Factors Regulating the Activity of LINE1 Retrotransposons
Abstract
:1. Introduction
2. L1 Structure
3. Retrotransposition Mechanism
4. L1 Evolution
4.1. LINE Evolution in Deuterostomes and Non-Mammals
4.2. LINE Evolution in Mammals
4.3. LINE Evolution in Primates
4.4. LINE Evolution in Ancient and Modern Humans
4.5. LINE Evolution and Host Regulation
5. Regulation of L1 Activity
5.1. Regulation of L1 in the Early Stages of Embryogenesis
5.2. Regulation of L1 in Germ Cells in Mice
5.3. Other Factors Controlling L1 in the Embryonic Development of Mice
5.4. Features of L1 Regulation during Human Embryonic Development
5.5. L1 Regulation in Somatic Cells
5.5.1. L1 Regulation by Decreasing Chromatin Availability in Somatic Cells
5.5.2. Post-Transcriptional L1 Regulation in Somatic Cells
5.5.3. RNA Interference and miRNA Factors
5.5.4. Antiviral Factors
Interferon-Induced Factors Inhibiting the Formation of L1 RNP
Deaminases
5.5.5. L1 Integration into the Genome and DNA Repair Mechanism
5.5.6. Regulation of L1 by Cell Cycle Factors
5.5.7. Positive Regulation of L1
6. Factors Affecting Changes in L1 Regulation in Neuropsychiatric Diseases
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [Green Version]
- Mills, R.E.; Bennett, E.A.; Iskow, R.C.; Devine, S.E. Which Transposable Elements Are Active in the Human Genome? Trends Genet. 2007, 23, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Sassaman, D.M.; Dombroski, B.A.; Moran, J.V.; Kimberland, M.L.; Naas, T.P.; DeBerardinis, R.J.; Gabriel, A.; Swergold, G.D.; Kazazian, H.H. Many Human L1 Elements Are Capable of Retrotransposition. Nat. Genet. 1997, 16, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H. Hot L1s Account for the Bulk of Retrotransposition in the Human Population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef] [Green Version]
- Penzkofer, T.; Jäger, M.; Figlerowicz, M.; Badge, R.; Mundlos, S.; Robinson, P.N.; Zemojtel, T. L1Base 2: More Retrotransposition-Active LINE-1s, More Mammalian Genomes. Nucleic Acids Res. 2017, 45, D68–D73. [Google Scholar] [CrossRef]
- Dewannieux, M.; Esnault, C.; Heidmann, T. LINE-Mediated Retrotransposition of Marked Alu Sequences. Nat. Genet. 2003, 35, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Ostertag, E.M.; Goodier, J.L.; Zhang, Y.; Kazazian, H.H. SVA Elements Are Nonautonomous Retrotransposons That Cause Disease in Humans. Am. J. Hum. Genet. 2003, 73, 1444–1451. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Xing, J.; Grover, D.; Hedges, D.J.; Han, K.; Walker, J.A.; Batzer, M.A. SVA Elements: A Hominid-Specific Retroposon Family. J. Mol. Biol. 2005, 354, 994–1007. [Google Scholar] [CrossRef]
- Xing, J.; Zhang, Y.; Han, K.; Salem, A.H.; Sen, S.K.; Huff, C.D.; Zhou, Q.; Kirkness, E.F.; Levy, S.; Batzer, M.A.; et al. Mobile Elements Create Structural Variation: Analysis of a Complete Human Genome. Genome Res. 2009, 19, 1516–1526. [Google Scholar] [CrossRef] [Green Version]
- Mandal, P.K.; Ewing, A.D.; Hancks, D.C.; Kazazian, H.H. Enrichment of Processed Pseudogene Transcripts in L1-Ribonucleoprotein Particles. Hum. Mol. Genet. 2013, 22, 3730–3748. [Google Scholar] [CrossRef] [Green Version]
- Pavlícek, A.; Paces, J.; Zíka, R.; Hejnar, J. Length Distribution of Long Interspersed Nucleotide Elements (LINEs) and Processed Pseudogenes of Human Endogenous Retroviruses: Implications for Retrotransposition and Pseudogene Detection. Gene 2002, 300, 189–194. [Google Scholar] [CrossRef]
- Scott, A.F.; Schmeckpeper, B.J.; Abdelrazik, M.; Comey, C.T.; O’Hara, B.; Rossiter, J.P.; Cooley, T.; Heath, P.; Smith, K.D.; Margolet, L. Origin of the Human L1 Elements: Proposed Progenitor Genes Deduced from a Consensus DNA Sequence. Genomics 1987, 1, 113–125. [Google Scholar] [CrossRef]
- Mätlik, K.; Redik, K.; Speek, M. L1 Antisense Promoter Drives Tissue-Specific Transcription of Human Genes. J. Biomed. Biotechnol. 2006, 2006, 71753. [Google Scholar] [CrossRef] [Green Version]
- Swergold, G.D. Identification, Characterization, and Cell Specificity of a Human LINE-1 Promoter. Mol. Cell. Biol. 1990, 10, 6718–6729. [Google Scholar] [CrossRef]
- Moran, J.V.; Holmes, S.E.; Naas, T.P.; DeBerardinis, R.J.; Boeke, J.D.; Kazazian, H.H. High Frequency Retrotransposition in Cultured Mammalian Cells. Cell 1996, 87, 917–927. [Google Scholar] [CrossRef] [Green Version]
