Gene Amplification and the Extrachromosomal Circular DNA
Abstract
:1. Gene Amplification and the Extrachromosomal Circles in Human Cancer
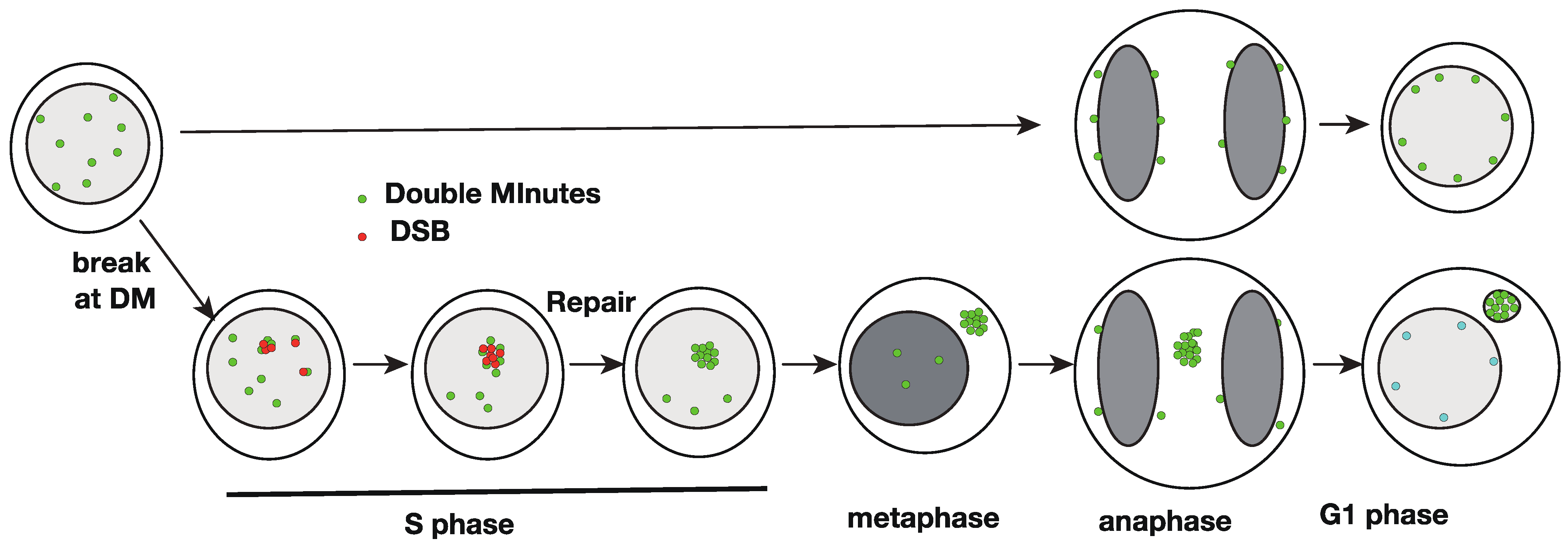
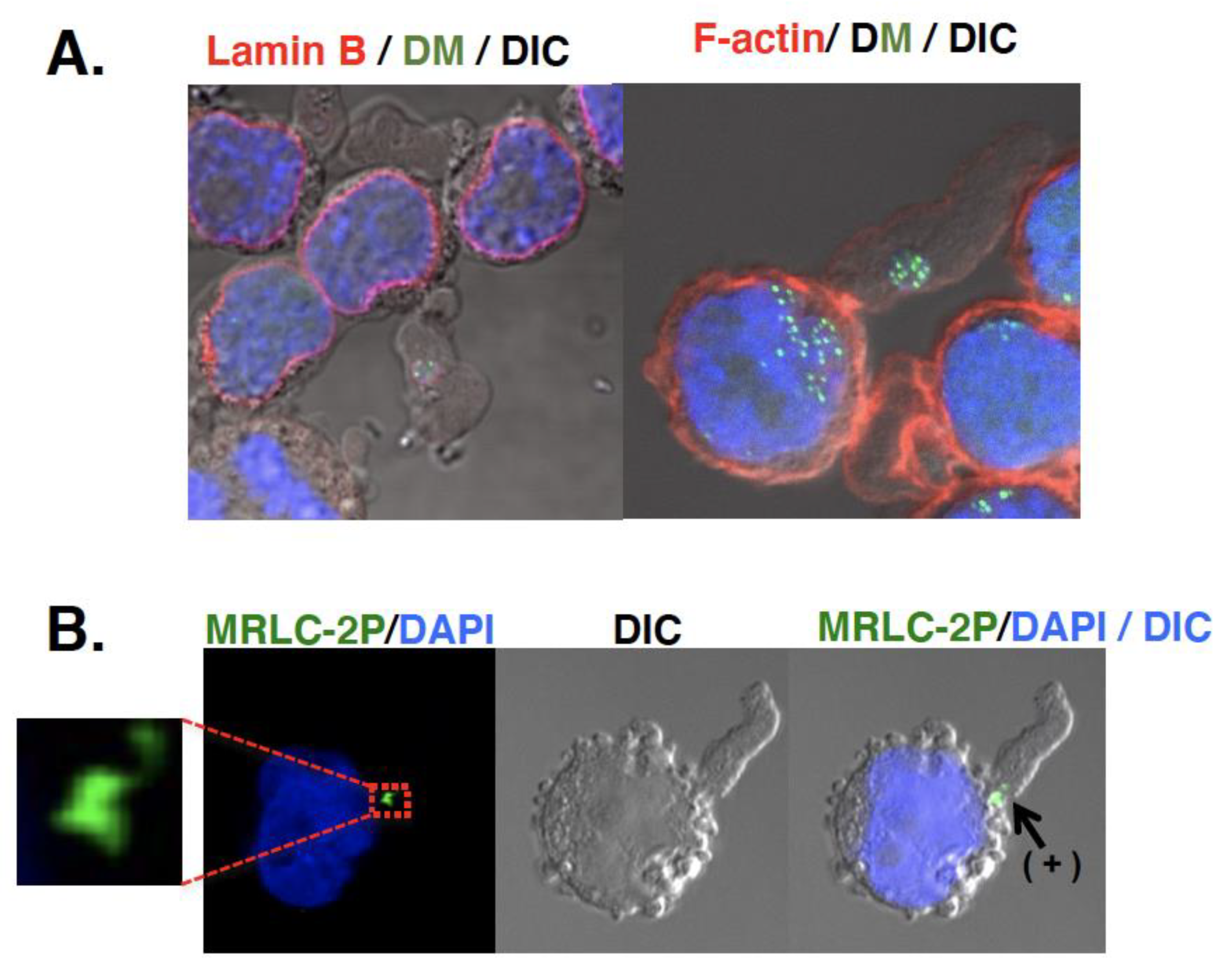
2. Intra-Cellular Behavior of the Extrachromosomal Circles
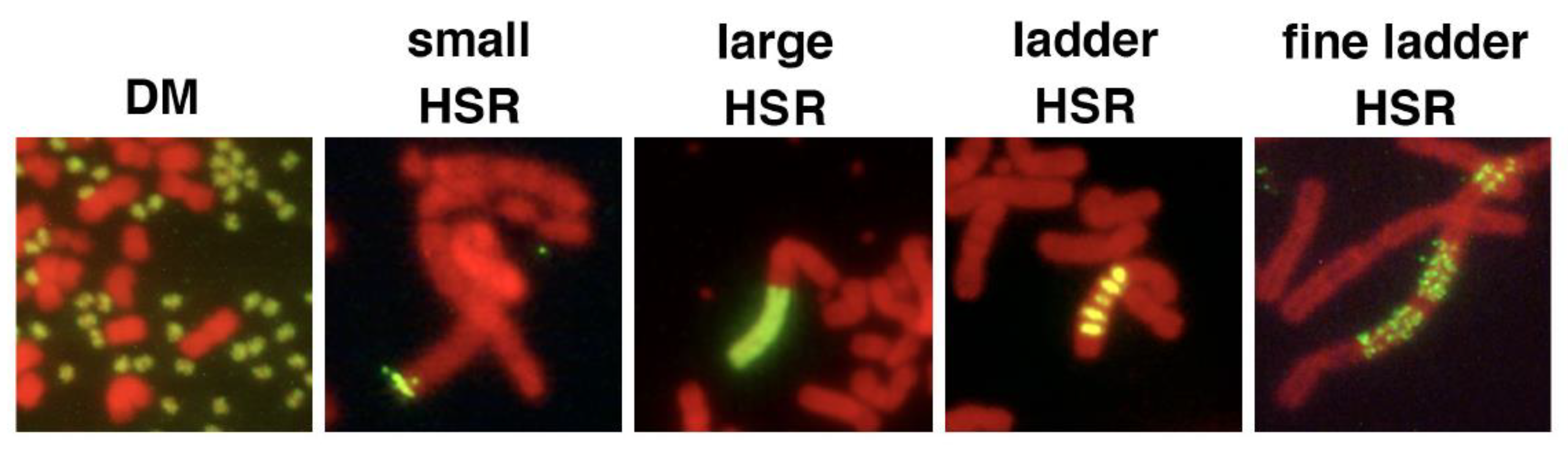
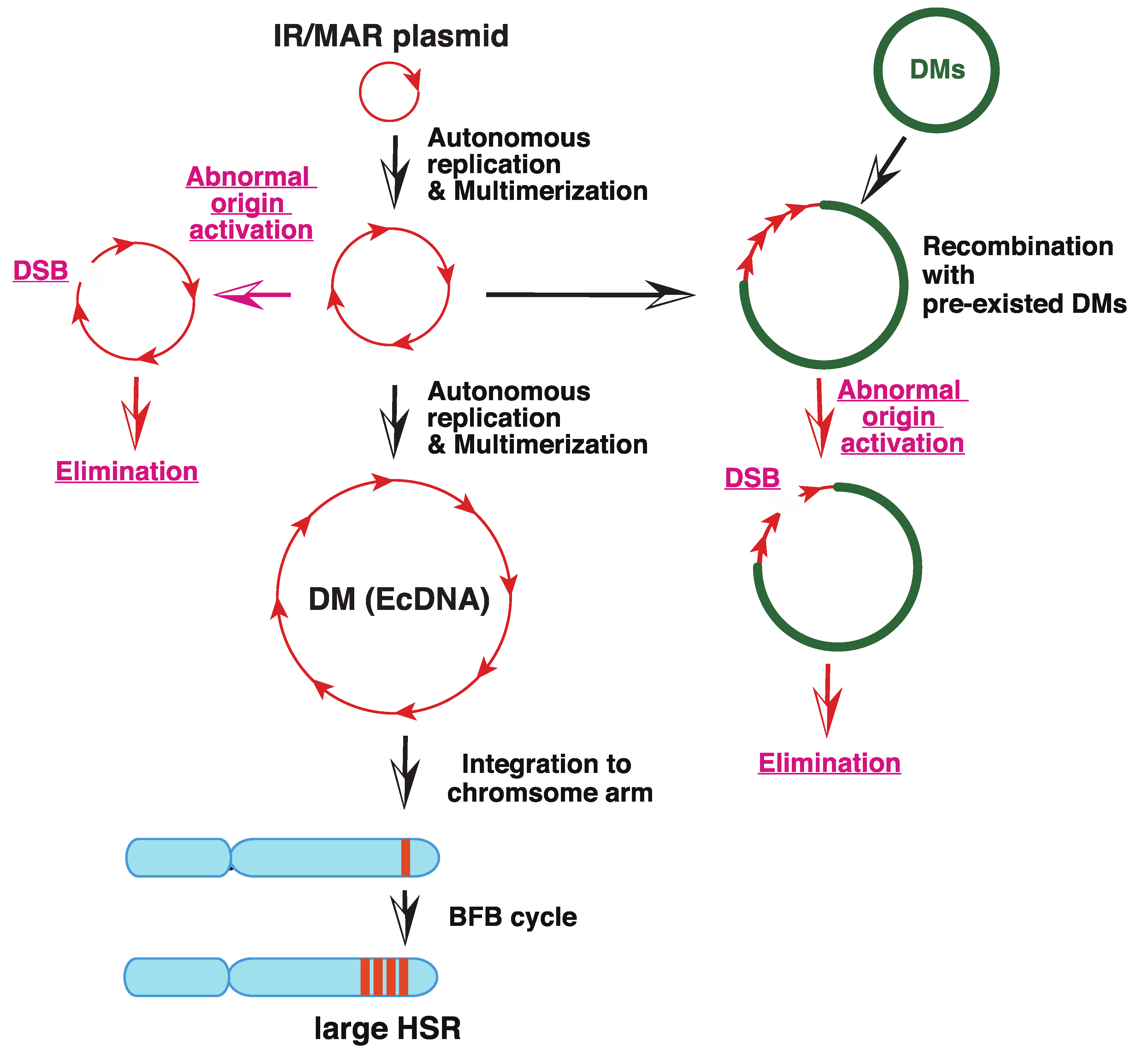
3. Generation of DMs/EcDNA and HSR from Small eccDNA
4. From Chromosome Arm to Gene Amplification
5. Applications of the Extrachromosomal Element-Mediated Gene Amplification
6. Future Task
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cowell, J.K. Double minutes and homogenously staining regions: Gene amplification in mammalian cells. Annu. Rev. Genet. 1982, 16, 21–59. [Google Scholar] [CrossRef]
- Wahl, G.M. The importance of circular DNA in mammalian gene amplification. Cancer Res. 1989, 49, 1333–1340. [Google Scholar]
- Von Hoff, D.D.; Forseth, B.; Clare, C.N.; Hansen, K.L.; VanDevanter, D. Double minutes arise from circular extrachromosomal DNA intermediates which integrate into chromosomal sites in human HL-60 leukemia cells. J. Clin. Investig. 1990, 85, 1887–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, N.; Ochi, T.; Itonaga, K. Replication timing of amplified genetic regions relates to intranuclear localization but not to genetic activity or G/R band. Exp. Cell Res. 2001, 268, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Hamkalo, B.A.; Farnham, P.J.; Johnston, R.; Schimke, R.T. Ultrastructural features of minute chromosomes in a methotrexate-resistant mouse 3T3 cell line. Proc. Natl. Acad. Sci. USA 1985, 82, 1026–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanDevanter, D.R.; Piaskowski, V.D.; Casper, J.T.; Douglass, E.C.; Von Hoff, D.D. Ability of circular extrachromosomal DNA molecules to carry amplified MYCN proto-oncogenes in human neuroblastomas in vivo. J. Natl. Cancer Inst. 1990, 82, 1815–1821. [Google Scholar] [CrossRef]
- Lin, C.C.; Meyne, J.; Sasi, R.; Moyzis, R.K. Apparent lack of telomere sequences on double minute chromosomes. Cancer Genet. Cytogenet. 1990, 48, 271–274. [Google Scholar] [CrossRef]
- Wu, S.; Turner, K.M.; Nguyen, N.; Raviram, R.; Erb, M.; Santini, J.; Luebeck, J.; Rajkumar, U.; Diao, Y.; Li, B.; et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 2019, 575, 699–703. [Google Scholar] [CrossRef]
- Gaubatz, J.W. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat. Res. 1990, 237, 271–292. [Google Scholar] [CrossRef]
- Paulsen, T.; Kumar, P.; Koseoglu, M.M.; Dutta, A. Discoveries of Extrachromosomal Circles of DNA in Normal and Tumor Cells. Trends Genet. 2018, 34, 270–278. [Google Scholar] [CrossRef]
- Shibata, Y.; Kumar, P.; Layer, R.; Willcox, S.; Gagan, J.R.; Griffith, J.D.; Dutta, A. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science 2012, 336, 82–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moller, H.D.; Mohiyuddin, M.; Prada-Luengo, I.; Sailani, M.R.; Halling, J.F.; Plomgaard, P.; Maretty, L.; Hansen, A.J.; Snyder, M.P.; Pilegaard, H.; et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat. Commun. 2018, 9, 1069. [Google Scholar] [CrossRef] [Green Version]
- Turner, K.M.; Deshpande, V.; Beyter, D.; Koga, T.; Rusert, J.; Lee, C.; Li, B.; Arden, K.; Ren, B.; Nathanson, D.A.; et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 2017, 543, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.W.; Bafna, V.; Mischel, P.S. Extrachromosomal oncogene amplification in tum.our pathogenesis and evolution. Nat. Rev. Cancer 2019, 19, 283–288. [Google Scholar] [CrossRef]
- Shimizu, N.; Hanada, N.; Utani, K.; Sekiguchi, N. Interconversion of intra- and extra-chromosomal sites of gene amplification by modulation of gene expression and DNA methylation. J. Cell Biochem. 2007, 102, 515–529. [Google Scholar] [CrossRef]
- Mitsuda, S.H.; Shimizu, N. Epigenetic Repeat-Induced Gene Silencing in the Chromosomal and Extrachromosomal Contexts in Human Cells. PLoS ONE 2016, 11, e0161288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; McGill, J.R.; Forseth, B.J.; Davidson, K.K.; Bradley, T.P.; Van Devanter, D.R. Elimination of extrachromosomally amplified MYC genes from human tumor cells reduces their tumorigenicity. Proc. Natl. Acad. Sci. USA 1992, 89, 8165–8169. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, N.; Nakamura, H.; Kadota, T.; Kitajima, K.; Oda, T.; Hirano, T. Loss of amplified c-myc genes in the spontaneously differentiated HL-60 cells. Cancer Res. 1994, 54, 3561–3567. [Google Scholar]
- Eckhardt, S.G.; Dai, A.; Davidson, K.K.; Forseth, B.J.; Wahl, G.M.; Von Hoff, D.D. Induction of differentiation in HL60 cells by the reduction of extrachromosomally amplified c-myc. Proc. Natl. Acad. Sci. USA 1994, 91, 6674–6678. [Google Scholar] [CrossRef] [Green Version]
- Haaf, T.; Schmid, M. Analysis of double minutes and double minute-like chromatin in human and murine tumor cells using antikinetochore antibodies. Cancer Genet. Cytogenet. 1988, 30, 73–82. [Google Scholar] [CrossRef]
- deCarvalho, A.C.; Kim, H.; Poisson, L.M.; Winn, M.E.; Mueller, C.; Cherba, D.; Koeman, J.; Seth, S.; Protopopov, A.; Felicella, M.; et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat. Genet. 2018, 50, 708–717. [Google Scholar] [CrossRef]
- Nathanson, D.A.; Gini, B.; Mottahedeh, J.; Visnyei, K.; Koga, T.; Gomez, G.; Eskin, A.; Hwang, K.; Wang, J.; Masui, K.; et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 2014, 343, 72–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levan, A.; Levan, G. Have double minutes functioning centromeres? Hereditas 1978, 88, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Otter, M.; Wahl, G.M. Mitotic segregation of viral and cellular acentric extrachromosomal molecules by chromosome tethering. J Cell Sci. 2001, 114, 49–58. [Google Scholar] [CrossRef]
- Tanaka, T.; Shimizu, N. Induced detachment of acentric chromatin from mitotic chromosomes leads to their cytoplasmic localization at G1 and the micronucleation by lamin reorganization at S phase. J. Cell Sci. 2000, 113, 697–707. [Google Scholar] [CrossRef]
- Shimizu, N.; Itoh, N.; Utiyama, H.; Wahl, G. Selective Entrapment of Extrachromosomally Amplified DNA by Nuclear Budding and Micronucleation during S-phase. J. Cell Biol. 1998, 140, 1307–1320. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, N.; Kanda, T.; Wahl, G.M. Selective capture of acentric fragments by micronuclei provides a rapid method for purifying extrachromosomally amplified DNA. Nat. Genet. 1996, 12, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, N.; Misaka, N.; Utani, K. Nonselective DNA damage induced by a replication inhibitor results in the selective elimination of extrachromosomal double minutes from human cancer cells. Genes Chromosom. Cancer 2007, 46, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Oobatake, Y.; Shimizu, N. Double-strand breakage in the extrachromosomal double minutes triggers their aggregation in the nucleus, micronucleation, and morphological transformation. Genes Chromosom. Cancer 2019, 59, 133–143. [Google Scholar] [CrossRef]
- Utani, K.; Okamoto, A.; Shimizu, N. Generation of micronuclei during interphase by coupling between cytoplasmic membrane blebbing and nuclear budding. PLoS ONE 2011, 6, e27233. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, N.; Kamezaki, F.; Shigematsu, S. Tracking of microinjected DNA in live cells reveals the intracellular behavior and elimination of extrachromosomal genetic material. Nucleic Acids Res. 2005, 33, 6296–6307. [Google Scholar] [CrossRef] [Green Version]
- de Noronha, C.M.; Sherman, M.P.; Lin, H.W.; Cavrois, M.V.; Moir, R.D.; Goldman, R.D.; Greene, W.C. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science 2001, 294, 1105–1108. [Google Scholar] [CrossRef]
- Shah, P.; Wolf, K.; Lammerding, J. Bursting the Bubble—Nuclear Envelope Rupture as a Path to Genomic Instability? Trends Cell Biol. 2017, 27, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Chen, Z.J. cGAS in action: Expanding roles in immunity and inflammation. Science 2019, 363, eaat8657. [Google Scholar] [CrossRef]
- Von Hoff, H.D. New mechanisms of gene amplification in drug resistance (the episome model). Cancer Treat. Res. 1991, 57, 1–11. [Google Scholar]
- Shimizu, N.; Miura, Y.; Sakamoto, Y.; Tsutsui, K. Plasmids with a mammalian replication origin and a matrix attachment region initiate the event similar to gene amplification. Cancer Res. 2001, 61, 6987–6990. [Google Scholar] [PubMed]
- Shimizu, N.; Hashizume, T.; Shingaki, K.; Kawamoto, J.K. Amplification of plasmids containing a mammalian replication initiation region is mediated by controllable conflict between replication and transcription. Cancer Res. 2003, 63, 5281–5290. [Google Scholar]
- Hashizume, T.; Shimizu, N. Dissection of mammalian replicators by a novel plasmid stability assay. J. Cell. Biochem. 2007, 101, 552–565. [Google Scholar] [CrossRef]
- Okada, N.; Shimizu, N. Dissection of the β-Globin Replication-Initiation Region Reveals Specific Requirements for Replicator Elements during Gene Amplification. PLoS ONE 2013, 8, e77350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, N.; Shingaki, K.; Kaneko-Sasaguri, Y.; Hashizume, T.; Kanda, T. When, where and how the bridge breaks: Anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp. Cell Res. 2005, 302, 233–243. [Google Scholar] [CrossRef]
- Tanaka, S.S.; Mitsuda, S.H.; Shimizu, N. How a Replication Origin and Matrix Attachment Region Accelerate Gene Amplification under Replication Stress in Mammalian Cells. PLoS ONE 2014, 9, e103439. [Google Scholar] [CrossRef]
- Araki, Y.; Hamafuji, T.; Noguchi, C.; Shimizu, N. Efficient Recombinant Production in Mammalian Cells Using a Novel IR/MAR Gene Amplification Method. PLoS ONE 2012, 7, e41787. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, R.; Utani, K.; Thakur, B.; Ishine, K.; Aladjem, M.I.; Shimizu, N. SIRT1 stabilizes extrachromosomal gene amplification and contributes to repeat-induced gene silencing. J. Biol. Chem. 2021, 296, 100356. [Google Scholar] [CrossRef] [PubMed]
- Hamlin, J.L. Initiation of replication in mammalian chromosomes. Crit. Rev. Eukaryot. Gene Expr. 1992, 2, 359–381. [Google Scholar]
- L’Abbate, A.; Macchia, G.; D’Addabbo, P.; Lonoce, A.; Tolomeo, D.; Trombetta, D.; Kok, K.; Bartenhagen, C.; Whelan, C.; Palumbo, O.; et al. Genomic organization and evolution of double minutes/homogeneously staining regions with MYC amplification in human cancer. Nucleic Acids Res. 2014, 42, 9131–9145. [Google Scholar] [CrossRef] [Green Version]
- Koche, R.P.; Rodriguez-Fos, E.; Helmsauer, K.; Burkert, M.; MacArthur, I.C.; Maag, J.; Chamorro, R.; Munoz-Perez, N.; Puiggròs, M.; Garcia, H.D.; et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat. Genet. 2020, 52, 29–34. [Google Scholar] [CrossRef]
- Morton, A.R.; Dogan-Artun, N.; Faber, Z.J.; MacLeod, G.; Bartels, C.F.; Piazza, M.S.; Allan, K.C.; Mack, S.C.; Wang, X.; Gimple, R.C.; et al. Functional Enhancers Shape Extrachromosomal Oncogene Amplifications. Cell 2019, 179, 1330–1341.e13. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, N.; Kapoor, R.; Naniwa, S.; Sakamaru, N.; Yamada, T.; Yamamura, Y.K.; Utani, K.-I. Generation and maintenance of acentric stable double minutes from chromosome arms in inter-species hybrid cells. BMC Mol. Cell Biol. 2019, 20, 2. [Google Scholar] [CrossRef] [PubMed]
- Utani, K.; Fu, H.; Jang, S.M.; Marks, A.B.; Smith, O.K.; Zhang, Y.; Redon, C.E.; Shimizu, N.; Aladjem, M.I. Phosphorylated SIRT1 associates with replication origins to prevent excess replication initiation and preserve genomic stability. Nucleic Acids Res. 2017, 45, 7807–7824. [Google Scholar] [CrossRef] [Green Version]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef]
- Meyerson, M.; Pellman, D. Cancer genomes evolve by pulverizing single chromosomes. Cell 2011, 144, 9–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef]
- Zhang, C.Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatch, E.M.; Fischer, A.H.; Deerinck, T.J.; Hetzer, M.W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 2013, 154, 47–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lusk, C.P.; King, M.C. Rotten to the Core: Why Micronuclei Rupture. Dev. Cell 2018, 47, 265–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamoto, A.; Utani, K.I.; Shimizu, N. DNA replication occurs in all lamina positive micronuclei, but never in lamina negative micronuclei. Mutagenesis 2012, 27, 323–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Utani, K.; Kawamoto, J.K.; Shimizu, N. Micronuclei bearing acentric extrachromosomal chromatin are transcriptionally competent and may perturb the cancer cell phenotype. Mol. Cancer Res. 2007, 5, 695–704. [Google Scholar] [CrossRef] [Green Version]
- Ly, P.; Cleveland, D.W. Rebuilding Chromosomes After Catastrophe: Emerging Mechanisms of Chromothripsis. Trends Cell Biol. 2017, 27, 917–930. [Google Scholar] [CrossRef]
- Asoshina, M.; Myo, G.; Tada, N.; Tajino, K.; Shimizu, N. Targeted amplification of a sequence of interest in artificial chromosome in mammalian cells. Nucleic Acids Res. 2019, 47, 5998–6006. [Google Scholar] [CrossRef]
- Ohira, T.; Miyauchi, K.; Uno, N.; Shimizu, N.; Kazuki, Y.; Oshimura, M.; Kugoh, H. An efficient protein production system via gene amplification on a human artificial chromosome and the chromosome transfer to CHO cells. Sci. Rep. 2019, 9, 16954. [Google Scholar] [CrossRef]
- Garrick, D.; Fiering, S.; Martin, D.I.; Whitelaw, E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998, 18, 56–59. [Google Scholar] [CrossRef]
- Hsieh, J.; Fire, A. Recognition and silencing of repeated DNA. Annu. Rev. Genet. 2000, 34, 187–204. [Google Scholar] [CrossRef]
- Reddy, B.D.; Wang, Y.; Niu, L.; Higuchi, E.C.; Marguerat, S.B.; Bahler, J.; Smith, G.R.; Jia, S. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev. 2011, 25, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Kondo, Y.; Issa, J.P. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J. Biol. Chem. 2003, 278, 27658–27662. [Google Scholar] [CrossRef] [Green Version]
- McBurney, M.W.; Mai, T.; Yang, X.; Jardine, K. Evidence for repeat-induced gene silencing in cultured Mammalian cells: Inactivation of tandem repeats of transfected genes. Expr. Cell Res. 2002, 274, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Henikoff, S. Conspiracy of silence among repeated transgenes. Bioessays 1998, 20, 532–535. [Google Scholar] [CrossRef]
- Ogaki, Y.; Fukuma, M.; Shimizu, N. Repeat induces not only gene silencing, but also gene activation in mammalian cells. PLoS ONE 2020, 15, e0235127. [Google Scholar] [CrossRef] [PubMed]
- Ohsaki, K.; Ohgaki, Y.; Shimizu, N. Amplification of a transgene within a long array of replication origins favors higher gene expression in animal cells. PLoS ONE 2017, 12, e0175585. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, N.; Shimura, T.; Tanaka, T. Selective elimination of acentric double minutes from cancer cells through the extrusion of micronuclei. Mutat. Res. 2000, 448, 81–90. [Google Scholar]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Sin, S.T.K.; Jiang, P.; Deng, J.; Ji, L.; Cheng, S.H.; Dutta, A.; Leung, T.Y.; Chan, K.C.A.; Chiu, R.W.K.; Lo, Y.M.D. Identification and characterization of extrachromosomal circular DNA in maternal plasma. Proc. Natl. Acad. Sci. USA 2020, 117, 1658–1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimizu, N. Gene Amplification and the Extrachromosomal Circular DNA. Genes 2021, 12, 1533. https://doi.org/10.3390/genes12101533
Shimizu N. Gene Amplification and the Extrachromosomal Circular DNA. Genes. 2021; 12(10):1533. https://doi.org/10.3390/genes12101533
Chicago/Turabian StyleShimizu, Noriaki. 2021. "Gene Amplification and the Extrachromosomal Circular DNA" Genes 12, no. 10: 1533. https://doi.org/10.3390/genes12101533
APA StyleShimizu, N. (2021). Gene Amplification and the Extrachromosomal Circular DNA. Genes, 12(10), 1533. https://doi.org/10.3390/genes12101533




