aromatoleum is to be retained as a separate genus, it could be divided into two genera (the non-diazotrophic Aromatoleum sensu stricto and the mainly diazotrophic Aromatoleum Group 2 or “CIB-Group”), and three additional genera could be created comprising Az. pumilus, Az. taiwanensis, and the single strain lineage Azoarcus strain KH32C (which is so far undescribed to species level), respectively; thus creating six genera within Azoarcus-Aromatoleum. In terms of ecology, retaining the umbrella name Azoarcus might be justified, as with the exception of a few plant-associated strains in Azoarcus (s.s.), across the entire Azoarcus-Aromatoleum group as most strains/species are found in soil and water (often contaminated with petroleum or related compounds), sewage sludge, and seawater. On the other hand, if metabolism/lifestyle is considered to be the primary factor in describing bacterial genera, the ability of Aromatoleum to utilize nitrate as a terminal electron acceptor for the anaerobic degradation of aromatic compounds such as benzoate and the inability of Azoarcus (s.s.) to do this makes for an obvious division between Azoarcus and Aromatoleum as proposed by Rabus et al. [21]. However, it could be argued that if Azoarcus strains like Az. communis Swub3T can be induced to perform the anaerobic degradation of benzoate by the relatively simple insertion of the bzd gene cassette [65], then even this phenotype is not so clear as a distinguishing feature. Additional complications are created by the presence of the aforementioned “intermediate” groups. Azoarcus sp. KH32C possesses the anaerobic benzoate degradation phenotype, but it clearly does not belong to Aromatoleum. Moreover, Az. pumilus and Az. taiwanensis are both facultative anaerobes [11,23], and neither possess nif genes, suggesting a closer affiliation to the EbN1 group of Aromatoleum, but genomically, they are not close to either Azoarcus or Aromatoleum (this study). Similarly, the Azoarcus strains CC-YHH838 and CC-YHH848 from Taiwan occupy a peripheral position within the genus Azoarcus, and their metabolism (aerobic vs. anaerobic) is so-far undescribed, but they do appear to have fairly typical Azoarcus-type nif genes (Figure 6).
Clearly, more work is required to resolve the taxonomy of Azoarcus-Aromatoleum, both in terms of sequencing more genomes, and in terms of examining lifestyles and habitats of this highly diverse and potentially useful group of micro-organisms.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/12/1/71/s1, File S1. SWeeP Analyses. File S2. Growth conditions for assessment of nitrogenase activity using the acetylene reduction assay. Table S1. Accession numbers of sequences used in phylogenies, abbreviations used, and original sources of strains (a), the genome sequencing statistics for strains sequenced in this study (b) and accession numbers of sequences used in the analysis of nif and other functional genes (c). Table S2. ANI percentage values for (a) the 32 genomes of Azoarcus and Aromatoleum strains and (b) the genomes of Azoarcus, Aromatoleum plus Thauera strains. Table S3. Percentage similarity values between 16S rRNA sequences extracted from genomes of (a) Azoarcus and Aromatoleum strains and (b) Azoarcus, Aromatoleum, Thauera, Azospira, Dechloromonas, Zoogloea ramigera NBRC 15342, Rhodocyclus tenuis DSM109T, and Rubrivivax galatinosus CBS strains. Figure S1. Clustering of Azoarcus, Aromatoleum, and Thauera (Branch 9468) in the global bacterial tree previously presented in the