- Kolosha, V.O.; Martin, S.L. In Vitro Properties of the First ORF Protein from Mouse LINE-1 Support Its Role in Ribonucleoprotein Particle Formation during Retrotransposition. Proc. Natl. Acad. Sci. USA 1997, 94, 10155–10160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naufer, M.N.; Furano, A.V.; Williams, M.C. Protein-Nucleic Acid Interactions of LINE-1 ORF1p. Semin. Cell Dev. Biol. 2019, 86, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.T.; Sokolowski, M.; Roy-Engel, A.M.; Smither, M.E.; Belancio, V.P. Identification of Charged Amino Acids Required for Nuclear Localization of Human L1 ORF1 Protein. Mob. DNA 2019, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Mathias, S.L.; Scott, A.F.; Kazazian, H.H.; Boeke, J.D.; Gabriel, A. Reverse Transcriptase Encoded by a Human Transposable Element. Science 1991, 254, 1808–1810. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Moran, J.V.; Kazazian, H.H.; Boeke, J.D. Human L1 Retrotransposon Encodes a Conserved Endonuclease Required for Retrotransposition. Cell 1996, 87, 905–916. [Google Scholar] [CrossRef] [Green Version]
- Macia, A.; Muñoz-Lopez, M.; Cortes, J.L.; Hastings, R.K.; Morell, S.; Lucena-Aguilar, G.; Marchal, J.A.; Badge, R.M.; Garcia-Perez, J.L. Epigenetic Control of Retrotransposon Expression in Human Embryonic Stem Cells. Mol. Cell. Biol. 2011, 31, 300–316. [Google Scholar] [CrossRef] [Green Version]
- Wheelan, S.J.; Aizawa, Y.; Han, J.S.; Boeke, J.D. Gene-Breaking: A New Paradigm for Human Retrotransposon-Mediated Gene Evolution. Genome Res. 2005, 15, 1073–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denli, A.M.; Narvaiza, I.; Kerman, B.E.; Pena, M.; Benner, C.; Marchetto, M.C.N.; Diedrich, J.K.; Aslanian, A.; Ma, J.; Moresco, J.J.; et al. Primate-Specific ORF0 Contributes to Retrotransposon-Mediated Diversity. Cell 2015, 163, 583–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adney, E.M.; Ochmann, M.T.; Sil, S.; Truong, D.M.; Mita, P.; Wang, X.; Kahler, D.J.; Fenyö, D.; Holt, L.J.; Boeke, J.D. Comprehensive Scanning Mutagenesis of Human Retrotransposon LINE-1 Identifies Motifs Essential for Function. Genetics 2019, 213, 1401–1414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, D.D.; Korman, M.H.; Jakubczak, J.L.; Eickbush, T.H. Reverse Transcription of R2Bm RNA Is Primed by a Nick at the Chromosomal Target Site: A Mechanism for Non-LTR Retrotransposition. Cell 1993, 72, 595–605. [Google Scholar] [CrossRef]
- Cost, G.J.; Feng, Q.; Jacquier, A.; Boeke, J.D. Human L1 Element Target-Primed Reverse Transcription in Vitro. EMBO J. 2002, 21, 5899–5910. [Google Scholar] [CrossRef] [PubMed]
- Athanikar, J.N.; Badge, R.M.; Moran, J.V. A YY1-Binding Site Is Required for Accurate Human LINE-1 Transcription Initiation. Nucleic Acids Res. 2004, 32, 3846–3855. [Google Scholar] [CrossRef] [Green Version]
- Dai, L.; Taylor, M.S.; O’Donnell, K.A.; Boeke, J.D. Poly(A) Binding Protein C1 Is Essential for Efficient L1 Retrotransposition and Affects L1 RNP Formation. Mol. Cell. Biol. 2012, 32, 4323–4336. [Google Scholar] [CrossRef] [Green Version]
- Doucet, A.J.; Hulme, A.E.; Sahinovic, E.; Kulpa, D.A.; Moldovan, J.B.; Kopera, H.C.; Athanikar, J.N.; Hasnaoui, M.; Bucheton, A.; Moran, J.V.; et al. Characterization of LINE-1 Ribonucleoprotein Particles. PLoS Genet. 2010, 6, e1001150. [Google Scholar] [CrossRef] [Green Version]
- Horn, A.V.; Celic, I.; Dong, C.; Martirosyan, I.; Han, J.S. A Conserved Role for the ESCRT Membrane Budding Complex in LINE Retrotransposition. PLoS Genet. 2017, 13, e1006837. [Google Scholar] [CrossRef] [Green Version]
- Mita, P.; Wudzinska, A.; Sun, X.; Andrade, J.; Nayak, S.; Kahler, D.J.; Badri, S.; LaCava, J.; Ueberheide, B.; Yun, C.Y.; et al. LINE-1 Protein Localization and Functional Dynamics during the Cell Cycle. eLife 2018, 7, e30058. [Google Scholar] [CrossRef]
- Macia, A.; Widmann, T.J.; Heras, S.R.; Ayllon, V.; Sanchez, L.; Benkaddour-Boumzaouad, M.; Muñoz-Lopez, M.; Rubio, A.; Amador-Cubero, S.; Blanco-Jimenez, E.; et al. Engineered LINE-1 Retrotransposition in Nondividing Human Neurons. Genome Res. 2017, 27, 335–348. [Google Scholar] [CrossRef] [Green Version]
- Cost, G.J.; Boeke, J.D. Targeting of Human Retrotransposon Integration Is Directed by the Specificity of the L1 Endonuclease for Regions of Unusual DNA Structure. Biochemistry 1998, 37, 18081–18093. [Google Scholar] [CrossRef]
- Flasch, D.A.; Macia, Á.; Sánchez, L.; Ljungman, M.; Heras, S.R.; García-Pérez, J.L.; Wilson, T.E.; Moran, J.V. Genome-Wide de Novo L1 Retrotransposition Connects Endonuclease Activity with Replication. Cell 2019, 177, 837–851.e28. [Google Scholar] [CrossRef] [PubMed]
