Genome-Wide Association Studies for Milk Somatic Cell Score in Romanian Dairy Cattle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Phenotypes
2.2. Sampling and Genotyping
2.3. Quality Control (QC)
2.4. Association Analysis
3. Results
3.1. Descriptive Statistics
3.2. Quality Control
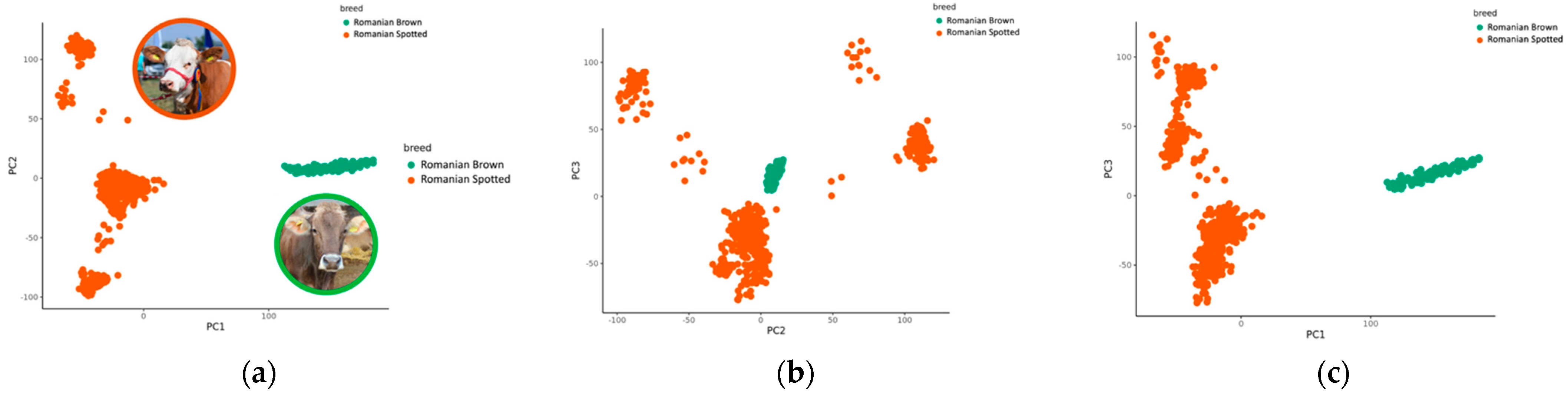
3.3. Principal Component Analysis
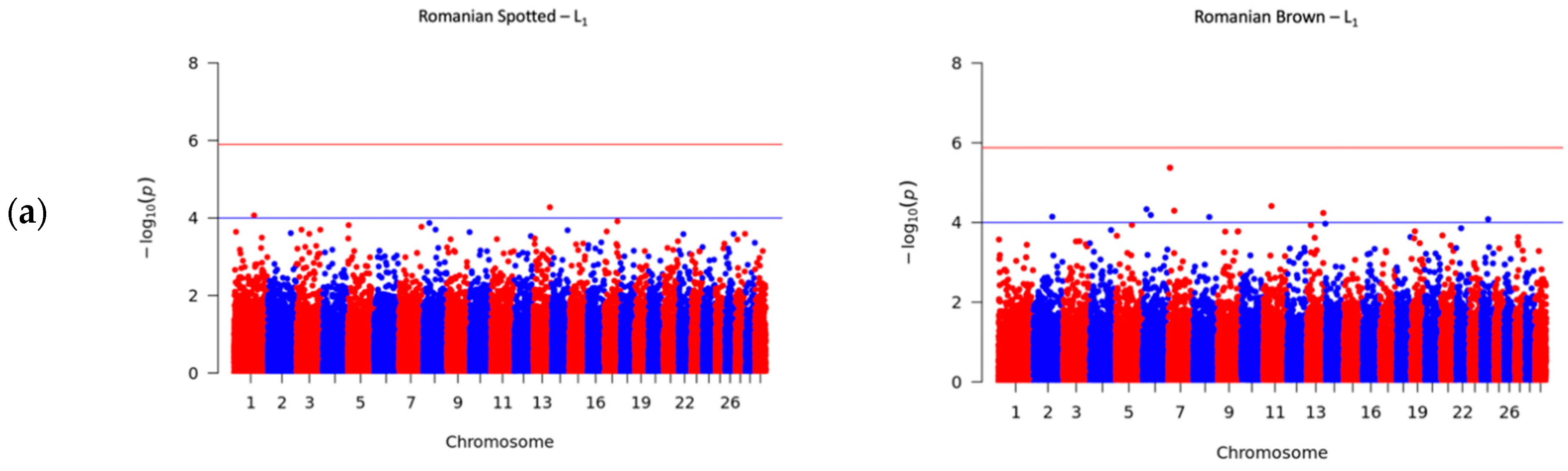
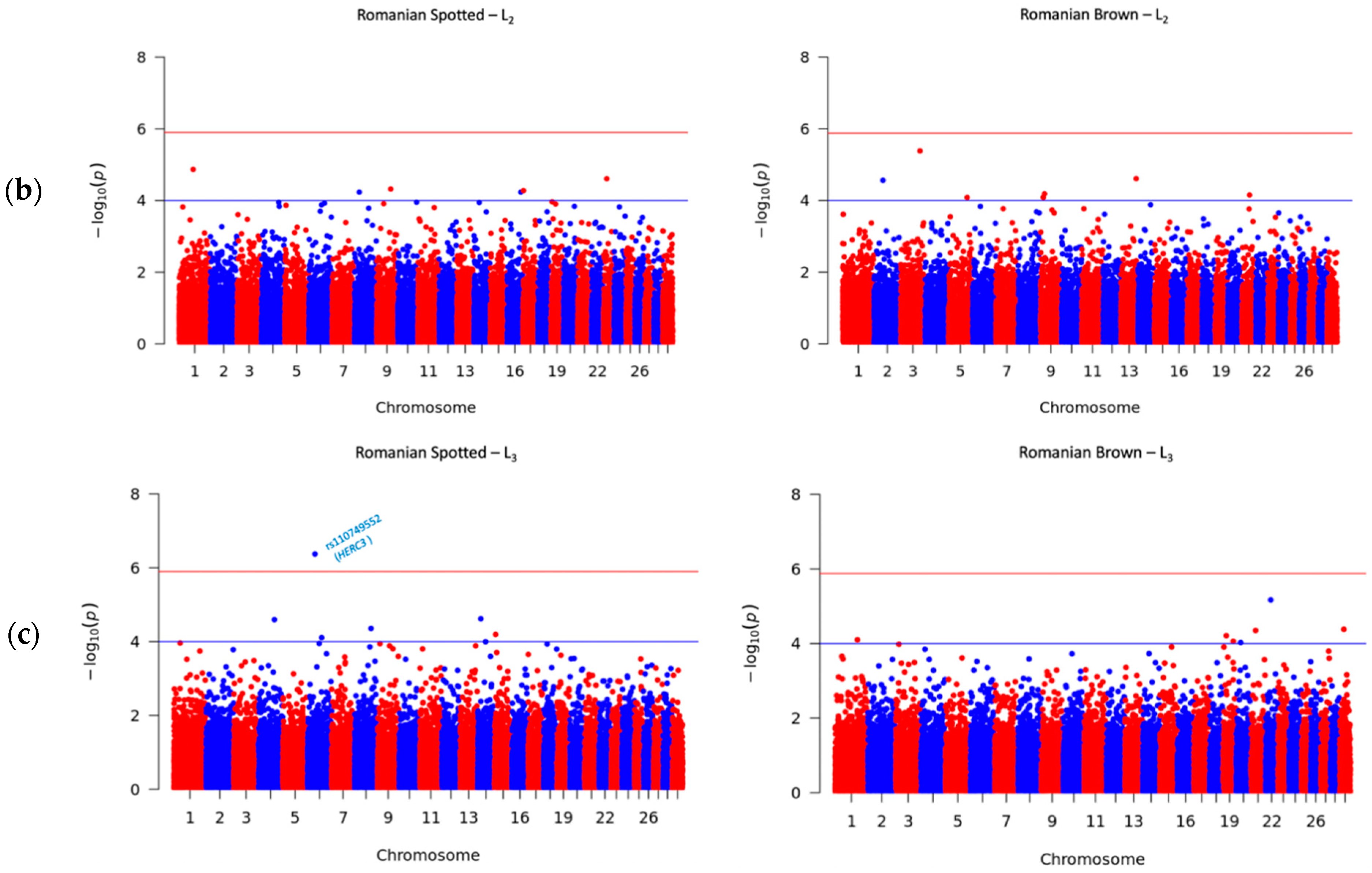
3.4. Significant SNPs Associated with SCS
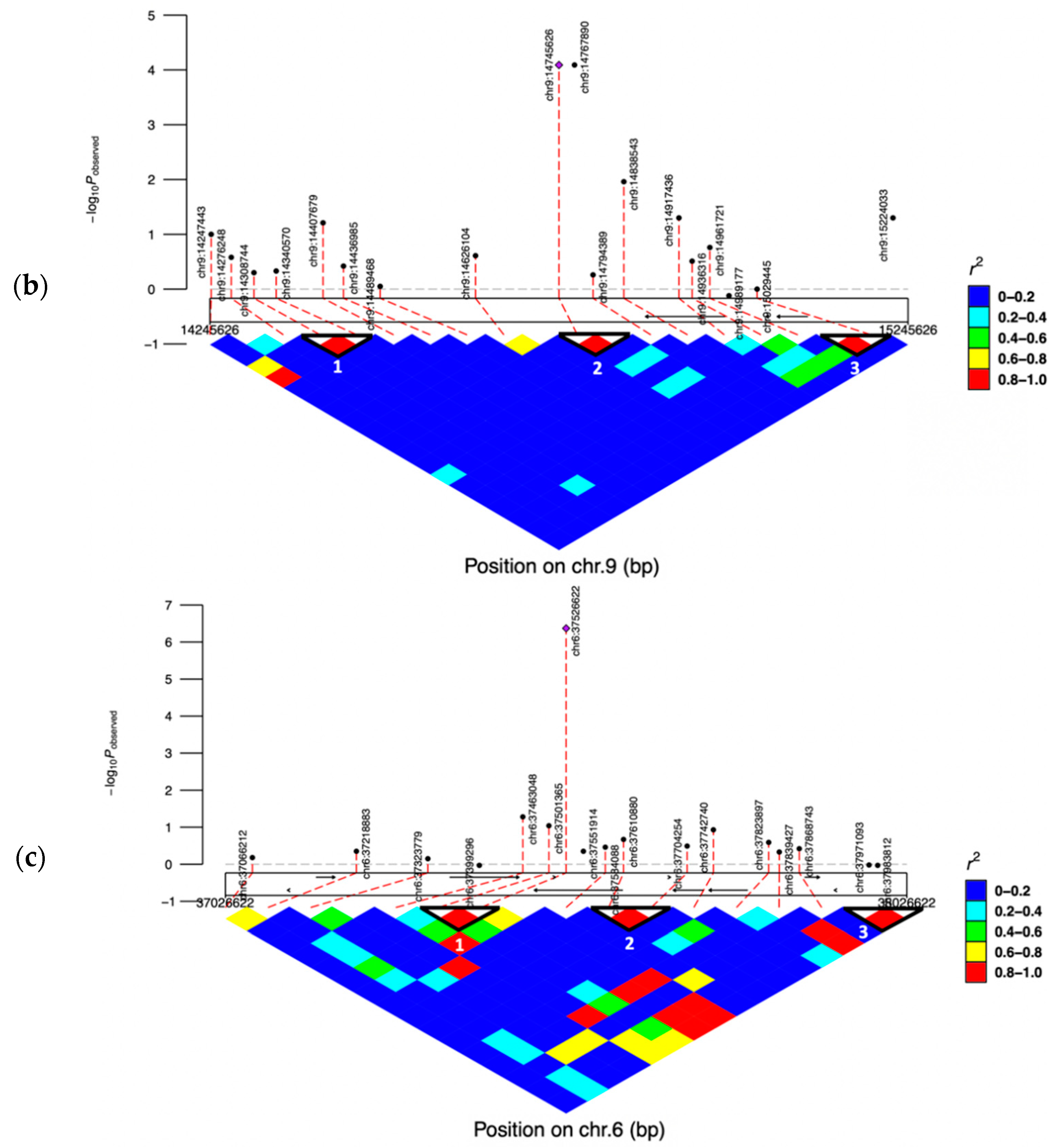
3.5. Linkage Disequilibrium (LD) Blocks of the Significant SNPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rupp, R.; Boichard, D. Genetics of resistance to mastitis in dairy cattle. Vet. Res. 2003, 34, 671–688. [Google Scholar] [CrossRef] [Green Version]
- Aarestrup, F.M.; Jensen, N.E.; Ostergård, H. Analysis of associations between major histocompatibility complex (BoLA) class I haplotypes and subclinical mastitis of dairy cows. J. Dairy Sci. 1995, 78, 1684–1692. [Google Scholar] [CrossRef]
- Dietz, A.B.; Cohen, N.D.; Timms, L.; Kehrli, M.E., Jr. Bovine lymphocyte antigen class II alleles as risk factors for high somatic cell counts in milk of lactating dairy cows. J. Dairy Sci. 1997, 80, 406–412. [Google Scholar] [CrossRef]
- Kelm, S.C.; Detilleux, J.C.; Freeman, A.E.; Kehrli, M.E., Jr.; Dietz, A.B.; Fox, L.K.; Butler, J.E.; Kasckovics, I.; Kelley, D.H. Genetic association between parameters of innate immunity and measures of mastitis in periparturient Holstein cattle. J. Dairy Sci. 1997, 80, 1767–1775. [Google Scholar] [CrossRef]
- Gilliespie, B.E.; Jayarao, B.M.; Dowlen, H.H.; Oliver, S.P. Analysis and frequency of bovine lymphocyte antigen DRB3.2 alleles in Jersey cows. J. Dairy Sci. 1999, 82, 2049–2053. [Google Scholar] [CrossRef]
- Takeshima, S.; Nakai, Y.; Ohta, M.; Aida, Y. Characterization of DRB3 alleles in the MHC of Japanese Shorthorn Cattle by polymerase chain reaction-sequence-based typing. J. Dairy Sci. 2002, 85, 1630–1632. [Google Scholar] [CrossRef]
- Zimin, A.V.; Delcher, A.L.; Florea, L.; Kelley, D.R.; Schatz, M.C.; Puiu, D.; Hanrahan, F.; Pertea, G.; Van Tassell, C.P.; Sonstegard, T.S.; et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009, 10, R42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meredith, B.K.; Kearney, F.J.; Finlay, E.K.; Bradley, D.G.; Fahey, A.G.; Berry, D.P.; Lynn, D.J. Genome-wide associations for milk production and somatic cell score in Holstein-Friesian cattle in Ireland. BMC Genet. 2012, 13, 21. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Liu, L.; Chen, C.J.; Zhang, M.; Lu, X.; Zhang, Z.; Huang, X.; Shi, Y. Genome-wide association study of milk and reproductive traits in dual-purpose Xinjiang Brown cattle. BMC Genom. 2019, 20, 827. [Google Scholar] [CrossRef] [Green Version]
- Meredith, B.K.; Berry, D.P.; Kearney, F.; Finlay, E.K.; Fahey, A.G.; Bradley, D.G.; Lynn, D.J. A genome-wide association study for somatic cell score using the Illumina high-density bovine beadchip identifies several novel QTL potentially related to mastitis susceptibility. Front. Genet. 2013, 4, 229. [Google Scholar] [CrossRef] [Green Version]
- Strillacci, M.G.; Frigo, E.; Schiavini, F.; Samoré, A.B.; Canavesi, F.; Vevey, M.; Cozzi, M.C.; Soller, M.; Lipkin, E.; Bagnato, A. Genome-wide association study for somatic cell score in Valdostana Red Pied cattle breed using pooled DNA. BMC Genet. 2014, 15, 106. [Google Scholar] [CrossRef] [Green Version]
- Nani, J.P.; Raschia, M.A.; Poli, M.A.; Calvinho, L.F.; Amadio, A.F. Genome-wide association study for somatic cell score in Argentinean dairy cattle. Livest. Sci. 2015, 175, 1–9. [Google Scholar] [CrossRef]
- The Animal Quantitative Trait Loci (QTL) Database. Available online: https://www.animalgenome.org/cgi-bin/QTLdb/BT/index (accessed on 20 May 2021).
- Breeds of Livestock, Department of Animal Science. Breeds of Livestock—Baltata Romaneasca Cattle. Available online: http://afs.okstate.edu/breeds/cattle/baltataromaneasca/index.html/ (accessed on 25 August 2021).
- Wiggans, G.R.; Shook, G.E. A lactation measure of somatic-cell count. J. Dairy Sci. 1987, 70, 2666–2672. [Google Scholar] [CrossRef]
- Interbull. Code of Practice. Available online: https://interbull.org/ib/codeofpractice (accessed on 25 August 2021).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Morgan, A.P. argyle: An R package for analysis of Illumina genotyping arrays. G3 2015, 6, 281–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D.H. BLUPF90 and related programs (BGF90). In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002; Volume 28. [Google Scholar]
- Lopes, F.B. DRP R Package. Available online: https://github.com/camult/DRP (accessed on 1 August 2021).
- Garrick, D.J.; Taylor, J.F.; Fernando, R.L. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 2009, 41, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. TPG 2011, 4, 250–255. [Google Scholar] [CrossRef] [Green Version]
- Turner, S. qqman: QQ and Manhattan Plots for GWAS Data; R Package Version 0.1.4; 2017. Available online: https://cran.r-project.org/package=qqman (accessed on 4 August 2021).
- He, F.; Ding, S.; Wang, H.; Qin, F. IntAssoPlot: An R Package for Integrated Visualization of Genome-Wide Association Study Results With Gene Structure and Linkage Disequilibrium Matrix. Front. Genet. 2020, 11, 260. [Google Scholar] [CrossRef] [Green Version]
- Obenchain, V.; Lawrence, M.; Carey, V.; Gogarten, S.; Shannon, P.; Morgan, M. VariantAnnotation: A Bioconductor package for exploration and annotation of genetic variants. Bioinformatics 2014, 30, 2076–2078. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.; Carey, V. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef]
- Parker Gaddis, K.L.; Null, D.J.; Cole, J.B. Explorations in genome-wide association studies and network analyses with dairy cattle fertility traits. J. Dairy Sci. 2016, 99, 6420–6435. [Google Scholar] [CrossRef] [PubMed]
- Welderufael, B.G.; Løvendahl, P.; de Koning, D.J.; Janss, L.L.G.; Fikse, W.F. Genome-Wide Association Study for Susceptibility to and Recoverability from Mastitis in Danish Holstein Cows. Front. Genet. 2018, 9, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boettcher, P. Breeding for improvement of functional traits in dairy cattle. Ital. J. Anim. Sci. 2005, 4, 7–16. [Google Scholar] [CrossRef]
- Alhussien, M.N.; Dang, A.K. Milk somatic cells, factors influencing their release, future prospects, and practical utility in dairy animals: An overview. Vet. World 2018, 11, 562–577. [Google Scholar] [CrossRef]
- Alam, M.; Cho, C.I.; Choi, T.J.; Park, B.; Choi, J.G.; Choy, Y.H.; Lee, S.S.; Cho, K.H. Estimation of Genetic Parameters for Somatic Cell Scores of Holsteins Using Multi-trait Lactation Models in Korea. Asian-Australas J. Anim. Sci. 2015, 28, 303–310. [Google Scholar] [CrossRef]
- Zavadilová, L.; Wolf, J.; Štípková, M.; Němcová, E.; Jamrozik, J. Genetic parameters for somatic cell score in the first three lactations of Czech Holstein and Fleckvieh breeds using a random regression model. Czech J. Anim. Sci. 2011, 56, 251–260. [Google Scholar] [CrossRef] [Green Version]
- Martins, A.M.; Silvestre, A.M.; Petim-Batista, M.F.; Colaço, J.A. Somatic cell score genetic parameter estimates of dairy cattle in Portugal using fractional polynomials. J. Anim. Sci. 2011, 89, 1281–1285. [Google Scholar] [CrossRef]
- Heins, B.J.; Hansen, L.B. Short communication: Fertility, somatic cell score, and production of Normande×Holstein, Montbéliarde×Holstein, and Scandinavian Red × Holstein crossbreds versus pure Holsteins during their first 5 lactations. J. Dairy Sci. 2012, 95, 918–924. [Google Scholar] [CrossRef]
- Wei, C.; Luo, H.P.; Wang, Y.C.; Huang, X.X.; Zhang, M.H.; Zhang, X.X.; Wang, D.; Ge, J.J.; Xu, L.; Jiang, H.; et al. Analyses of the genetic relationships between lactose, somatic cell score, and growth traits in Simmental cattle. Animal 2021, 15, 100027. [Google Scholar] [CrossRef]
- Mancin, E.; Sartori, C.; Guzzo, N.; Tuliozi, B.; Mantovani, R. Selection Response Due to Different Combination of Antagonistic Milk, Beef, and Morphological Traits in the Alpine Grey Cattle Breed. Animals 2021, 11, 1340. [Google Scholar] [CrossRef]
- Rincón, F.J.; Zambrano, A.J.; Echeverri, J. Estimation of genetic and phenotypic parameters for production traits in Holstein and Jersey from Colombia. Rev. MVZ Córdoba 2015, 20, 4962–4973. [Google Scholar] [CrossRef] [Green Version]
- DeGroot, B.J.; Keown, J.F.; Van Vleck, L.D.; Kachman, S.D. Estimates of genetic parameters for Holstein cows for test-day yield traits with a random regression cubic spline model. Genet. Mol. Res. 2007, 6, 434–444. [Google Scholar] [PubMed]
- Negussie, E.; Strandén, I.; Mäntysaari, E.A. Genetic association of clinical mastitis with test-day somatic cell score and milk yield during first lactation of Finnish Ayrshire cows. J. Dairy Sci. 2008, 91, 1189–1197. [Google Scholar] [CrossRef]
- Sartori, C.; Guzzo, N.; Mantovani, R. Genetic correlations of fighting ability with somatic cells and longevity in cattle. Animal 2020, 14, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.S.; Pinto, L.F.B.; Valloto, A.A.; Horst, J.A.; Pedrosa, V.B. Genetic parameters between somatic cell score and production traits for Holstein cattle in Southern Brazil. Rev. Colomb. Cienc. Pecu. 2020, 33, 60–70. [Google Scholar] [CrossRef]
- Samoré, A.B.; Groen, A.F.; Boettcher, P.J.; Jamrozik, J.; Canavesi, F.; Bagnato, A. Genetic correlation patterns between somatic cell score and protein yield in the Italian Holstein-Friesian population. J. Dairy Sci. 2008, 91, 4013–4021. [Google Scholar] [CrossRef] [PubMed]
- Torshizi, M.E.; Farhangfar, H. The use of dijkstra mechanistic model for genetic analysis of the lactation curve characteristics and their relationships with age at first calving and somatic cell score of Iranian dairy cows. Acta Sci. Anim. Sci. 2020, 42, e50181. [Google Scholar] [CrossRef]
- Abdel-Shafy, H.; Bortfeldt, R.H.; Tetens, J.; Brockmann, G.A. Single nucleotide polymorphism and haplotype effects associated with somatic cell score in German Holstein cattle. Genet. Sel. Evol. 2014, 46, 35. [Google Scholar] [CrossRef] [Green Version]
- Kurz, J.P.; Yang, Z.; Weiss, R.B.; Wilson, D.J.; Rood, K.A.; Liu, G.E.; Wang, Z. A genome-wide association study for mastitis resistance in phenotypically well-characterized Holstein dairy cattle using a selective genotyping approach. Immunogenetics 2019, 71, 35–47. [Google Scholar] [CrossRef]
- Pan, D.; Zhang, S.; Jiang, J.; Jiang, L.; Zhang, Q.; Liu, J. Genome-wide detection of selective signature in Chinese Holstein. PLoS ONE 2013, 8, e60440. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Cheng, Z.; Zhang, S.; Werling, D.; Wathes, D. Combining Genome Wide Association Studies and Differential Gene Expression Data Analyses Identifies Candidate Genes Affecting Mastitis Caused by Two Different Pathogens in the Dairy Cow. Open J. Anim. Sci. 2015, 5, 358–393. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ma, P.; Liu, J.; Zhang, Q.; Zhang, Y.; Ding, X.; Jiang, L.; Wang, Y.; Zhang, Y.; Sun, D. Genome-wide association study in Chinese Holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. 2015, 16, 111. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Liu, J.; Sun, D.; Ma, P.; Ding, X.; Yu, Y.; Zhanh, Q. Genome Wide Association Studies for Milk Production Traits in Chinese Holstein Population. PLoS ONE 2010, 5, e13661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, M.P.; Govignon-Gion, A.; Croiseau, P.; Fritz, S.; Hozé, C.; Miranda, G.; Martin, P.; Barbat-Leterrier, A.; Letaïef, R.; Rocha, D.; et al. Within-breed and multi-breed GWAS on imputed whole-genome sequence variants reveal candidate mutations affecting milk protein composition in dairy cattle. Genet. Sel. Evol. 2017, 49, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porto-Neto, L.R.; Reverter, A.; Prayaga, K.C.; Chan, E.K.; Johnston, D.J.; Hawken, R.J.; Fordyce, G.; Garcia, J.F.; Sonstegard, T.S.; Bolormaa, S.; et al. The Genetic Architecture of Climatic Adaptation of Tropical Cattle. PLoS ONE 2014, 9, e113284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Kalaldeh, M.; Gibson, J.; Lee, S.H.; Gondro, C.; van der Werf, J.H.J. Detection of genomic regions underlying resistance to gastrointestinal parasites in Australian sheep. Genet. Sel. Evol. 2019, 51, 37. [Google Scholar] [CrossRef] [Green Version]
- Wolf, M.J.; Yin, T.; Neumann, G.B.; Korkuc, P.; Brockmann, G.A.; König, S.; May, K. Genome-Wide Association Study Using Whole-Genome Sequence Data for Fertility, Health Indicator, and Endoparasite Infection Traits in German Black Pied Cattle. Genes 2021, 12, 1163. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, D.; Zhang, S.; Wang, S.; Wu, X.; Zhang, Q.; Liu, L.; Li, Y.; Qiao, L. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. PLoS ONE 2014, 9, e96186. [Google Scholar] [CrossRef] [Green Version]
- Turner, L.B.; Harrison, B.E.; Bunch, R.J.; Porto Neto, L.R.; Li, Y.; Barendse, W. A genome wide association study of tick burden and milk composition in cattle. Anim. Prod. Sci. 2010, 50, 235–245. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Ma, L.; Prakapenka, D.; VanRaden, P.M.; Cole, J.B.; Da, Y. A Large-Scale Genome-Wide Association Study in U.S. Holstein Cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef]
- Sun, Z.; Pang, S.; Cui, Y.; Yan, B. Genetic and Functional Variants Analysis of the GATA6 Gene Promoter in Acute Myocardial Infarction. Front. Genet. 2019, 10, 1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jawasreh, K.; Al-Omari, H.D. Duplication in ECR near HMX1 and a SNP in GATA6 Genes Regulate Microtia in Awassi Sheep. Genes 2020, 11, 597. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Duan, A.; Liang, S.; Ma, X.; Deng, T. Genomic Identification, Evolution, and Expression Analysis of Collagen Genes Family in Water Buffalo during Lactation. Genes 2020, 11, 515. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; He, G.; Zhang, W.; Xu, T.; Qi, H.; Li, J.; Zhang, Y.; Gao, M.Q. Stromal fibroblasts derived from mammary gland of bovine with mastitis display inflammation-specific changes. Sci. Rep. 2016, 6, 27462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilie, D.E.; Cean, A.; Cziszter, L.T.; Gavojdian, D.; Ivan, A.; Kusza, S. Microsatellite and Mitochondrial DNA Study of Native Eastern European Cattle Populations: The Case of the Romanian Grey. PLoS ONE 2015, 10, e0138736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizeranschi, A.E.; Neamț, R.I.; Mihali, C.V.; Goilean, V.; Carabaş, M.; Ilie, D.E. Comparative Analysis of Romanian and Swiss Bovine Populations Using Whole Genome Sequencing and SNP Microarrays. Sci. Pap. Anim. Sci. Biotechnol. 2020, 53, 93–99. [Google Scholar]

| Breed | Trait | N | Mean | SD | Min | Max | h2 | VP | VA |
|---|---|---|---|---|---|---|---|---|---|
| RS | LSCS1 | 11,081 | 2.81 | 1.72 | −1.64 | 8.84 | 0.08 | 2.9916 | 0.2564 |
| LSCS2 | 7479 | 3.06 | 1.89 | −2.06 | 8.82 | 0.10 | 3.4975 | 0.3686 | |
| LSCS3 | 5735 | 3.27 | 1.88 | −2.06 | 8.83 | 0.11 | 3.5347 | 0.3966 | |
| RB | LSCS1 | 3462 | 4.53 | 1.92 | −0.84 | 8.82 | 0.03 | 2.7739 | 0.0857 |
| LSCS2 | 3072 | 4.51 | 1.86 | −1.36 | 8.84 | 0.07 | 2.9304 | 0.2067 | |
| LSCS3 | 2501 | 4.62 | 1.87 | −1.64 | 8.82 | 0.06 | 3.0061 | 0.2048 |
| Parity 1 | Informative SNP 2 | SNP rsID | Chr:Position 3 | A1 | A2 | SNP Effect 4 | –log10 (p) | Nearest Gene 5 | Distance (bp) |
|---|---|---|---|---|---|---|---|---|---|
| L1 | |||||||||
| AX-115117070 | NA | 13:78383148 | G | A | −4.74 × 10−8 | 4.28 | NA | NA | |
| AX-117085597 | rs137805472 | 1:95119031 | C | T | 5.34 × 10−8 | 4.07 | NA | NA | |
| L2 | |||||||||
| AX-106721594 | NA | 1:71370844 | T | C | 3.07 × 10−8 | 4.87 | ZDHHC19 | 25909 | |
| AX-106755404 | NA | 23:17013312 | A | G | 2.21 × 10−8 | 4.61 | NA | NA | |
| AX-106740205 | rs109232438 | 9:71884731 | C | T | 5.01 × 10−8 | 4.32 | NA | NA | |
| AX-171465786 | rs110140732 | 17:3030264 | G | A | 9.27 × 10−8 | 4.28 | NA | NA | |
| AX-117088706 | rs43209122 | 16:70728849 | A | G | −2.50 × 10−8 | 4.23 | NA | NA | |
| AX-106761299 | NA | 8:25435054 | C | T | −1.70 × 10−8 | 4.23 | NA | NA | |
| L3 | |||||||||
| AX-106761943 | rs110749552 | 6:37526622 | G | A | 5.47 × 10−8 | 6.37 | HERC3 | within | |
| AX-106728871 | rs29021886 | 14:17674401 | C | A | −1.20 × 10−8 | 4.62 | NA | NA | |
| AX-124381671 | NA | 4:79254806 | A | G | 1.20 × 10−8 | 4.6 | NA | NA | |
| AX-106740778 | rs42627158 | 8:81747455 | C | T | −7.00 × 10−8 | 4.36 | DAPK1 | 459784 | |
| AX-115112140 | NA | 15:5927942 | A | G | 3.73 × 10−8 | 4.19 | MMP7 | 462217 | |
| AX-185121504 | rs209378984 | 6:70369168 | A | G | 5.97 × 10−8 | 4.11 | NA | NA | |
| AX-115108867 | NA | 14:41169239 | A | G | 6.76 × 10−9 | 4 | NA | NA |
| Parity 1 | Informative SNP 2 | SNP rsID | Chr:Position 3 | A1 | A2 | SNP Effect 4 | –log10 (p) | Nearest Gene 5 | Distance (bp) |
|---|---|---|---|---|---|---|---|---|---|
| L1 | |||||||||
| AX-106741653 | NA | 7:8272794 | C | A | 6.14 × 10−9 | 5.37 | AKAP8 | 514640 | |
| AX-115114947 | NA | 7:8276425 | G | A | 6.14 × 10−9 | 5.37 | AKAP8 | 511009 | |
| AX-106739297 | rs108991944 | 11:37261133 | A | C | 1.54 × 10−9 | 4.41 | CLHC1 | 526855 | |
| AX-124375018 | rs42894728 | 6:20276795 | G | A | 7.39 × 10−9 | 4.33 | NA | NA | |
| AX-171466608 | rs42228650 | 7:27601185 | G | A | 5.65 × 10−10 | 4.29 | MEGF10 | 219169 | |
| AX-106727011 | NA | 13:76008898 | G | A | −3.96 × 10−9 | 4.24 | NA | NA | |
| AX-117090125 | rs110642171 | 6:39688028 | A | G | −2.65 × 10−9 | 4.19 | NA | NA | |
| AX-117084655 | rs109189476 | 2:88162563 | C | T | 6.31 × 10−10 | 4.14 | SATB2 | 86169 | |
| AX-106731475 | NA | 8:75798051 | T | C | 2.67 × 10−9 | 4.14 | NA | NA | |
| AX-124381310 | NA | 24:34453446 | A | G | −1.98 × 10−9 | 4.08 | GATA6 | 96211 | |
| L2 | |||||||||
| AX-124381305 | rs43358795 | 3:97560648 | A | G | −3 × 10−10 | 5.38 | SPATA6 | 809734 | |
| AX-169513997 | rs133252983 | 13:77447650 | G | T | 5.07 × 10−9 | 4.61 | NA | NA | |
| AX-117080179 | rs110709131 | 2:46155190 | G | A | 3.47 × 10−9 | 4.56 | NA | NA | |
| AX-117083483 | rs133489631 | 2:46126141 | T | C | 3.47 × 10−9 | 4.56 | NA | NA | |
| AX-124350324 | rs42797639 | 9:20734430 | C | T | −9.3 × 10−10 | 4.19 | NA | NA | |
| AX-124377880 | rs41981703 | 21:40381539 | A | C | 1.02 × 10−8 | 4.15 | NA | NA | |
| AX-106735825 | rs43585636 | 9:14745626 | C | A | 3.02 × 10−9 | 4.09 | COL12A1 | 123884 | |
| AX-117085949 | rs43585209 | 9:14767890 | A | G | 3.02 × 10−9 | 4.09 | COL12A1 | 101620 | |
| AX-106726354 | NA | 5:94517415 | A | G | 5.58 × 10−9 | 4.08 | EPS8 | 156339 | |
| L3 | |||||||||
| AX-115109333 | NA | 22:26294371 | A | G | 6.59 × 10−9 | 5.17 | NA | NA | |
| AX-124375014 | rs110955314 | 29:19740081 | C | A | −1.57 × 10−8 | 4.38 | LUZP2 | 519687 | |
| AX-106753982 | NA | 21:23675656 | A | G | −6.09 × 10−9 | 4.35 | RAMAC | 30303 | |
| AX-106735391 | rs110359025 | 19:16700029 | C | T | −7.19 × 10−9 | 4.21 | NA | NA | |
| AX-106735614 | rs43256975 | 1:108154057 | A | G | −3.37 × 10−9 | 4.1 | IL12A | 131225 | |
| AX-115115579 | rs110825674 | 19:50547082 | T | C | −9.06 × 10−9 | 4.06 | NA | NA | |
| AX-106730586 | rs109619272 | 20:22823334 | A | G | 4.48 × 10−9 | 4.03 | ANKRD55 | 184056 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilie, D.E.; Mizeranschi, A.E.; Mihali, C.V.; Neamț, R.I.; Goilean, G.V.; Georgescu, O.I.; Zaharie, D.; Carabaș, M.; Huțu, I. Genome-Wide Association Studies for Milk Somatic Cell Score in Romanian Dairy Cattle. Genes 2021, 12, 1495. https://doi.org/10.3390/genes12101495
Ilie DE, Mizeranschi AE, Mihali CV, Neamț RI, Goilean GV, Georgescu OI, Zaharie D, Carabaș M, Huțu I. Genome-Wide Association Studies for Milk Somatic Cell Score in Romanian Dairy Cattle. Genes. 2021; 12(10):1495. https://doi.org/10.3390/genes12101495
Chicago/Turabian StyleIlie, Daniela Elena, Alexandru Eugeniu Mizeranschi, Ciprian Valentin Mihali, Radu Ionel Neamț, George Vlad Goilean, Ovidiu Ionuț Georgescu, Daniela Zaharie, Mihai Carabaș, and Ioan Huțu. 2021. "Genome-Wide Association Studies for Milk Somatic Cell Score in Romanian Dairy Cattle" Genes 12, no. 10: 1495. https://doi.org/10.3390/genes12101495
APA StyleIlie, D. E., Mizeranschi, A. E., Mihali, C. V., Neamț, R. I., Goilean, G. V., Georgescu, O. I., Zaharie, D., Carabaș, M., & Huțu, I. (2021). Genome-Wide Association Studies for Milk Somatic Cell Score in Romanian Dairy Cattle. Genes, 12(10), 1495. https://doi.org/10.3390/genes12101495