study by De Pierri et al. [47] (with 10,324 whole genomes), and available at: https://sourceforge.net/projects/spacedwordsprojection/. Figure S2. Whole genome comparison of 67 genomes analysed in this study based on SWeeP vectors. (a) Two principal components projection for k-means clusters (k = 4) generated with SWeeP vectors; k1, k2, k3, and k4 are related to groups in the neighbour joining phylogeny presented in (b). Figure S3. Clustal Omega phylogenetic Inference of 16S rRNA analysis for all 67 genomes in this study. Figure S4. Core gene cluster comparisons for all 67 genomes. As observed in the complete genome analysis, Azospira and Dechloromonas were separated from the other genomes in the NJ phylogeny based on SWeeP projection and the ordered vectors distance matrix heatmap. Figure S5. NJ phylogeny and heatmap illustrating the comparison of 67 genomes in this study based on SWeeP projection of the nif gene cluster set for each organism. Analogous relationships between Aromatoleum-Azospira and Azoarcus-Dechloromonas were observed, confirming the nifH gene analysis. Figure S6. Comparative analysis of the gene neighbourhoods of nif regulatory genes carried out with PATRIC 3.6.6 [72]. (A) Search conducted with NifL (ANQ83604.1) from Azoarcus olearius strain DQS4 as reference. nifL, its protein product and homologs are shown in red, with nifA homologs (labelled as either 2 or 12 according to product annotation) in green. rnfA1, rnfB1, and rnfC1 are depicted in brown, yellow, and mauve, respectively. The gene depicted in light green (number 13) from Azoarcus sp.CC-YHH848 is also a homolog of rnfC1. The gene depicted in light blue at the left of the figure is rnfD1. (B) Search conducted with NifA (AKU10620.1) from Aromatoleum sp. CIB as reference. nifA, its protein product and homologs are shown in red. Genes shown in light green to the left of nifA encode hypothetical proteins. nifB is depicted in brown. Genes located to the right of nifA, depicted in dark blue (number 4) and cyan (number 6) are adjacent to nifA in seven of the strains shown and encode a nucleoside diphosphate kinase and a 23S RNA adenine methyl transferase, respectively.
Author Contributions
R.T.R., C.R.D.P., M.M., M.B.B., M.C., E.d.S., F.d.O.P., R.A.D. and E.K.J. conceived and designed the analyses; R.T.R., C.R.D.P., M.M. and M.B.B. analysed the data and constructed the figures; M.M. and M.B.B. designed and performed the N-fixation experiments; R.T.R., C.R.D.P., M.M., M.B.B., M.C., M.J., H.F., L.M.C., E.d.S., F.d.O.P., W.-M.C., P.S.P., R.A.D. and E.K.J. provided strains, genomes, and sequencing support; R.T.R., C.R.D.P., M.M., M.B.B., M.C., M.J., H.F., L.M.C., F.B., E.d.S., F.d.O.P., W.-M.C., P.S.P., R.A.D. and E.K.J. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the BBSRC-Newton Fund (grant numbers BB/N013476/1 and BB/N003608/1). MC was funded by grant BIO2016-79736-R from the Ministry of Economy and Competitiveness of Spain.
Data Availability Statement
The data presented in this study are available in the Supplementary Material.
Acknowledgments