- Sultana, T.; van Essen, D.; Siol, O.; Bailly-Bechet, M.; Philippe, C.; Zine El Aabidine, A.; Pioger, L.; Nigumann, P.; Saccani, S.; Andrau, J.-C.; et al. The Landscape of L1 Retrotransposons in the Human Genome Is Shaped by Pre-Insertion Sequence Biases and Post-Insertion Selection. Mol. Cell 2019, 74, 555–570.e7. [Google Scholar] [CrossRef] [PubMed]
- Doucet, A.J.; Wilusz, J.E.; Miyoshi, T.; Liu, Y.; Moran, J.V. A 3′ Poly(A) Tract Is Required for LINE-1 Retrotransposition. Mol. Cell 2015, 60, 728–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulpa, D.A.; Moran, J.V. Cis-Preferential LINE-1 Reverse Transcriptase Activity in Ribonucleoprotein Particles. Nat. Struct. Mol. Biol. 2006, 13, 655–660. [Google Scholar] [CrossRef]
- Monot, C.; Kuciak, M.; Viollet, S.; Mir, A.A.; Gabus, C.; Darlix, J.-L.; Cristofari, G. The Specificity and Flexibility of L1 Reverse Transcription Priming at Imperfect T-Tracts. PLoS Genet. 2013, 9, e1003499. [Google Scholar] [CrossRef] [Green Version]
- Gibson, B.A.; Kraus, W.L. New Insights into the Molecular and Cellular Functions of Poly(ADP-Ribose) and PARPs. Nat. Rev. Mol. Cell. Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef]
- Miyoshi, T.; Makino, T.; Moran, J.V. Poly(ADP-Ribose) Polymerase 2 Recruits Replication Protein A to Sites of LINE-1 Integration to Facilitate Retrotransposition. Mol. Cell 2019, 75, 1286–1298.e12. [Google Scholar] [CrossRef]
- Chen, R.; Wold, M.S. Replication Protein A: Single-Stranded DNA’s First Responder: Dynamic DNA-Interactions Allow Replication Protein A to Direct Single-Strand DNA Intermediates into Different Pathways for Synthesis or Repair. Bioessays 2014, 36, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.S.; Altukhov, I.; Molloy, K.R.; Mita, P.; Jiang, H.; Adney, E.M.; Wudzinska, A.; Badri, S.; Ischenko, D.; Eng, G.; et al. Dissection of Affinity Captured LINE-1 Macromolecular Complexes. eLife 2018, 7, e30094. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.S.; LaCava, J.; Mita, P.; Molloy, K.R.; Huang, C.R.L.; Li, D.; Adney, E.M.; Jiang, H.; Burns, K.H.; Chait, B.T.; et al. Affinity Proteomics Reveals Human Host Factors Implicated in Discrete Stages of LINE-1 Retrotransposition. Cell 2013, 155, 1034–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregersen, L.H.; Schueler, M.; Munschauer, M.; Mastrobuoni, G.; Chen, W.; Kempa, S.; Dieterich, C.; Landthaler, M. MOV10 Is a 5′ to 3′ RNA Helicase Contributing to UPF1 MRNA Target Degradation by Translocation along 3′ UTRs. Mol. Cell 2014, 54, 573–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Marchetto, M.C.N.; Muotri, A.R.; Mu, Y.; Carson, C.T.; Macia, A.; Moran, J.V.; Gage, F.H. Ataxia Telangiectasia Mutated (ATM) Modulates Long Interspersed Element-1 (L1) Retrotransposition in Human Neural Stem Cells. Proc. Natl. Acad. Sci. USA 2011, 108, 20382–20387. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, J.; Yamaguchi, K.; Kajikawa, M.; Ichiyanagi, K.; Adachi, N.; Koyama, H.; Takeda, S.; Okada, N. Genetic Evidence That the Non-Homologous End-Joining Repair Pathway Is Involved in LINE Retrotransposition. PLoS Genet. 2009, 5, e1000461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zingler, N.; Willhoeft, U.; Brose, H.-P.; Schoder, V.; Jahns, T.; Hanschmann, K.-M.O.; Morrish, T.A.; Löwer, J.; Schumann, G.G. Analysis of 5′ Junctions of Human LINE-1 and Alu Retrotransposons Suggests an Alternative Model for 5′-End Attachment Requiring Microhomology-Mediated End-Joining. Genome Res. 2005, 15, 780–789. [Google Scholar] [CrossRef] [Green Version]
- Kopera, H.C.; Moldovan, J.B.; Morrish, T.A.; Garcia-Perez, J.L.; Moran, J.V. Similarities between Long Interspersed Element-1 (LINE-1) Reverse Transcriptase and Telomerase. Proc. Natl. Acad. Sci. USA 2011, 108, 20345–20350. [Google Scholar] [CrossRef] [Green Version]
- Morrish, T.A.; Garcia-Perez, J.L.; Stamato, T.D.; Taccioli, G.E.; Sekiguchi, J.; Moran, J.V. Endonuclease-Independent LINE-1 Retrotransposition at Mammalian Telomeres. Nature 2007, 446, 208–212. [Google Scholar] [CrossRef]
- Morrish, T.A.; Gilbert, N.; Myers, J.S.; Vincent, B.J.; Stamato, T.D.; Taccioli, G.E.; Batzer, M.A.; Moran, J.V. DNA Repair Mediated by Endonuclease-Independent LINE-1 Retrotransposition. Nat. Genet. 2002, 31, 159–165. [Google Scholar] [CrossRef]
- Ohshima, K. Parallel Relaxation of Stringent RNA Recognition in Plant and Mammalian L1 Retrotransposons. Mol. Biol. Evol. 2012, 29, 3255–3259. [Google Scholar] [CrossRef] [Green Version]
- Ivancevic, A.M.; Kortschak, R.D.; Bertozzi, T.; Adelson, D.L. LINEs between Species: Evolutionary Dynamics of LINE-1 Retrotransposons across the Eukaryotic Tree of Life. Genome Biol. Evol. 2016, 8, 3301–3322. [Google Scholar] [CrossRef] [Green Version]
- Kordis, D.; Lovsin, N.; Gubensek, F. Phylogenomic Analysis of the L1 Retrotransposons in Deuterostomia. Syst. Biol. 2006, 55, 886–901. [Google Scholar] [CrossRef] [Green Version]
- Chalopin, D.; Naville, M.; Plard, F.; Galiana, D.; Volff, J.-N. Comparative Analysis of Transposable Elements Highlights Mobilome Diversity and Evolution in Vertebrates. Genome Biol. Evol. 2015, 7, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Sotero-Caio, C.G.; Platt, R.N.; Suh, A.; Ray, D.A. Evolution and Diversity of Transposable Elements in Vertebrate Genomes. Genome Biol. Evol. 2017, 9, 161–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, F.; Han, M.; Peng, Z. Evolution and Diversity of Transposable Elements in Fish Genomes. Sci. Rep. 2019, 9, 15399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metcalfe, C.J.; Filée, J.; Germon, I.; Joss, J.; Casane, D. Evolution of the Australian Lungfish (Neoceratodus Forsteri) Genome: A Major Role for CR1 and L2 LINE Elements. Mol. Biol. Evol. 2012, 29, 3529–3539. [Google Scholar] [CrossRef] [Green Version]
- Nikaido, M.; Noguchi, H.; Nishihara, H.; Toyoda, A.; Suzuki, Y.; Kajitani, R.; Suzuki, H.; Okuno, M.; Aibara, M.; Ngatunga, B.P.; et al. Coelacanth Genomes Reveal Signatures for Evolutionary Transition from Water to Land. Genome Res. 2013, 23, 1740–1748. [Google Scholar] [CrossRef] [Green Version]
- Luchetti, A.; Plazzi, F.; Mantovani, B. Evolution of Two Short Interspersed Elements in Callorhinchus milii (Chondrichthyes, Holocephali) and Related Elements in Sharks and the Coelacanth. Genome Biol. Evol. 2017, 9, 1406–1417. [Google Scholar] [CrossRef] [Green Version]
- Meyer, A.; Schloissnig, S.; Franchini, P.; Du, K.; Woltering, J.M.; Irisarri, I.; Wong, W.Y.; Nowoshilow, S.; Kneitz, S.; Kawaguchi, A.; et al. Giant Lungfish Genome Elucidates the Conquest of Land by Vertebrates. Nature 2021, 590, 284–289. [Google Scholar] [CrossRef]
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant Shark Genome Provides Unique Insights into Gnathostome Evolution. Nature 2014, 505, 174–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.J.; Kuraku, S.; Holt, C.; Sauka-Spengler, T.; Jiang, N.; Campbell, M.S.; Yandell, M.D.; Manousaki, T.; Meyer, A.; Bloom, O.E.; et al. Sequencing of the Sea Lamprey (Petromyzon Marinus) Genome Provides Insights into Vertebrate Evolution. Nat. Genet. 2013, 45, 415–421. [Google Scholar] [CrossRef]
- Alföldi, J.; Di Palma, F.; Grabherr, M.; Williams, C.; Kong, L.; Mauceli, E.; Russell, P.; Lowe, C.B.; Glor, R.E.; Jaffe, J.D.; et al. The Genome of the Green Anole Lizard and a Comparative Analysis with Birds and Mammals. Nature 2011, 477, 587–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boissinot, S.; Sookdeo, A. The Evolution of LINE-1 in Vertebrates. Genome Biol. Evol. 2016, 8, 3485–3507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suh, A.; Churakov, G.; Ramakodi, M.P.; Platt, R.N.; Jurka, J.; Kojima, K.K.; Caballero, J.; Smit, A.F.; Vliet, K.A.; Hoffmann, F.G.; et al. Multiple Lineages of Ancient CR1 Retroposons Shaped the Early Genome Evolution of Amniotes. Genome Biol. Evol. 2014, 7, 205–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasquesi, G.I.M.; Adams, R.H.; Card, D.C.; Schield, D.R.; Corbin, A.B.; Perry, B.W.; Reyes-Velasco, J.; Ruggiero, R.P.; Vandewege, M.W.; Shortt, J.A.; et al. Squamate Reptiles Challenge Paradigms of Genomic Repeat Element Evolution Set by Birds and Mammals. Nat. Commun. 2018, 9, 2774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boissinot, S.; Bourgeois, Y.; Manthey, J.D.; Ruggiero, R.P. The Mobilome of Reptiles: Evolution, Structure, and Function. Cytogenet. Genome Res. 2019, 157, 21–33. [Google Scholar] [CrossRef]
- Gemmell, N.J.; Rutherford, K.; Prost, S.; Tollis, M.; Winter, D.; Macey, J.R.; Adelson, D.L.; Suh, A.; Bertozzi, T.; Grau, J.H.; et al. The Tuatara Genome Reveals Ancient Features of Amniote Evolution. Nature 2020, 584, 403–409. [Google Scholar] [CrossRef]
- Shedlock, A.M. Phylogenomic Investigation of CR1 LINE Diversity in Reptiles. Syst. Biol. 2006, 55, 902–911. [Google Scholar] [CrossRef] [Green Version]
- Suh, A. The Specific Requirements for CR1 Retrotransposition Explain the Scarcity of Retrogenes in Birds. J. Mol. Evol. 2015, 81, 18–20. [Google Scholar] [CrossRef]
- Kapusta, A.; Suh, A. Evolution of Bird Genomes-a Transposon’s-Eye View. Ann. N. Y. Acad. Sci. 2017, 1389, 164–185. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.F.; Tóth, G.; Riggs, A.D.; Jurka, J. Ancestral, Mammalian-Wide Subfamilies of LINE-1 Repetitive Sequences. J. Mol. Biol. 1995, 246, 401–417. [Google Scholar] [CrossRef] [PubMed]
- Warren, W.C.; Hillier, L.W.; Marshall Graves, J.A.; Birney, E.; Ponting, C.P.; Grützner, F.; Belov, K.; Miller, W.; Clarke, L.; Chinwalla, A.T.; et al. Genome Analysis of the Platypus Reveals Unique Signatures of Evolution. Nature 2008, 453, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, T.S.; Wakefield, M.J.; Aken, B.; Amemiya, C.T.; Chang, J.L.; Duke, S.; Garber, M.; Gentles, A.J.; Goodstadt, L.; Heger, A.; et al. Genome of the Marsupial Monodelphis Domestica Reveals Innovation in Non-Coding Sequences. Nature 2007, 447, 167–177. [Google Scholar] [CrossRef] [Green Version]
- Polychronopoulos, D.; King, J.W.D.; Nash, A.J.; Tan, G.; Lenhard, B. Conserved Non-Coding Elements: Developmental Gene Regulation Meets Genome Organization. Nucleic Acids Res. 2017, 45, 12611–12624. [Google Scholar] [CrossRef]
- Wichman, H.A.; Scott, L.; Howell, E.K.; Martinez, A.R.; Yang, L.; Baker, R.J. Flying Around in the Genome: Characterization of LINE-1 in Chiroptera. Spec. Publ. Tex. Tech. Univ. Mus. 2019, 71, 379–392. [Google Scholar]
- Platt, R.N.; Ray, D.A. A Non-LTR Retroelement Extinction in Spermophilus Tridecemlineatus. Gene 2012, 500, 47–53. [Google Scholar] [CrossRef]
- Sookdeo, A.; Hepp, C.M.; Boissinot, S. Contrasted Patterns of Evolution of the LINE-1 Retrotransposon in Perissodactyls: The History of a LINE-1 Extinction. Mob. DNA 2018, 9, 12. [Google Scholar] [CrossRef]
- Yang, L.; Scott, L.; Wichman, H.A. Tracing the History of LINE and SINE Extinction in Sigmodontine Rodents. Mob. DNA 2019, 10, 22. [Google Scholar] [CrossRef] [Green Version]
- Blumenstiel, J.P. Birth, School, Work, Death, and Resurrection: The Life Stages and Dynamics of Transposable Element Proliferation. Genes 2019, 10, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouse Genome Sequencing Consortium; Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; et al. Initial Sequencing and Comparative Analysis of the Mouse Genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef]
- Gibbs, R.A.; Weinstock, G.M.; Metzker, M.L.; Muzny, D.M.; Sodergren, E.J.; Scherer, S.; Scott, G.; Steffen, D.; Worley, K.C.; Burch, P.E.; et al. Genome Sequence of the Brown Norway Rat Yields Insights into Mammalian Evolution. Nature 2004, 428, 493–521. [Google Scholar] [CrossRef] [Green Version]
- Glazko, G.V.; Nei, M. Estimation of Divergence Times for Major Lineages of Primate Species. Mol. Biol. Evol. 2003, 20, 424–434. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, H.J.; Ho, S.Y.W.; Barnes, I.; Groves, C. Estimating the Phylogeny and Divergence Times of Primates Using a Supermatrix Approach. BMC Evol. Biol. 2009, 9, 259. [Google Scholar] [CrossRef] [Green Version]
- Konkel, M.K.; Walker, J.A.; Batzer, M.A. LINEs and SINEs of Primate Evolution. Evol. Anthropol. 2010, 19, 236–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Liang, P. Comparative Genomics Analysis Reveals High Levels of Differential Retrotransposition among Primates from the Hominidae and the Cercopithecidae Families. Genome Biol. Evol. 2019, 11, 3309–3325. [Google Scholar] [CrossRef] [Green Version]
- Boissinot, S.; Roos, C.; Furano, A.V. Different Rates of LINE-1 (L1) Retrotransposon Amplification and Evolution in New World Monkeys. J. Mol. Evol. 2004, 58, 122–130. [Google Scholar] [CrossRef]
- Sookdeo, A.; Ruiz-García, M.; Schneider, H.; Boissinot, S. Contrasting Rates of LINE-1 Amplification among New World Primates of the Atelidae Family. Cytogenet. Genome Res. 2018, 154, 217–228. [Google Scholar] [CrossRef]
- Han, K.; Konkel, M.K.; Xing, J.; Wang, H.; Lee, J.; Meyer, T.J.; Huang, C.T.; Sandifer, E.; Hebert, K.; Barnes, E.W.; et al. Mobile DNA in Old World Monkeys: A Glimpse through the Rhesus Macaque Genome. Science 2007, 316, 238–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Tang, W.; Liang, P.; Han, K. A Comprehensive Analysis of Chimpanzee (Pan Troglodytes)-Specific LINE-1 Retrotransposons. Gene 2019, 693, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.D.; Zamudio-Hurtado, A.; Clawson, H.; Kent, W.J.; Haussler, D.; Salama, S.R.; Haeussler, M. The UCSC Repeat Browser Allows Discovery and Visualization of Evolutionary Conflict across Repeat Families. Mob. DNA 2020, 11, 13. [Google Scholar] [CrossRef] [Green Version]
- Castellano, D.; Munch, K. Population Genomics in the Great Apes. Methods Mol. Biol. 2020, 2090, 453–463. [Google Scholar] [CrossRef] [Green Version]
- Jeon, S.; Kim, S.; Oh, M.H.; Liang, P.; Tang, W.; Han, K. A Comprehensive Analysis of Gorilla-Specific LINE-1 Retrotransposons. Genes Genom. 2021, 43, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cordaux, R.; Han, K.; Wang, J.; Hedges, D.J.; Liang, P.; Batzer, M.A. Different Evolutionary Fates of Recently Integrated Human and Chimpanzee LINE-1 Retrotransposons. Gene 2007, 390, 18–27. [Google Scholar] [CrossRef] [Green Version]
- Beck, C.R.; Collier, P.; Macfarlane, C.; Malig, M.; Kidd, J.M.; Eichler, E.E.; Badge, R.M.; Moran, J.V. LINE-1 Retrotransposition Activity in Human Genomes. Cell 2010, 141, 1159–1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordaux, R.; Batzer, M.A. The Impact of Retrotransposons on Human Genome Evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, S.R.; Doucet, A.J.; Kopera, H.C.; Moldovan, J.B.; Garcia-Perez, J.L.; Moran, J.V. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiol. Spectr. 2015, 3, MDNA3-0061-2014. [Google Scholar] [CrossRef] [Green Version]
- Gardner, E.J.; Lam, V.K.; Harris, D.N.; Chuang, N.T.; Scott, E.C.; Pittard, W.S.; Mills, R.E.; 1000 Genomes Project Consortium; Devine, S.E. The Mobile Element Locator Tool (MELT): Population-Scale Mobile Element Discovery and Biology. Genome Res. 2017, 27, 1916–1929. [Google Scholar] [CrossRef] [Green Version]
- Guichard, E.; Peona, V.; Malagoli Tagliazucchi, G.; Abitante, L.; Jagoda, E.; Musella, M.; Ricci, M.; Rubio-Roldán, A.; Sarno, S.; Luiselli, D.; et al. Impact of Non-LTR Retrotransposons in the Differentiation and Evolution of Anatomically Modern Humans. Mob. DNA 2018, 9, 28. [Google Scholar] [CrossRef] [Green Version]
- Rishishwar, L.; Tellez Villa, C.E.; Jordan, I.K. Transposable Element Polymorphisms Recapitulate Human Evolution. Mob. DNA 2015, 6, 21. [Google Scholar] [CrossRef] [Green Version]
- Watkins, W.S.; Feusier, J.E.; Thomas, J.; Goubert, C.; Mallick, S.; Jorde, L.B. The Simons Genome Diversity Project: A Global Analysis of Mobile Element Diversity. Genome Biol. Evol. 2020, 12, 779–794. [Google Scholar] [CrossRef]
- Ito, J.; Gifford, R.J.; Sato, K. Retroviruses Drive the Rapid Evolution of Mammalian APOBEC3 Genes. Proc. Natl. Acad. Sci. USA 2020, 117, 610–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uriu, K.; Kosugi, Y.; Ito, J.; Sato, K. The Battle between Retroviruses and APOBEC3 Genes: Its Past and Present. Viruses 2021, 13, 124. [Google Scholar] [CrossRef] [PubMed]
- Hayward, J.A.; Tachedjian, M.; Cui, J.; Cheng, A.Z.; Johnson, A.; Baker, M.L.; Harris, R.S.; Wang, L.-F.; Tachedjian, G. Differential Evolution of Antiretroviral Restriction Factors in Pteropid Bats as Revealed by APOBEC3 Gene Complexity. Mol. Biol. Evol. 2018, 35, 1626–1637. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Emerman, M.; Malik, H.S.; McLaughlin, R.N. Retrocopying Expands the Functional Repertoire of APOBEC3 Antiviral Proteins in Primates. eLife 2020, 9, e58436. [Google Scholar] [CrossRef]
- Uriu, K.; Kosugi, Y.; Suzuki, N.; Ito, J.; Sato, K. Elucidation of the Complicated Scenario of Primate APOBEC3 Gene Evolution. J. Virol. 2021, 95, e00144-21. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Dudley, J.P. APOBECs and Virus Restriction. Virology 2015, 479–480, 131–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parhad, S.S.; Theurkauf, W.E. Rapid Evolution and Conserved Function of the PiRNA Pathway. Open Biol. 2019, 9, 180181. [Google Scholar] [CrossRef] [Green Version]
- Castro-Diaz, N.; Ecco, G.; Coluccio, A.; Kapopoulou, A.; Yazdanpanah, B.; Friedli, M.; Duc, J.; Jang, S.M.; Turelli, P.; Trono, D. Evolutionally Dynamic L1 Regulation in Embryonic Stem Cells. Genes Dev. 2014, 28, 1397–1409. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Luque, F.J.; Kempen, M.-J.H.C.; Gerdes, P.; Vargas-Landin, D.B.; Richardson, S.R.; Troskie, R.-L.; Jesuadian, J.S.; Cheetham, S.W.; Carreira, P.E.; Salvador-Palomeque, C.; et al. LINE-1 Evasion of Epigenetic Repression in Humans. Mol. Cell 2019, 75, 590–604.e12. [Google Scholar] [CrossRef]
- Douse, C.H.; Tchasovnikarova, I.A.; Timms, R.T.; Protasio, A.V.; Seczynska, M.; Prigozhin, D.M.; Albecka, A.; Wagstaff, J.; Williamson, J.C.; Freund, S.M.V.; et al. TASOR Is a Pseudo-PARP That Directs HUSH Complex Assembly and Epigenetic Transposon Control. Nat. Commun. 2020, 11, 4940. [Google Scholar] [CrossRef] [PubMed]
- Marchetto, M.C.N.; Narvaiza, I.; Denli, A.M.; Benner, C.; Lazzarini, T.A.; Nathanson, J.L.; Paquola, A.C.M.; Desai, K.N.; Herai, R.H.; Weitzman, M.D.; et al. Differential L1 Regulation in Pluripotent Stem Cells of Humans and Apes. Nature 2013, 503, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Muotri, A.R.; Chu, V.T.; Marchetto, M.C.N.; Deng, W.; Moran, J.V.; Gage, F.H. Somatic Mosaicism in Neuronal Precursor Cells Mediated by L1 Retrotransposition. Nature 2005, 435, 903–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, J.; Karnik, R.; Gu, H.; Ziller, M.J.; Clement, K.; Tsankov, A.M.; Akopian, V.; Gifford, C.A.; Donaghey, J.; Galonska, C.; et al. Targeted Disruption of DNMT1, DNMT3A and DNMT3B in Human Embryonic Stem Cells. Nat. Genet. 2015, 47, 469–478. [Google Scholar] [CrossRef] [Green Version]
- Hatanaka, Y.; Inoue, K.; Oikawa, M.; Kamimura, S.; Ogonuki, N.; Kodama, E.N.; Ohkawa, Y.; Tsukada, Y.; Ogura, A. Histone Chaperone CAF-1 Mediates Repressive Histone Modifications to Protect Preimplantation Mouse Embryos from Endogenous Retrotransposons. Proc. Natl. Acad. Sci. USA 2015, 112, 14641–14646. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Fu, X.; Zhang, M.; He, F.; Li, W.; Abdul, M.M.; Zhou, J.; Sun, L.; Chang, C.; Li, Y.; et al. Transposable Elements Are Regulated by Context-Specific Patterns of Chromatin Marks in Mouse Embryonic Stem Cells. Nat. Commun. 2019, 10, 34. [Google Scholar] [CrossRef]
- Silverman, R.H. Viral Encounters with 2′,5′-Oligoadenylate Synthetase and RNase L during the Interferon Antiviral Response. J. Virol. 2007, 81, 12720–12729. [Google Scholar] [CrossRef] [Green Version]
- Yao, Q.; Cao, G.; Li, M.; Wu, B.; Zhang, X.; Zhang, T.; Guo, J.; Yin, H.; Shi, L.; Chen, J.; et al. Ribonuclease Activity of MARF1 Controls Oocyte RNA Homeostasis and Genome Integrity in Mice. Proc. Natl. Acad. Sci. USA 2018, 115, 11250–11255. [Google Scholar] [CrossRef] [Green Version]
- Orecchini, E.; Frassinelli, L.; Galardi, S.; Ciafrè, S.A.; Michienzi, A. Post-Transcriptional Regulation of LINE-1 Retrotransposition by AID/APOBEC and ADAR Deaminases. Chromosome Res. 2018, 26, 45–59. [Google Scholar] [CrossRef]
- Goodier, J.L.; Pereira, G.C.; Cheung, L.E.; Rose, R.J.; Kazazian, H.H. The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition. PLoS Genet. 2015, 11, e1005252. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, A.; Wittmann, S.; Thomas, D.; Shepard, C.N.; Kim, B.; Ferreirós, N.; Gramberg, T. The SAMHD1-Mediated Block of LINE-1 Retroelements Is Regulated by Phosphorylation. Mob. DNA 2018, 9, 11. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Byun, H.-M.; Zhong, J.; Motta, V.; Barupal, J.; Zheng, Y.; Dou, C.; Zhang, F.; McCracken, J.P.; Diaz, A.; et al. Effects of Short-Term Exposure to Inhalable Particulate Matter on DNA Methylation of Tandem Repeats. Environ. Mol. Mutagen. 2014, 55, 322–335. [Google Scholar] [CrossRef] [Green Version]
- Gasior, S.L.; Roy-Engel, A.M.; Deininger, P.L. ERCC1/XPF Limits L1 Retrotransposition. DNA Repair. (Amst.) 2008, 7, 983–989. [Google Scholar] [CrossRef] [Green Version]
- Servant, G.; Streva, V.A.; Derbes, R.S.; Wijetunge, M.I.; Neeland, M.; White, T.B.; Belancio, V.P.; Roy-Engel, A.M.; Deininger, P.L. The Nucleotide Excision Repair Pathway Limits L1 Retrotransposition. Genetics 2017, 205, 139–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pizarro, J.G.; Cristofari, G. Post-Transcriptional Control of LINE-1 Retrotransposition by Cellular Host Factors in Somatic Cells. Front. Cell Dev. Biol. 2016, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Mita, P.; Sun, X.; Fenyö, D.; Kahler, D.J.; Li, D.; Agmon, N.; Wudzinska, A.; Keegan, S.; Bader, J.S.; Yun, C.; et al. BRCA1 and S Phase DNA Repair Pathways Restrict LINE-1 Retrotransposition in Human Cells. Nat. Struct. Mol. Biol. 2020, 27, 179–191. [Google Scholar] [CrossRef]
- Aravin, A.A.; Hannon, G.J.; Brennecke, J. The Piwi-PiRNA Pathway Provides an Adaptive Defense in the Transposon Arms Race. Science 2007, 318, 761–764. [Google Scholar] [CrossRef] [Green Version]
- Hancks, D.C.; Kazazian, H.H. Roles for Retrotransposon Insertions in Human Disease. Mob. DNA 2016, 7, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohlrausch, F.B.; Berteli, T.S.; Wang, F.; Navarro, P.A.; Keefe, D.L. Control of LINE-1 Expression Maintains Genome Integrity in Germline and Early Embryo Development. Reprod. Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Günesdogan, U.; Zylicz, J.J.; Hackett, J.A.; Cougot, D.; Bao, S.; Lee, C.; Dietmann, S.; Allen, G.E.; Sengupta, R.; et al. PRMT5 Protects Genomic Integrity during Global DNA Demethylation in Primordial Germ Cells and Preimplantation Embryos. Mol. Cell 2014, 56, 564–579. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Liu, W.; Chen, J.; Liu, S.; Wang, M.; Yang, L.; Chen, C.; Qi, M.; Xu, Y.; Qiao, Z.; et al. Nuclear Exosome Targeting Complex Core Factor Zcchc8 Regulates the Degradation of LINE1 RNA in Early Embryos and Embryonic Stem Cells. Cell Rep. 2019, 29, 2461–2472.e6. [Google Scholar] [CrossRef] [Green Version]
