Temporal Transcriptome Analysis Reveals Dynamic Expression Profiles of Gametes and Embryonic Development in Japanese Flounder (Paralichthys olivaceus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
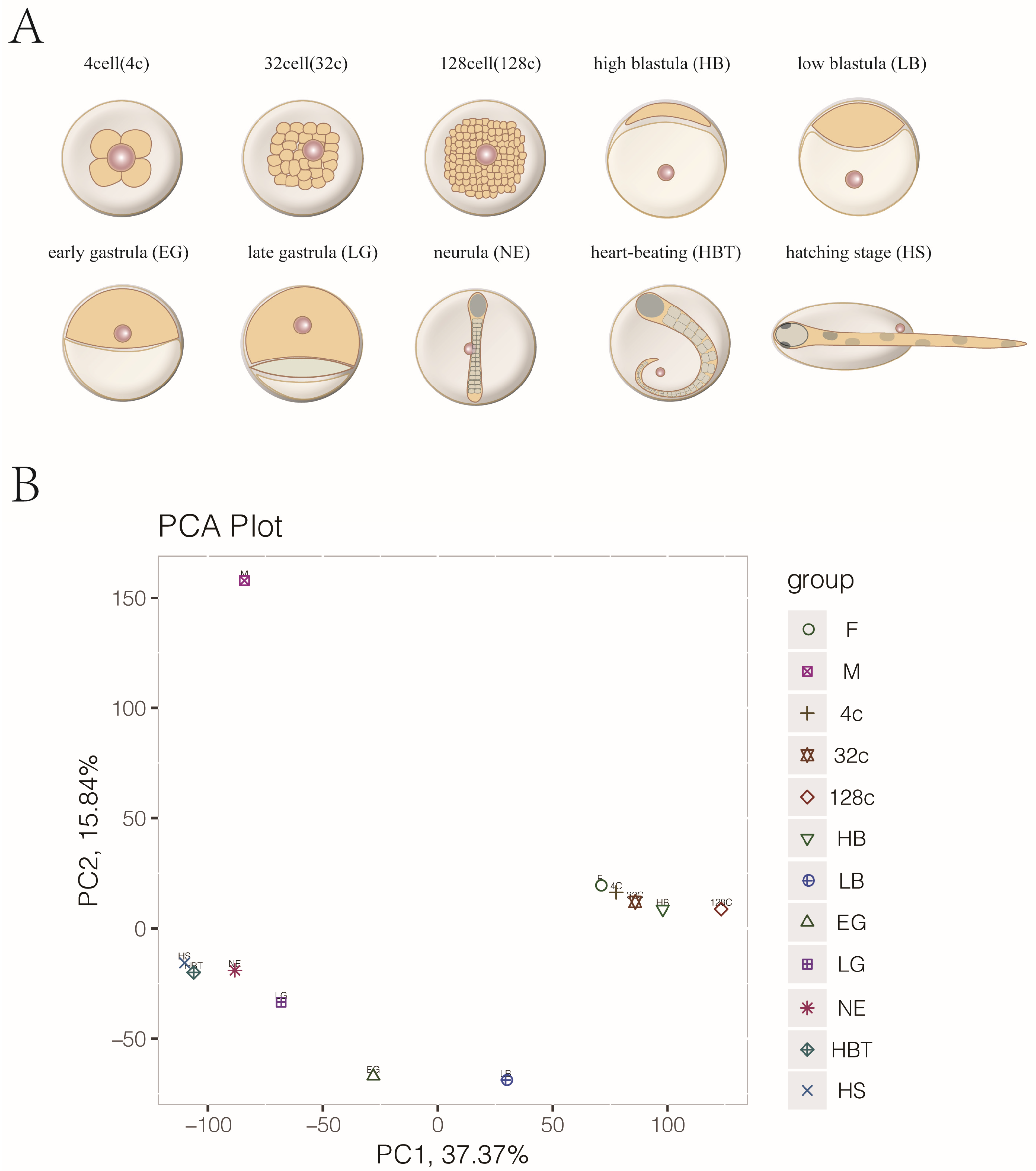
2.2. Embryo Preparation and Sample Collection
2.3. RNA-seq Library Preparation
2.4. Data Filtering and Genome Mapping
2.5. Gene Expression and Transcriptome-Wide Time-Series Analyses and Identification of Differential Expression Genes (DEGs)
2.6. Identification of the Maternal and Zygotic Genes
3. Results
3.1. Transcriptome Sequencing, Cluster Analysis of Gametes and Embryo Development in Japanese Flounder
3.2. The Performance of Gene Expression during Embryo Development
3.3. Pattern of Maternal mRNA products Degradation in Japanese Flounder Embryo
3.4. Identification of Stage of ZGA and Its Association with Expression Change
3.5. The Roles of Wnt Signaling Pathway in Embryo Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clift, D.; Schuh, M. Restarting Life: Fertilization and the Transition from Meiosis to Mitosis. Nat. Rev. Mol. Cell Biol. 2013, 14, 549–562. [Google Scholar] [CrossRef]
- Jukam, D.; Shariati, S.A.M.; Skotheim, J.M. Zygotic Genome Activation in Vertebrates. Dev. Cell 2017, 42, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Tadros, W.; Lipshitz, H.D. The Maternal-to-Zygotic Transition: A Play in Two Acts. Development 2009, 136, 3033–3042. [Google Scholar] [CrossRef] [Green Version]
- Laue, K.; Rajshekar, S.; Courtney, A.J.; Lewis, Z.A.; Goll, M.G. The Maternal to Zygotic Transition Regulates Genome-Wide Heterochromatin Establishment in the Zebrafish Embryo. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Schulz, K.N.; Harrison, M.M. Mechanisms Regulating Zygotic Genome Activation. Nat. Rev. Genet. 2019, 20, 221–234. [Google Scholar] [CrossRef]
- Alizadeh, Z.; Kageyama, S.-I.; Aoki, F. Degradation of Maternal Mrna in Mouse Embryos: Selective Degradation of Specific Mrnas after Fertilization. Mol. Reprod. Dev. Inc. Gamete Res. 2005, 72, 281–290. [Google Scholar] [CrossRef]
- Goddard, M.J.; Pratt, H.P.M. Control of Events During Early Cleavage of the Mouse Embryo: An Analysis of the ‘2-Cell Block’. Development 1983, 73, 111–133. [Google Scholar] [CrossRef]
- Liu, B.; Xu, Q.; Wang, Q.; Feng, S.; Lai, F.; Wang, P.; Zheng, F.; Xiang, Y.; Wu, J.; Nie, J.; et al. The Landscape of RNA Pol Ii Binding Reveals a Stepwise Transition During Zga. Nature 2020, 587, 139–144. [Google Scholar] [CrossRef]
- Tadros, W.; Houston, S.; Bashirullah, A.; Cooperstock, R.L.; Semotok, J.L.; Reed, B.H.; Lipshitz, H.D. Regulation of Maternal Transcript Destabilization During Egg Activation in Drosophila. Genetics 2003, 164, 989–1001. [Google Scholar] [CrossRef]
- Baroux, C.; Autran, D.; Gillmor, C.; Grimanelli, D.; Grossniklaus, U. The Maternal to Zygotic Transition in Animals and Plants. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 89–100. [Google Scholar] [CrossRef]
- Lee, M.T.; Bonneau, A.R.; Takacs, C.M.; Bazzini, A.A.; DiVito, K.R.; Fleming, E.S.; Giraldez, A.J. Nanog, Pou5f1 and Soxb1 Activate Zygotic Gene Expression During the Maternal-to-Zygotic Transition. Nature 2013, 503, 360–364. [Google Scholar] [CrossRef] [Green Version]
- Aanes, H.; Winata, C.L.; Lin, C.H.; Chen, J.P.; Srinivasan, K.G.; Lee, S.G.P.; Lim, A.Y.M.; Hajan, H.S.; Collas, P.; Bourque, G. Zebrafish Mrna Sequencing Deciphers Novelties in Transcriptome Dynamics During Maternal to Zygotic Transition. Genome Res. 2011, 21, 1328–1338. [Google Scholar] [CrossRef] [Green Version]
- Heyn, P.; Kircher, M.; Dahl, A.; Kelso, J.; Tomancak, P.; Kalinka, A.T.; Neugebauer, K.M. The Earliest Transcribed Zygotic Genes Are Short, Newly Evolved, and Different across Species. Cell Rep. 2014, 6, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferg, M.; Sanges, R.; Gehrig, J.; Kiss, J.; Bauer, M.; Lovas, A.; Szabo, M.; Yang, L.; Straehle, U.; Pankratz, M.J. The Tata-Binding Protein Regulates Maternal Mrna Degradation and Differential Zygotic Transcription in Zebrafish. EMBO J. 2007, 26, 3945–3956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikegami, R.; Hunter, P.; Yager, T.D. Developmental Activation of the Capability to Undergo Checkpoint-Induced Apoptosis in the Early Zebrafish Embryo. Dev. Biol. 1999, 209, 409–433. [Google Scholar] [CrossRef] [Green Version]
- Schier, A.F.; Talbot, W.S. Molecular Genetics of Axis Formation in Zebrafish. Annu. Rev. Genet. 2005, 39, 561–613. [Google Scholar] [CrossRef] [Green Version]
- Satija, R.; Farrell, J.; Gennert, D.; Schier, A.F.; Regev, A. Spatial Reconstruction of Single-Cell Gene Expression Data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Mathavan, S.; Lee, S.G.P.; Mak, A.; Miller, L.; Murthy, K.R.K.; Govindarajan, K.R.; Tong, Y.; Wu, Y.L.; Lam, S.H.; Yang, H.; et al. Transcriptome Analysis of Zebrafish Embryogenesis Using Microarrays. PLoS Genet. 2005, 1, e29. [Google Scholar] [CrossRef] [Green Version]
- Gentsch, G.E.; Owens, N.D.; Smith, J.C. The Spatiotemporal Control of Zygotic Genome Activation. Iscience 2019, 16, 485–498. [Google Scholar] [CrossRef]
- Bellipanni, G.; Varga, M.; Maegawa, S.; Imai, Y.; Kelly, C.; Myers, A.P.; Chu, F.; Talbot, W.S.; Weinberg, E.S. Essential and Opposing Roles of Zebrafish Β-Catenins in the Formation of Dorsal Axial Structures and Neurectoderm. Development 2006, 133, 1299–1309. [Google Scholar] [CrossRef] [Green Version]
- Melby, A.E.; Beacha, C.; Mullinsb, M.; Kimelman, D. Patterning the Early Zebrafish by the Opposing Actions of Bozozok and Vox/Vent. Dev. Biol. 2000, 224, 275–285. [Google Scholar] [CrossRef] [Green Version]
- Kelly, C.; Chin, A.; Leatherman, J.; Kozlowski, D.; Weinberg, E. Maternally Controlled (beta)-Catenin-Mediated Signaling Is Required for Organizer Formation in the Zebrafish. Development 2000, 127, 3899–3911. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ploper, D.; Sosa, E.; Colozza, G.; Moriyama, Y.; Benitez, M.D.J.; Zhang, K.; Merkurjev, D.; De Robertis, E.M. Spemann Organizer Transcriptome Induction by Early beta-Catenin, Wnt, Nodal, and Siamois Signals in Xenopus Laevis. Proc. Natl. Acad. Sci. 2017, 114, E3081–E90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, C.; Bao, B.; Xie, Z.; Chen, X.; Li, B.; Jia, X.; Yao, Q.; Orti, G.; Li, W.; Li, X.; et al. The Genome and Transcriptome of Japanese Flounder Provide Insights into Flatfish Asymmetry. Nat. Genet. 2017, 49, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.J.; Wang, C.A.; Zhu, X.C.; Liu, Y.X.; Zhang, X.Y.; Hou, J.L.; Tang, N. Embryonic Development of Gynogenetic Diploid and Triploid Japanese Flounder Paralichthys Olivaceus. J. Dalian Fish. Univ. 2008, 23, 161–167. [Google Scholar]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yuxin, C.; Li, Z.; et al. Soapnuke: A Mapreduce Acceleration-Supported Software for Integrated Quality Control and Preprocessing of High-Throughput Sequencing Data. Gigascience 2017, 7, gix120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with Hisat2 and Hisat-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with Deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Kumar, L.; Futschik, M.E. Mfuzz: A Software Package for Soft Clustering of Microarray Data. Bioinformation 2007, 2, 5. [Google Scholar] [CrossRef]
- Sha, Q.-Q.; Zheng, W.; Wu, Y.-W.; Li, S.; Guo, L.; Zhang, S.; Lin, G.; Ou, X.-H.; Fan, H.-Y. Dynamics and Clinical Relevance of Maternal Mrna Clearance During the Oocyte-to-Embryo Transition in Humans. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Chen, H.; Einstein, L.C.; Little, S.C.; Good, M. Spatiotemporal Patterning of Zygotic Genome Activation in a Model Vertebrate Embryo. Dev. Cell 2019, 49, 852–866.e7. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.H.; Au, K.F.; Yablonovitch, A.L.; Wills, A.E.; Chuang, J.; Baker, J.C.; Wong, W.H.; Li, J.B. RNA Sequencing Reveals a Diverse and Dynamic Repertoire of the Xenopus Tropicalis Transcriptome over Development. Genome Res. 2013, 23, 201–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, R.J.; Collins, J.E.; Sealy, I.M.; Wali, N.; Dooley, C.M.; Digby, Z.; Stemple, D.L.; Murphy, D.N.; Billis, K.; Hourlier, T. A High-Resolution Mrna Expression Time Course of Embryonic Development in Zebrafish. Elife 2017, 6, e30860. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.T.; Bonneau, A.R.; Giraldez, A.J. Zygotic Genome Activation During the Maternal-to-Zygotic Transition. Annu. Rev. Cell Dev. Biol. 2014, 30, 581–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Renzis, S.; Elemento, O.; Tavazoie, S.; Wieschaus, E.F. Unmasking Activation of the Zygotic Genome Using Chromosomal Deletions in the Drosophila Embryo. PLoS Biol. 2007, 5, e117. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Jia, L.; Shi, Z.; Zhang, J.; Li, W. Gene Expression Patterns Regulating Embryogenesis Based on the Integrated De Novo Transcriptome Assembly of the Japanese Flounder. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 22, 58–66. [Google Scholar] [CrossRef]
- Giraldez, A.J.; Mishima, Y.; Rihel, J.; Grocock, R.J.; Van Dongen, S.; Inoue, K.; Enright, A.; Schier, A.F. Zebrafish Mir-430 Promotes Deadenylation and Clearance of Maternal Mrnas. Science 2006, 312, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.; Ji, S.-Y.; Dang, Y.-J.; Sha, Q.-Q.; Yuan, Y.-F.; Zhou, J.-J.; Yan, L.-Y.; Qiao, J.; Tang, F.; Fan, H.-Y. Oocyte-Expressed Yes-Associated Protein Is a Key Activator of the Early Zygotic Genome in Mouse. Cell Res. 2016, 26, 275–287. [Google Scholar] [CrossRef] [Green Version]
- Hirasawa, R.; Chiba, H.; Kaneda, M.; Tajima, S.; Li, E.; Jaenisch, R.; Sasaki, H. Maternal and Zygotic Dnmt1 Are Necessary and Sufficient for the Maintenance of DNA Methylation Imprints During Preimplantation Development. Genes Dev. 2008, 22, 1607–1616. [Google Scholar] [CrossRef] [Green Version]
- Dooley, C.M.; Wali, N.; Sealy, I.M.; White, R.J.; Stemple, D.L.; Collins, J.E.; Busch-Nentwich, E.M. The Gene Regulatory Basis of Genetic Compensation During Neural Crest Induction. PLoS Genet. 2019, 15, e1008213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miles, L.B.; Darido, C.; Kaslin, J.; Heath, J.K.; Jane, S.M.; Dworkin, S. Mis-Expression of Grainyhead-Like Transcription Factors in Zebrafish Leads to Defects in Enveloping Layer (Evl) Integrity, Cellular Morphogenesis and Axial Extension. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dougan, S.T.; Warga, R.M.; Kane, D.A.; Schier, A.F.; Talbot, W.S. The Role of the Zebrafish Nodal-Related Genes Squint and Cyclops in Patterning of Mesendoderm. Development 2003, 130, 1837–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larabell, C.A.; Torres, M.A.; Rowning, B.A.; Yost, C.; Miller, J.R.; Wu, M.; Kimelman, D.; Moon, R. Establishment of the Dorso-Ventral Axis in Xenopus Embryos Is Presaged by Early Asymmetries in Β-Catenin That Are Modulated by the Wnt Signaling Pathway. J. Cell Biol. 1997, 136, 1123–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumanas, S.; Strege, P.; Heasman, J.; Ekker, S.C. The Putative Wnt Receptor Xenopus Frizzled-7 Functions Upstream of beta-Catenin in Vertebrate Dorsoventral Mesoderm Patterning. Development 2000, 127, 1981–1990. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.-W.; Tadjuidje, E.; Tao, Q.; Wylie, C.; Heasman, J. Wnt5a and Wnt11 Interact in a Maternal Dkk1-Regulated Fashion to Activate Both Canonical and Non-Canonical Signaling in Xenopus Axis Formation. Development 2008, 135, 3719–3729. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, X.; Wang, Q.; Hou, J.; Liu, K.; Feng, B.; Shao, C. Temporal Transcriptome Analysis Reveals Dynamic Expression Profiles of Gametes and Embryonic Development in Japanese Flounder (Paralichthys olivaceus). Genes 2021, 12, 1561. https://doi.org/10.3390/genes12101561
Hao X, Wang Q, Hou J, Liu K, Feng B, Shao C. Temporal Transcriptome Analysis Reveals Dynamic Expression Profiles of Gametes and Embryonic Development in Japanese Flounder (Paralichthys olivaceus). Genes. 2021; 12(10):1561. https://doi.org/10.3390/genes12101561
Chicago/Turabian StyleHao, Xiancai, Qian Wang, Jilun Hou, Kaiqiang Liu, Bo Feng, and Changwei Shao. 2021. "Temporal Transcriptome Analysis Reveals Dynamic Expression Profiles of Gametes and Embryonic Development in Japanese Flounder (Paralichthys olivaceus)" Genes 12, no. 10: 1561. https://doi.org/10.3390/genes12101561
APA StyleHao, X., Wang, Q., Hou, J., Liu, K., Feng, B., & Shao, C. (2021). Temporal Transcriptome Analysis Reveals Dynamic Expression Profiles of Gametes and Embryonic Development in Japanese Flounder (Paralichthys olivaceus). Genes, 12(10), 1561. https://doi.org/10.3390/genes12101561