Unless noted otherwise, genome sequencing was provided by MicrobesNG (http://www.microbesng.uk), which is supported by the BBSRC (grant number BB/L024209/1).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Reinhold, B.; Hurek, T.; Niemann, E.-G.; Fendrik, I. Close association of Azospirillum and diazotrophic rods with different root zones of Kallar Grass. Appl. Environ. Microbiol. 1986, 52, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, B.; Hurek, T.; Fendrik, I.; Pot, B.; Gillis, M.; Kersters, K.; Thielemans, S.; De Ley, J. Azospirillum halopraeferens sp. nov., a nitrogen-fixing organism associated with roots of Kallar Grass (Leptochloa fusca (L.) Kunth). Int. J. Syst. Evol. Microbiol. 1987, 37, 43–51. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T.; Gillis, M.; Hoste, B.; Vancanneyt, M.; Kersters, K.; Ley, J. Azoarcus gen. nov., nitrogen-fixing proteobacteria associated with roots of Kallar Grass (Leptochloa fusca (L.) Kunth), and description of two species, Azoarcus indigens sp. nov. and Azoarcus communis sp. nov. Int. J. Syst. Evol. Microbiol. 1993, 43. [Google Scholar] [CrossRef]
- Chen, M.H.; Sheu, S.Y.; James, E.K.; Young, C.C.; Chen, W.M. Azoarcus olearius sp. nov., a nitrogen-fixing bacterium isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 3755–3761. [Google Scholar] [CrossRef] [PubMed]
- Faoro, H.; Rene Menegazzo, R.; Battistoni, F.; Gyaneshwar, P.; do Amaral, F.P.; Taule, C.; Rausch, S.; Goncalves Galvao, P.; de Los Santos, C.; Mitra, S.; et al. The oil-contaminated soil diazotroph Azoarcus olearius DQS-4(T) is genetically and phenotypically similar to the model grass endophyte Azoarcus sp. BH72. Environ. Microbiol. Rep. 2017, 9, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Anders, H.J.; Kaetzke, A.; Kampfer, P.; Ludwig, W.; Fuchs, G. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 1995, 45, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Fries, M.R.; Chee-Sanford, J.C.; Tiedje, J.M. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth on toluene and description of Azoarcus tolulyticus sp. nov. Int. J. Syst. Bacteriol. 1995, 45, 500–506. [Google Scholar] [CrossRef]
- Springer, N.; Ludwig, W.; Philipp, B.; Schink, B. Azoarcus anaerobius sp. nov., a resorcinol-degrading, strictly anaerobic, denitrifying bacterium. Int. J. Syst. Bacteriol. 1998, 48, 953–956. [Google Scholar] [CrossRef]
- Song, B.; Haggblom, M.M.; Zhou, J.; Tiedje, J.M.; Palleroni, N.J. Taxonomic characterization of denitrifying bacteria that degrade aromatic compounds and description of Azoarcus toluvorans sp. nov. and Azoarcus toluclasticus sp. nov. Int. J. Syst. Bacteriol. 1999, 3, 1129–1140. [Google Scholar] [CrossRef]
- Mechichi, T.; Stackebrandt, E.; Gad’on, N.; Fuchs, G. Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions, and description of Thauera phenylacetica sp. nov., Thauera aminoaromatica sp. nov., and Azoarcus buckelii sp. nov. Arch. Microbiol. 2002, 178, 26–35. [Google Scholar] [CrossRef]
- Lee, D.-J.; Wong, B.-T.; Adav, S.S. Azoarcus taiwanensis sp. nov., a denitrifying species isolated from a hot spring. Appl. Microbiol. Biotechnol. 2014, 98, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Rabus, R.; Kube, M.; Heider, J.; Beck, A.; Heitmann, K.; Widdel, F.; Reinhardt, R. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 2005, 183, 27–36. [Google Scholar] [CrossRef]
- Nishizawa, T.; Tago, K.; Oshima, K.; Hattori, M.; Ishii, S.; Otsuka, S.; Senoo, K. Complete genome sequence of the denitrifying and N2O-reducing bacterium Azoarcus sp. strain KH32C. J. Bacteriol. 2012, 194, 06618-11. [Google Scholar] [CrossRef]
- Fernández-Llamosas, H.; Prandoni, N.; Fernández-Pascual, M.; Fajardo, S.; Morcillo, C.; Díaz, E.; Carmona, M. Azoarcus sp. CIB, an anaerobic biodegrader of aromatic compounds shows an endophytic lifestyle. PLoS ONE 2014, 9, e110771. [Google Scholar] [CrossRef]
- Junghare, M.; Patil, Y.; Schink, B. Draft genome sequence of a nitrate-reducing, o-phthalate degrading bacterium, Azoarcus sp. strain PA01(T). Stand Genom. Sci. 2015, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Li, F.; Ye, L.; Li, M. Complete genome sequence of Azoarcus sp. strain DD4, a gram-negative propanotroph that degrades 1,4-dioxane and 1,1-dichloroethylene. Microbiol. Resour. Announc. 2019, 8, e00775-19. [Google Scholar] [CrossRef] [PubMed]
- Reinhold-Hurek, B.; Hurek, T. Reassessment of the taxonomic structure of the diazotrophic genus Azoarcus sensu lato and description of three new genera and new species, Azovibrio restrictus gen. nov., sp. nov., Azospira oryzae gen. nov., sp. nov. and Azonexus fungiphilus gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 2, 649–659. [Google Scholar] [CrossRef]
- Loy, A.; Schulz, C.; Lücker, S.; Schöpfer-Wendels, A.; Stoecker, K.; Baranyi, C.; Lehner, A.; Wagner, M. 16S rRNA gene-based oligonucleotide microarray for environmental monitoring of the betaproteobacterial order “Rhodocyclales”. Appl. Environ. Microbiol. 2005, 71, 1373–1386. [Google Scholar] [CrossRef]
- Hurek, T.; Reinhold-Hurek, B. Azoarcus sp. strain BH72 as a model for nitrogen-fixing grass endophytes. J. Bacteriol. 2003, 106, 169–178. [Google Scholar] [CrossRef]
- Martin-Moldes, Z.; Zamarro, M.T.; Del Cerro, C.; Valencia, A.; Gomez, M.J.; Arcas, A.; Udaondo, Z.; Garcia, J.L.; Nogales, J.; Carmona, M.; et al. Whole-genome analysis of Azoarcus sp. strain CIB provides genetic insights to its different lifestyles and predicts novel metabolic features. Syst. Appl. Microbiol. 2015, 38, 462–471. [Google Scholar] [CrossRef]
- Rabus, R.; Wohlbrand, L.; Thies, D.; Meyer, M.; Reinhold-Hurek, B.; Kampfer, P. Aromatoleum gen. nov., a novel genus accommodating the phylogenetic lineage including Azoarcus evansii and related species, and proposal of Aromatoleum aromaticum sp. nov., Aromatoleum petrolei sp. nov., Aromatoleum bremense sp. nov., Aromatoleum toluolicum sp. nov. and Aromatoleum diolicum sp. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 982–997. [Google Scholar] [CrossRef] [PubMed]
- Hurek, T.; Egener, T.; Reinhold-Hurek, B. Divergence in nitrogenases of Azoarcus spp., Proteobacteria of the beta subclass. J. Bacteriol. 1997, 179, 4172–4178. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.-Y.; Yu, X.-Y.; Yu, X.-D.; Zhao, Z.; Chen, C.; Wang, R.-J.; Wu, M.; Zhang, X.-Q. Azoarcus pumilus sp. nov., isolated from seawater in Sanya, China. Int. J. Syst. Evol. Microbiol. 2019, 69, 1459–1464. [Google Scholar] [CrossRef] [PubMed]
- Beukes, C.W.; Palmer, M.; Manyaka, P.; Chan, W.Y.; Avontuur, J.R.; van Zyl, E.; Huntemann, M.; Clum, A.; Pillay, M.; Palaniappan, K.; et al. Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Estrada-de Los Santos, P.; Palmer, M.; Chavez-Ramirez, B.; Beukes, C.; Steenkamp, E.T.; Briscoe, L.; Khan, N.; Maluk, M.; Lafos, M.; Humm, E.; et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 2018, 9, 389. [Google Scholar] [CrossRef]
- Avontuur, J.R.; Palmer, M.; Beukes, C.W.; Chan, W.Y.; Coetzee, M.P.A.; Blom, J.; Stepkowski, T.; Kyrpides, N.C.; Woyke, T.; Shapiro, N.; et al. Genome-informed Bradyrhizobium taxonomy: Where to from here? Syst. Appl. Microbiol. 2019, 42, 427–439. [Google Scholar] [CrossRef]
- Ormeño-Orrillo, E.; Martínez-Romero, E. A genomotaxonomy view of the Bradyrhizobium genus. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 January 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap. Available online: https://sourceforge.net/p/bbmap/activity/?page=1&limit=100#5a6a7e4634309d1dd873dd30 (accessed on 20 April 2020).
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling genomes and mini-metagenomes from highly chimeric reads. In Proceedings of the Annual International Conference on Research in Computational Molecular Biology, Beijing, China, 7–10 April 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 158–170. [Google Scholar]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Rabus, R.; Kube, M.; Beck, A.; Widdel, F.; Reinhardt, R. Genes involved in the anaerobic degradation of ethylbenzene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 2002, 178, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Kube, M.; Heider, J.; Amann, J.; Hufnagel, P.; Kühner, S.; Beck, A.; Reinhardt, R.; Rabus, R. Genes involved in the anaerobic degradation of toluene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 2004, 181, 182–194. [Google Scholar] [CrossRef]
- Kühner, S.; Wöhlbrand, L.; Fritz, I.; Wruck, W.; Hultschig, C.; Hufnagel, P.; Kube, M.; Reinhardt, R.; Rabus, R. Substrate-dependent regulation of anaerobic degradation pathways for toluene and ethylbenzene in a denitrifying bacterium, strain EbN1. J. Bacteriol. 2005, 187, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Devanadera, A.; Vejarano, F.; Zhai, Y.; Suzuki-Minakuchi, C.; Ohtsubo, Y.; Tsuda, M.; Kasai, Y.; Takahata, Y.; Okada, K.; Nojiri, H. Complete genome sequence of an anaerobic benzene-degrading bacterium, Azoarcus sp. Strain DN11. Microbiol. Resour. Announc. 2019, 8, e01699-18. [Google Scholar] [CrossRef] [PubMed]
- Zorraquino, V.; Toubiana, D.; Yan, D.; Blumwald, E. Draft genome sequence of the nitrogen-fixing endophyte Azoarcus communis SWub3. Microbiol. Resour. Announc. 2018, 7, e01080-18. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Cha, S.H.; Suk, H.Y.; Park, N.-H.; Woo, J.-H. Complete genome sequences of Azoarcus sp. TSPY31 and TSNA42 potentially having biosynthetic ability to produce indigo. Korean J. Microbiol. 2018, 54, 283–285. [Google Scholar] [CrossRef]
- Krause, A.; Ramakumar, A.; Bartels, D.; Battistoni, F.; Bekel, T.; Boch, J.; Bohm, M.; Friedrich, F.; Hurek, T.; Krause, L.; et al. Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 2006, 24, 1385–1391. [Google Scholar] [CrossRef]
- Carlström, C.I.; Lucas, L.N.; Rohde, R.A.; Haratian, A.; Engelbrektson, A.L.; Coates, J.D. Characterization of an anaerobic marine microbial community exposed to combined fluxes of perchlorate and salinity. Appl. Microbiol. Biotechnol. 2016, 100, 9719–9732. [Google Scholar] [CrossRef]
- Lin, S.-Y.; Hameed, A.; Tsai, C.-F.; Huang, G.-H.; Hsu, Y.-H.; Young, C.-C. Description of Azoarcus nasutitermitis sp. nov. and Azoarcus rhizosphaerae sp. nov., two nitrogen-fixing species isolated from termite nest and rhizosphere of Ficus religiosa. Antonie Van Leeuwenhoek 2020, 113, 933–946. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- De Lima Nichio, B.T.; de Oliveira, A.M.R.; de Pierri, C.R.; Santos, L.G.C.; Lejambre, A.Q.; Vialle, R.A.; da Rocha Coimbra, N.A.; Guizelini, D.; Marchaukoski, J.N.; Pedrosa, F.O.; et al. RAFTS(3)G: An efficient and versatile clustering software to analyses in large protein datasets. BMC Bioinform. 2019, 20, 392. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, P.C.; Fang, Z.; Mason, S.W.; Setubal, J.C.; Dixon, R. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genom. 2012, 13, 1471–2164. [Google Scholar] [CrossRef] [PubMed]
- Vialle, R.A.; de Oliveira Pedrosa, F.; Weiss, V.A.; Guizelini, D.; Tibaes, J.H.; Marchaukoski, J.N.; de Souza, E.M.; Raittz, R.T. RAFTS3: Rapid Alignment-Free Tool for Sequence Similarity Search. bioRxiv 2016, 055269. [Google Scholar] [CrossRef]
- De Pierri, C.R.; Voyceik, R.; Santos de Mattos, L.G.C.; Kulik, M.G.; Camargo, J.O.; Repula de Oliveira, A.M.; de Lima Nichio, B.T.; Marchaukoski, J.N.; da Silva Filho, A.C.; Guizelini, D.; et al. SWeeP: Representing large biological sequences datasets in compact vectors. Sci. Rep. 2020, 10, 91. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Dilworth, M.J. Acetylene reduction by nitrogen-fixing preparations from Clostridium pasteurianum. Biochim. Biophys. Acta 1966, 127, 285–294. [Google Scholar] [CrossRef]
- Schöllhorn, R.; Burris, R.H. Acetylene as a competitive inhibitor of N-2 fixation. Proc. Natl. Acad. Sci. USA 1967, 58, 213–216. [Google Scholar] [CrossRef]
- Zhang, W.; Wen, C.-K. Preparation of ethylene gas and comparison of ethylene responses induced by ethylene, ACC, and ethephon. Plant Physiol. Biochem. 2010, 48, 45–53. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Yang, L.; Muhadesi, J.B.; Wang, M.M.; Wang, B.J.; Liu, S.J.; Jiang, C.Y. Thauera hydrothermalis sp. nov., a thermophilic bacterium isolated from hot spring. Int. J. Syst. Evol. Microbiol. 2018, 68, 3163–3168. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.-S.; Liang, Q.-Y.; Du, Z.-J. Thauera lacus sp. nov., isolated from a saline lake in Inner Mongolia. Int. J. Syst. Evol. Microbiol. 2019, 69, 3786–3791. [Google Scholar] [CrossRef] [PubMed]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.-J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glockner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzeby, J.; Amann, R.; Rossello-Mora, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Barco, R.A.; Garrity, G.M.; Scott, J.J.; Amend, J.P.; Nealson, K.H.; Emerson, D. A genus definition for bacteria and archaea based on a standard genome relatedness index. mBio 2020, 11. [Google Scholar] [CrossRef]
- Qin, Q.-L.; Xie, B.-B.; Zhang, X.-Y.; Chen, X.-L.; Zhou, B.-C.; Zhou, J.; Oren, A.; Zhang, Y.-Z. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014, 196, 2210–2215. [Google Scholar] [CrossRef]
- Kraft, B.; Strous, M.; Tegetmeyer, H.E. Microbial nitrate respiration—Genes, enzymes and environmental distribution. J. Biotechnol. 2011, 155, 104–117. [Google Scholar] [CrossRef]
- López Barragán, M.J.; Carmona, M.; Zamarro, M.T.; Thiele, B.; Boll, M.; Fuchs, G.; García, J.L.; Díaz, E. The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J. Bacteriol. 2004, 186, 5762–5774. [Google Scholar] [CrossRef]
- Valderrama, J.A.; Durante-Rodríguez, G.; Blázquez, B.; García, J.L.; Carmona, M.; Díaz, E. Bacterial degradation of benzoate: Cross-regulation between aerobic and anaerobic pathways. J. Biol. Chem. 2012, 287, 10494–10508. [Google Scholar] [CrossRef] [PubMed]
- Zamarro, M.T.; Barragán, M.J.L.; Carmona, M.; García, J.L.; Díaz, E. Engineering a bzd cassette for the anaerobic bioconversion of aromatic compounds. Microb. Biotechnol. 2017, 10, 1418–1425. [Google Scholar] [CrossRef]
- Salinero, K.K.; Keller, K.; Feil, W.S.; Feil, H.; Trong, S.; Di Bartolo, G.; Lapidus, A. Metabolic analysis of the soil microbe Dechloromonas aromatica str. RCB: Indications of a surprisingly complex life-style and cryptic anaerobic pathways for aromatic degradation. BMC Genom. 2009, 10, 1471–2164. [Google Scholar] [CrossRef] [PubMed]
- Engelhard, M.; Hurek, T.; Reinhold-Hurek, B. Preferential occurrence of diazotrophic endophytes, Azoarcus spp., in wild rice species and land races of Oryza sativa in comparison with modern races. Environ. Microbiol. 2000, 2, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Morett, E.; Segovia, L. The sigma 54 bacterial enhancer-binding protein family: Mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 1993, 175, 6067–6074. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ledbetter, R.N.; Garcia Costas, A.M.; Lubner, C.E.; Mulder, D.W.; Tokmina-Lukaszewska, M.; Artz, J.H.; Patterson, A.; Magnuson, T.y.S.; Jay, Z.J.; Duan, H.; et al. The electron bifurcating FixABCX protein complex from Azotobacter vinelandii: Generation of low-potential reducing equivalents for nitrogenase catalysis. Biochemistry 2017, 56, 4177–4190. [Google Scholar] [CrossRef] [PubMed]
- Bueno Batista, M.; Dixon, R. Manipulating nitrogen regulation in diazotrophic bacteria for agronomic benefit. Biochem. Soc. Trans. 2019, 47, 603–614. [Google Scholar] [CrossRef]
- Fernández-Llamosas, H.; Ibero, J.; Thijs, S.; Imperato, V.; Vangronsveld, J.; Díaz, E.; Carmona, M. Enhancing the rice seedlings growth promotion abilities of Azoarcus sp. CIB by heterologous expression of ACC deaminase to improve performance of plants exposed to cadmium stress. Microorganisms 2020, 8, 1453. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).