- Lees-Murdock, D.J.; Walsh, C.P. DNA Methylation Reprogramming in the Germ Line. Adv. Exp. Med. Biol. 2008, 626, 1–15. [Google Scholar] [CrossRef]
- Smith, Z.D.; Chan, M.M.; Mikkelsen, T.S.; Gu, H.; Gnirke, A.; Regev, A.; Meissner, A. A Unique Regulatory Phase of DNA Methylation in the Early Mammalian Embryo. Nature 2012, 484, 339–344. [Google Scholar] [CrossRef] [Green Version]
- De Iaco, A.; Coudray, A.; Duc, J.; Trono, D. DPPA2 and DPPA4 Are Necessary to Establish a 2C-like State in Mouse Embryonic Stem Cells. EMBO Rep. 2019, 20, e47382. [Google Scholar] [CrossRef]
- Kazazian, H.H.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370. [Google Scholar] [CrossRef]
- Protasova, M.S.; Gusev, F.E.; Grigorenko, A.P.; Kuznetsova, I.L.; Rogaev, E.I.; Andreeva, T.V. Quantitative Analysis of L1-Retrotransposons in Alzheimer’s Disease and Aging. Biochemistry 2017, 82, 962–971. [Google Scholar] [CrossRef]
- Kano, H.; Godoy, I.; Courtney, C.; Vetter, M.R.; Gerton, G.L.; Ostertag, E.M.; Kazazian, H.H. L1 Retrotransposition Occurs Mainly in Embryogenesis and Creates Somatic Mosaicism. Genes Dev. 2009, 23, 1303–1312. [Google Scholar] [CrossRef] [Green Version]
- Baillie, J.K.; Barnett, M.W.; Upton, K.R.; Gerhardt, D.J.; Richmond, T.A.; De Sapio, F.; Brennan, P.M.; Rizzu, P.; Smith, S.; Fell, M.; et al. Somatic Retrotransposition Alters the Genetic Landscape of the Human Brain. Nature 2011, 479, 534–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Yeo, G.W.; Mu, Y.; Lovci, M.T.; Morell, M.; O’Shea, K.S.; Moran, J.V.; Gage, F.H. L1 Retrotransposition in Human Neural Progenitor Cells. Nature 2009, 460, 1127–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, E.; Bestor, T.H.; Jaenisch, R. Targeted Mutation of the DNA Methyltransferase Gene Results in Embryonic Lethality. Cell 1992, 69, 915–926. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for de Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Gretarsson, K.H.; Hackett, J.A. Dppa2 and Dppa4 Counteract de Novo Methylation to Establish a Permissive Epigenome for Development. Nat. Struct. Mol. Biol. 2020, 27, 706–716. [Google Scholar] [CrossRef]
- Peaston, A.E.; Evsikov, A.V.; Graber, J.H.; de Vries, W.N.; Holbrook, A.E.; Solter, D.; Knowles, B.B. Retrotransposons Regulate Host Genes in Mouse Oocytes and Preimplantation Embryos. Dev. Cell 2004, 7, 597–606. [Google Scholar] [CrossRef]
- Fadloun, A.; Le Gras, S.; Jost, B.; Ziegler-Birling, C.; Takahashi, H.; Gorab, E.; Carninci, P.; Torres-Padilla, M.-E. Chromatin Signatures and Retrotransposon Profiling in Mouse Embryos Reveal Regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013, 20, 332–338. [Google Scholar] [CrossRef]
- Jachowicz, J.W.; Bing, X.; Pontabry, J.; Bošković, A.; Rando, O.J.; Torres-Padilla, M.-E. LINE-1 Activation after Fertilization Regulates Global Chromatin Accessibility in the Early Mouse Embryo. Nat. Genet. 2017, 49, 1502–1510. [Google Scholar] [CrossRef]
- Percharde, M.; Lin, C.-J.; Yin, Y.; Guan, J.; Peixoto, G.A.; Bulut-Karslioglu, A.; Biechele, S.; Huang, B.; Shen, X.; Ramalho-Santos, M. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell 2018, 174, 391–405.e19. [Google Scholar] [CrossRef] [Green Version]
- Bulut-Karslioglu, A.; De La Rosa-Velázquez, I.A.; Ramirez, F.; Barenboim, M.; Onishi-Seebacher, M.; Arand, J.; Galán, C.; Winter, G.E.; Engist, B.; Gerle, B.; et al. Suv39h-Dependent H3K9me3 Marks Intact Retrotransposons and Silences LINE Elements in Mouse Embryonic Stem Cells. Mol. Cell 2014, 55, 277–290. [Google Scholar] [CrossRef] [Green Version]
- Tachibana, M.; Sugimoto, K.; Nozaki, M.; Ueda, J.; Ohta, T.; Ohki, M.; Fukuda, M.; Takeda, N.; Niida, H.; Kato, H.; et al. G9a Histone Methyltransferase Plays a Dominant Role in Euchromatic Histone H3 Lysine 9 Methylation and Is Essential for Early Embryogenesis. Genes Dev. 2002, 16, 1779–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsui, T.; Leung, D.; Miyashita, H.; Maksakova, I.A.; Miyachi, H.; Kimura, H.; Tachibana, M.; Lorincz, M.C.; Shinkai, Y. Proviral Silencing in Embryonic Stem Cells Requires the Histone Methyltransferase ESET. Nature 2010, 464, 927–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi, M.M.; Goyal, P.; Maksakova, I.A.; