Genetic and Molecular Evaluation: Reporting Three Novel Mutations and Creating Awareness of Pycnodysostosis Disease
Abstract
:1. Introduction
2. Patients and Methods
3. Results
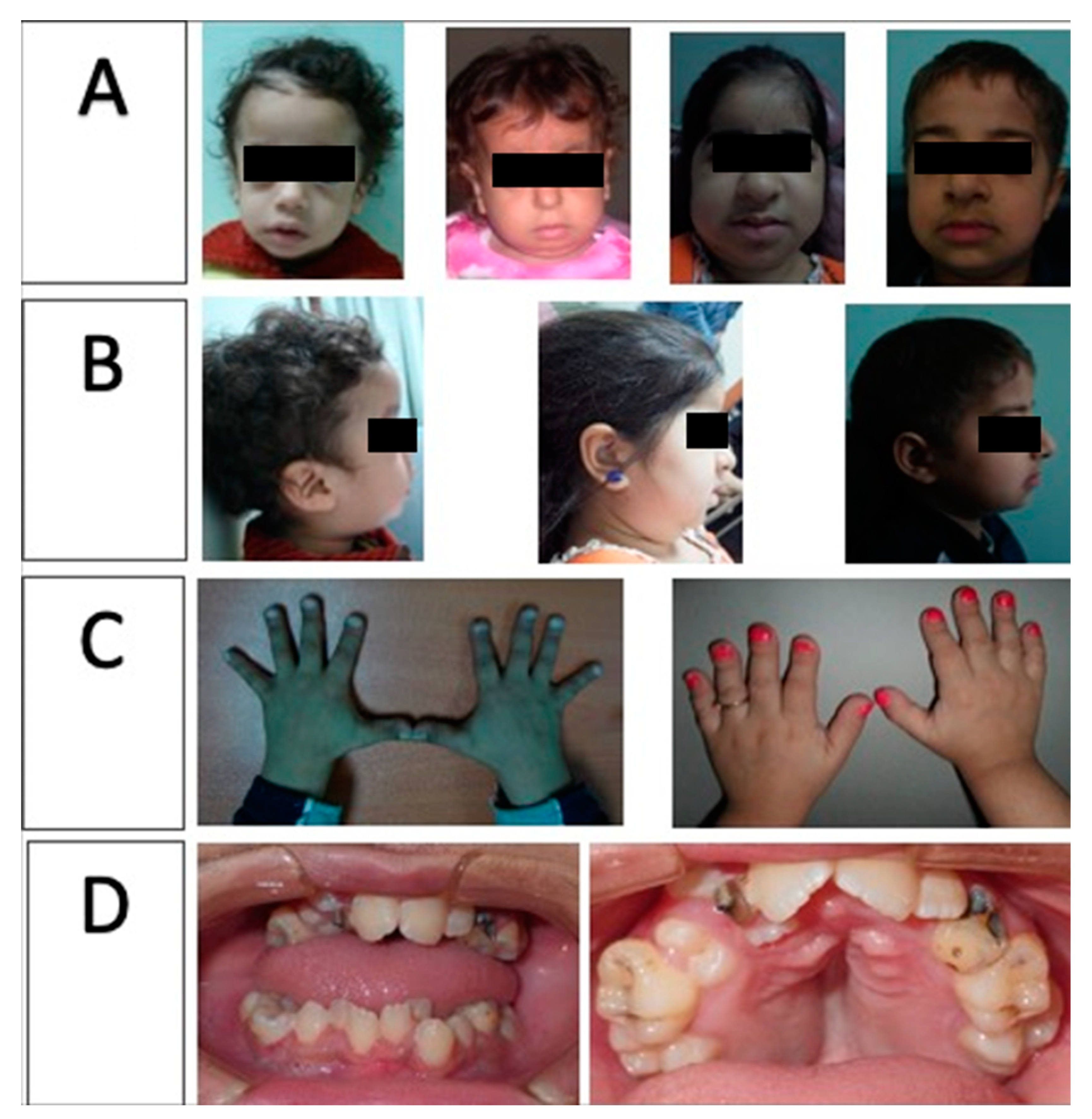
3.1. Phenotypic Results
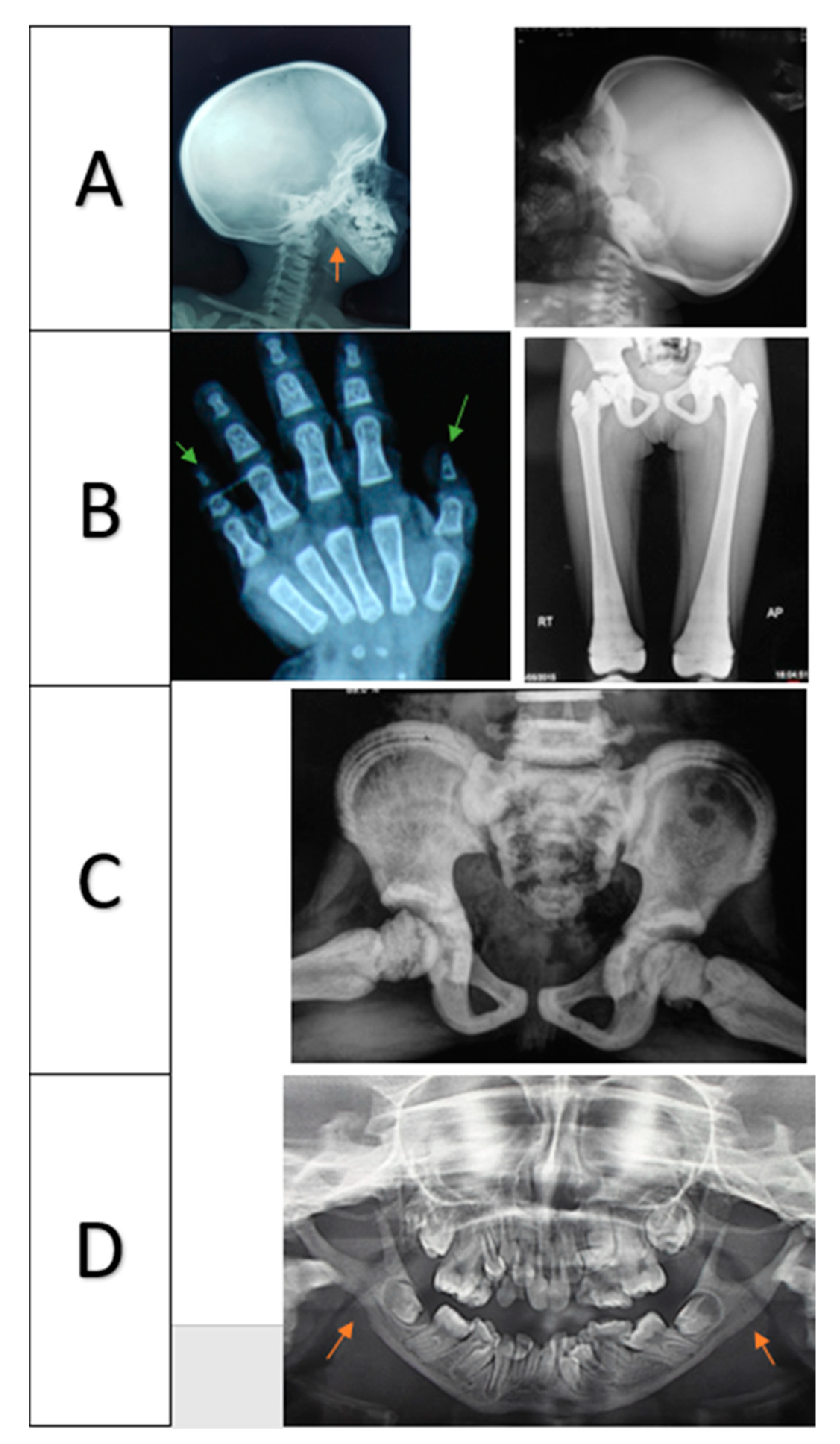
3.2. Radiologic Results
3.3. Molecular Results
3.4. Protein Function Prediction Analysis of c.509G>T
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bizaoui, V.; Michot, C.; Baujat, G.; Amouroux, C.; Baron, S.; Capri, Y.; Cohen-Solal, M.; Collet, C.; Dieux, A.; Geneviève, D.; et al. Pycnodysostosis: Natural history and management guidelines from 27 French cases and a literature review. Clin. Genet. 2019, 96, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Arman, A.; Bereket, A.; Coker, A.; Kiper, P.Ö.Ş.; Güran, T.; Özkan, B.; Atay, Z.; Akçay, T.; Haliloglu, B.; Boduroglu, K.; et al. Cathepsin K analysis in a pycnodysostosis cohort: Demographic, genotypic and phenotypic features. Orphanet J. Rare Dis. 2014, 9, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montanari, U. Acondroplasia e dysostosis cleidocranial digital. Chir. Org. Mov. 1923, 7, 379–391. [Google Scholar]
- Maroteaux, P.; Lamy, M. Pyknodysostosis. Presse Med. 1962, 70, 999–1002. [Google Scholar] [PubMed]
- Hodder, A.; Huntley, C.; Aronson, J.K.; Ramachandran, M. Pycnodysostosis and the making of an artist. Gene 2015, 555, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Cai, T.; Shi, S.; Wang, W.; Zhang, Y.; Mao, T.; Duan, X. Clinical and animal research findings in pycnodysostosis and gene mutations of cathepsin K from 1996 to 2011. Orphanet J. Rare Dis. 2011, 6, 20. [Google Scholar] [CrossRef] [Green Version]
- Bertola, D.; Aguena, M.; Yamamoto, G.; Ae Kim, C.; Passos-Bueno, M.R. Obesity in pycnodysostosis due to UPD1, Possible effect of an imprinted gene on chromosome 1. Am. J. Med. Genet. A. 2011, 155A, 1483–1486. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.T.; Ramadan, M.A.; Sherif, A.; Aziz Bedair, E.S.; Rizk, M.M. Pycnodysostosis: Clinical, radiologic, and endocrine evaluation and linear growth after growth hormone therapy. Metabolism 2001, 50, 905–911. [Google Scholar] [CrossRef]
- Alves, N.; Cantín, M. Clinical and radiographic maxillofacial features of pycnodysostosis. Int. J. Clin. Exp. Med. 2014, 7, 492–496. [Google Scholar]
- LeBlanc, S.; Savarirayan, R. Pycnodysostosis. 2020 Nov 5. In GeneReviews® [Internet]; Adam, M.P., Ardinger, H.H., Eds.; University of Washington: Seattle, WA, USA, 2020; pp. 1993–2021. [Google Scholar]
- Cortisse, N.; Forget, P.; Dresse, M.F.; Florkin, B.; Mascard, E.; Guinebretière, J.M.; Brugières, L.; Hoyoux, C. A Case of Osteosarcoma in a Patient with Pycnodysostosis. J. Pediatr. Hematol. Oncol. 2012, 34, 545–547. [Google Scholar] [CrossRef]
- Kshirsagar, V.; Ahmed, M.; Nagarsenkar, S.; Sahoo, K.; Shah, K.B. Ichthyosis vulgaris and pycnodysostosis: An unusual occurrence. Acta Med. Acad. 2012, 41, 214–218. [Google Scholar] [CrossRef] [Green Version]
- Testani, E.; Scarano, E.; Leoni, C.; Dittoni, S.; Losurdo, A.; Colicchio, S.; Gnoni, V.; Vollono, C.; Zampino, G.; Paludetti, G.; et al. Upper airway surgery of obstructive sleep apnea in pycnodysostosis: Case report and literature review. Am. J. Med. Genet. A 2014, 164A, 2029–2035. [Google Scholar] [CrossRef]
- Tinsa, F.; Hamouda, S.; Bellalah, M.; Bousnina, D.; Karboul, L.; Boussetta, K.; Bousnina, S. Unusual feature of pycnodysostosis: Pectus carinatum. Tunis. Med. 2014, 92, 180–181. [Google Scholar]
- Kyung, S.E.; Horton, J.C. Papilledema from craniosynostosis in pycnodysostosis. Pediatr. Neurol. 2015, 52, 128–129. [Google Scholar] [CrossRef] [Green Version]
- Aynaou, H.; Skiker, I.; Latrech, H. Short stature Revealing a Pycnodysostosis: A Case Report. J. Orthop. Case Rep. 2016, 6, 43–45. [Google Scholar] [PubMed]
- Verma, V.; Singh, R.K. A Case Report of Pycnodysostosis Associated with Multiple Pituitary Hormone Deficiencies and Response to Treatment. J. Clin. Res. Pediatr. Endocrinol. 2020, 12, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Otaify, G.A.; Abdel-Hamid, M.S.; Mehrez, M.I.; Aboul-Ezz, E.; Zaki, M.S.; Aglan, M.S.; Temtamy, S.A. Genetic study of eight Egyptian patients with pycnodysostosis: Identification of novel CTSK mutations and founder effect. Osteoporos. Int. 2018, 29, 1833–1841. [Google Scholar] [CrossRef]
- Donnarumma, M.; Regis, S.; Tappino, B.; Rosano, C.; Assereto, S.; Corsolini, F.; Di Rocco, M.; Filocamo, M. Molecular analysis and characterization of nine novel CTSK mutations in twelve patients affected by pycnodysostosis. Mutation in brief #961. Online. Hum. Mutat. 2007, 28, 524. [Google Scholar] [PubMed]
- Abdallah, E.M.; Matrawy, K.; Shwel, Y. Pycnodysostosis: Clinical and radiological features in two Egyptian families. J. Pediatr. Sci. 2012, 4, e124. [Google Scholar]
- Gelb, B.D.; Willner, J.P.; Dunn, T.M.; Kardon, N.B.; Verloes, A.; Poncin, J.; Desnick, R.J. Paternal uniparental disomy for chromosome 1 revealed by molecular analysis of a patient with pycnodysostosis. Am. J. Hum. Genet. 1998, 62, 848–854. [Google Scholar] [CrossRef] [Green Version]
- Afifi, H.H.; El-Ruby, M.O.; El-Bassyouni, H.T.; Ismail, S.I.; Aglan, M.S.; El-Harouni, A.A.; Mazen, I.M.; Zaki, M.S.; Bassiouni, R.I.; Hosny, L.A.; et al. The most encountered groups of genetic disorders in Giza Governorate, Egypt. Bratisl Lek Listy 2010, 111, 62–69. [Google Scholar] [PubMed]
- Doherty, M.A.; Langdahl, B.L.; Vogel, I.; Haagerup, A. Clinical and genetic evaluation of Danish patients with pycnodysostosis. Eur. J. Med. Genet. 2021, 64, 104135. [Google Scholar] [CrossRef] [PubMed]
- Pangrazio, A.; Puddu, A.; Oppo, M.; Valentini, M.; Zammataro, L.; Vellodi, A.; Gener, B.; Llano-Rivas, I.; Raza, J.; Atta, I.; et al. Exome sequencing identifies CTSK mutations in patients originally diagnosed as intermediate osteopetrosis. Bone 2014, 59, 122–126. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Sambandam, B. A case of pycnodysostosis presented with pathological femoral shaft fracture. Indian. J. Med. Res. 2014, 139, 180–181. [Google Scholar]
- Song, H.K.; Sohn, Y.B.; Choi, Y.J.; Chung, Y.-S.; Jang, J.-H. A case report of pycnodysostosis with atypical femur fracture diagnosed by next-generation sequencing of candidate genes. Medicine 2017, 96, e6367. [Google Scholar] [CrossRef]
- Roberts, T.; Stephen, L.; Beighton, P. Cleidocranial dysplasia: A review of the dental, historical, and practical implications with an overview of the South African experience. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2013, 115, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Jiang, T.; Liu, F.; Wang, W.-G.; Jiang, X.; Wen, X.; Hu, K.-J.; Xue, Y. Distribution of Cathepsin K in Late Stage of Tooth Germ Development and Its Function in Degrading Enamel Matrix Proteins in Mouse. PLoS ONE 2017, 12, e0169857. [Google Scholar]
- Stroup, G.B.; Kumar, S.; Jerome, C.P. Treatment with a potent cathepsin K inhibitor preserves cortical and trabecular bone mass in ovariectomized monkeys. Calcif. Tissue Int. 2009, 85, 344–355. [Google Scholar] [CrossRef]
- Cusick, T.; Chen, C.M.; Pennypacker, B.L.; Pickarski, M.; Kimmel, D.B.; Scott, B.B.; Duong, L.T. Odanacatib treatment increases hip bone mass and cortical thickness by preserving endocortical bone formation and stimulating periosteal bone formation in the ovariectomized adult rhesus monkey. J. Bone Miner. Res. 2012, 27, 524–537. [Google Scholar] [CrossRef]
- Langdahl, B.; Binkley, N.; Bone, H.; Gilchrist, N.; Resch, H.; Rodriguez Portales, J.; Denker, A.; Lombardi, A.; Le Bailly De Tilleghem, C.; Dasilva, C.; et al. Odanacatib in the treatment of postmenopausal women with low bone mineral density: Five years of continued therapy in a phase 2 study. J. Bone Miner. Res. 2012, 27, 2251–2258. [Google Scholar] [CrossRef]
- Bone, H.G.; Dempster, D.W.; Eisman, J.A.; Greenspan, S.L.; McClung, M.R.; Nakamura, T.; Papapoulos, S.; Shih, W.J.; Rybak-Feiglin, A.; Santora, A.C.; et al. Odanacatib for the treatment of postmenopausal osteoporosis: Development history and design and participant characteristics of LOFT, the Long-Term Odanacatib Fracture Trial. Osteoporos Int. 2015, 26, 699–712. [Google Scholar] [CrossRef] [Green Version]
- Cabal, A.; Williams, D.S.; Jayakar, R.Y.; Zhang, J.; Sardesai, S.; Duong, L.T. Long-term treatment with odanacatib maintains normal trabecular biomechanical properties in ovariectomized adult monkeys as demonstrated by micro-CT-based finite element analysis. Bone Rep. 2017, 6, 26–33. [Google Scholar] [CrossRef]
- Pirapaharan, D.C.; Søe, K.; Panwar, P.; Madsen, J.S.; Bergmann, M.L.; Overgaard, M.; Brömme, D.; Delaisse, J.M. A Mild Inhibition of Cathepsin K Paradoxically Stimulates the Resorptive Activity of Osteoclasts in Culture. Calcif. Tissue Int. 2019, 104, 92–101. [Google Scholar] [CrossRef]
- Mujawar, Q.; Naganoor, R.; Patil, H.; Thobbi, A.N.; Ukkali, S.; Malagi, N. Pycnodysostosis with unusual findings: A case report. Cases J. 2009, 2, 6544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramaiah, K.K.K.; George, G.B.; Padiyath, S.; Sethuraman, R.; Cherian, B. Pyknodysostosis: Report of a rare case with review of literature. Imaging Sci. Dent. 2011, 41, 177. [Google Scholar] [CrossRef] [Green Version]
- Hou, W.S.; Brömme, D.; Zhao, Y.; Mehler, E.; Dushey, C.; Weinstein, H.; Miranda, C.S.; Fraga, C.; Greig, F.; Carey, J.; et al. Characterization of novel cathepsin K mutations in the pro and mature polypeptide regions causing pycnodysostosis. J. Clin. Investig. 1999, 103, 731–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osimani, S.; Husson, I.; Passemard, S.; Elmaleh, M.; Perrin, L.; Quelin, C.; Marey, I.; Delalande, O.; Filocamo, M.; Verloes, A. Craniosynostosis: A rare complication of pycnodysostosis. Eur. J. Med. Genet. 2010, 53, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Haagerup, A.; Hertz, J.M.; Christensen, M.F.; Binderup, H.; Kruse, T.A. Cathepsin K gene mutations and 1q21 haplotypes in at patients with pycnodysostosis in an outbred population. Eur. J. Hum. Genet. 2000, 8, 431–436. [Google Scholar] [CrossRef] [Green Version]
- Araujo, T.F.; Ribeiro, E.M.; Arruda, A.P.; Moreno, C.A.; de Medeiros, P.F.V.; Minillo, R.M.; Melo, D.G.; Kim, C.A.; Doriqui, M.J.R.; Felix, T.M.; et al. Molecular analysis of the CTSK gene in a cohort of 33 Brazilian families with pycnodysostosis from a cluster in a Brazilian Northeast region. Eur. J. Med. Res. 2016, 21, 33. [Google Scholar] [CrossRef] [Green Version]
- Bae, J.S.; Kim, N.; Lee, C.; Kim, S.C.; Lee, H.R.; Song, H.R.; Park, K.B.; Kim, H.W.; Lee, S.H.; Kim, H.Y.; et al. Comprehensive genetic exploration of skeletal dysplasia using targeted exome sequencing. Genet. Med. 2016, 18, 563–569. [Google Scholar] [CrossRef] [Green Version]
- Sivaraman, J.; Lalumière, M.; Ménard, R.; Cygler, M. Crystal structure of wild-type human procathepsin K. Protein Sci. 1999, 8, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Nakata, K.; Yasui, N.; Matsui, Y.; Kataoka, E.; Hiroshima, K.; Shiba, R.; Ochi, T. Novel Mutations of the Cathepsin K Gene in Patients with Pycnodysostosis and Their Characterization. J. Clin. Endocrinol. Metab. 2000, 85, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Fratzl-Zelman, N.; Valenta, A.; Roschger, P.; Nader, A.; Gelb, B.D.; Fratzl, P.; Klaushofer, K. Decreased bone turnover and deterioration of bone structure in two cases of pycnodysostosis. J. Clin. Endocrinol. Metab. 2004, 89, 1538–1547. [Google Scholar] [CrossRef] [PubMed]



| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age in months | 9 | 15 | 9 | 7 | 6 | 10 | 6 | 8 | 6 | 7 | 9 | 17 | 39 | 7 | 6 | 4 | NA | 5 | 7 | 4 | 5 | |
| Sex | F | M | F | M | F | F | F | M | M | F | F | F | F | F | M | F | M | M | F | M | M | F |
| Consanguinity | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Short stature | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | NA | + | + | + | + |
| Frontal Bossing | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | NA | + | + | + | + |
| Open Fontanelle | + | + | + | + | -ve | + | + | + | + | + | -ve | -ve | -ve | -ve | + | + | + | NA | + | + | + | + |
| Brachydactyly | + | + | + | + | + | + | -ve | + | + | + | + | + | + | + | + | + | + | NA | + | + | + | NA |
| Micrognathia | + | + | + | + | - | + | + | + | -ve | + | + | + | + | + | + | + | + | NA | + | + | + | NA |
| Prominent nose | + | + | + | + | - | + | -ve | + | -ve | + | + | + | + | + | + | + | + | NA | + | + | + | NA |
| Dystrophic Flat Nails | + | + | + | + | - | + | + | -ve | -ve | + | + | + | + | + | + | + | + | NA | + | + | + | NA |
| Straight mandibular angle | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | NA | + | + | + | NA |
| Dental Caries | + | -ve | + | + | + | + | + | + | + | -ve | + | -ve | -ve | -ve | -ve | -ve | + | NA | + | + | + | + |
| Open Bite | + | -ve | + | + | + | + | -ve | + | + | + | + | + | + | + | + | + | + | NA | NA | NA | NA | NA |
| Hypodontia | + | -ve | -ve | -ve | + | + | -ve | + | + | + | + | -ve | -ve | -ve | -ve | + | -ve | NA | + | + | + | + |
| Previous Fractures | - | -ve | -ve | -ve | + | -ve | -ve | -ve | -ve | -ve | -ve | + (1) | (3X) | -ve | -ve | -ve | (9X) | NA | + (1) | -ve | -ve | + |
| Pancytopenia /BM depression | + | -ve | -ve | -ve | - | -ve | + | -ve | -ve | NA | NA | NA | NA | NA | NA | NA | NA | NA | -ve | -ve | -ve | NA |
| Sleep Apnea | -ve | -ve | + | -ve | -ve | -ve | -ve | NA | NA | NA | NA | NA | NA | NA | NA | NA | + | + | + | + | ||
| NO. of exon | 7 | 7 | 7 | 6 | 6 | 6 | 6 | 7 | 5 | 7 | 7 | 3 | 5 | 5 | 5 | 3 | 3 | 7 | NA | NA | NA | 7 |
| Location of CTSK cDNA | c.890G>A | c.890G>A | c.761_763delCCT | c.761_763delCCT | c.761_763delcct | c.761_763delCCT | c.436G>C | c.864_865delAA | c.509G>T | c.890G>A | c.890G>A | c.164A>C | c.436G>C | c.436G>C | c.433G>A | c.164A>C | c.164A>C | c.890G>A | NA | NA | NA | c.830C>T c.830C>T |
| Amino Acid Change | p.Ser297Asn | p.Ser297Asn | p.Ser255 | p.Ser255 | p.Ser255 | p.Ser255 | p.Gly146Arg | p.Asn289Glnfs6 | p.Cys170phe | p.5er297Asn | p.5er297Asn | p.Lys55Thr | p.Gly146R | p.G146Arg | p.Val145Meth | P.Lys55Thr | p.Lys55Thr | P.Ser297Asn | NA | NA | NA | p.Ala277Val |
| Mutation type | Missense | Missense | Inframe deletion | Inframe deletion | Inframe deletion | Inframe deletion | Missense | Frameshif | Missense | Missense | Missense | Missense | Missense | Missense | Missense | Missense | Missense | Missense | NA | NA | NA | Missense |
| F | R | |
|---|---|---|
| Exon 2 | CCAGCATCCTATCTAAACACAGG | GTCTCAGCCTTCCTGCCATG |
| Exon 3 | GATTGTGAGTTTCCTTTATTCTCC | GCATCAGCAGGGAACTAAAG |
| Exon 4 | GCTTTAGTTCCCTGCTGATGC | GGAAAAGGTCATGCCAGATTAC |
| Exon 5 | CACATGGAATTTCTTCAGGC | CATCATGCTGGGGAAGGAG |
| Exon 6 | GCTGCCTCTGTTAGTTCACTG | GACAGTGCTGTATAGGATCAGC |
| Exon 7 | GCTGATCCTATACAGCACTGTC | GAAAGGAATATCGGGAAGCTG |
| Exon 8 | GTGTACCATCAGTACCTCGCAC | CTCAGTATCACCACATCTGCTTC |
| cDNA Change | Protein Change | Mutation on Type | SIFT | Polyphen2 | Mutation Taster | ACMG-AMP | PhD-SNP | Mutation Assessor | PROVEAN | SNPs&GO | REVEL | MutPred |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.509G>T | p.Cys170phe | Missense | Damaging (0) | Probably Damaging (1) | Disease causing (1) | 3(PM2, PP3) | Disease (0.936, RI = 9) | High (4.69) | Deleterious (−10.223) | Disease (0.873, RI = 7) | Damaging effect (0.973) | 0.88 Damaging (0.887) Gain of catalytic residue at C170 (p = 0.0112) |
| c.761_763 delcct | p.Ser255 | Inframe deletion | NA | NA | Disease Ccusing | 5(PVS1, PM2, PP3 | NA | NA | NA | NA | NA | NA |
| c.864_865delAA | p.Asn289Glnfs6 | Deletion | NA | NA | Disease causing | 5(PVS1, PM2, PP3 | NA | NA | NA | NA | NA | NA |
| Patient | Exon | Domain | cDNA Change | Protein Change | Mutation Type | References |
|---|---|---|---|---|---|---|
| 1 | 7 | Mature domain | c.890G>A | p.Ser297Asn | Missense | Donnarumma et al. 2007 [19] |
| 2 | 7 | Mature domain | c.890G>A | p.Ser297Asn | Missense | Donnarumma et al. 2007 [19] |
| 3 | 6 | Mature domain | c.761_763delCCT | p.Ser255 | In-frame deletion | This study |
| 4 | 6 | Mature domain | c.761_763delCCT | p.Ser255 | In-frame deletion | This study |
| 5 | 6 | Mature domain | c.761_763delCCT | p.Ser255 | In-frame deletion | This study |
| 6 | 6 | Mature domain | c.761_763delCCT | p.Ser255 | In-frame deletion | This study |
| 7 | 5 | Mature domain | c.436G>C | p.Gly146Arg | Missense | Gelb et al. 1998 [21] |
| 8 | 7 | Mature domain | c.864_865delAA | p.Asn289Glnfs6 | Frameshift | This study |
| 9 | 6 | Mature domain | c.509G>T | p.Cys170phe | Missense | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sayed Amr, K.; El-Bassyouni, H.T.; Abdel Hady, S.; Mostafa, M.I.; Mehrez, M.I.; Coviello, D.; El-Kamah, G.Y. Genetic and Molecular Evaluation: Reporting Three Novel Mutations and Creating Awareness of Pycnodysostosis Disease. Genes 2021, 12, 1552. https://doi.org/10.3390/genes12101552
Sayed Amr K, El-Bassyouni HT, Abdel Hady S, Mostafa MI, Mehrez MI, Coviello D, El-Kamah GY. Genetic and Molecular Evaluation: Reporting Three Novel Mutations and Creating Awareness of Pycnodysostosis Disease. Genes. 2021; 12(10):1552. https://doi.org/10.3390/genes12101552
Chicago/Turabian StyleSayed Amr, Khalda, Hala T. El-Bassyouni, Sawsan Abdel Hady, Mostafa I. Mostafa, Mennat I. Mehrez, Domenico Coviello, and Ghada Y. El-Kamah. 2021. "Genetic and Molecular Evaluation: Reporting Three Novel Mutations and Creating Awareness of Pycnodysostosis Disease" Genes 12, no. 10: 1552. https://doi.org/10.3390/genes12101552
APA StyleSayed Amr, K., El-Bassyouni, H. T., Abdel Hady, S., Mostafa, M. I., Mehrez, M. I., Coviello, D., & El-Kamah, G. Y. (2021). Genetic and Molecular Evaluation: Reporting Three Novel Mutations and Creating Awareness of Pycnodysostosis Disease. Genes, 12(10), 1552. https://doi.org/10.3390/genes12101552






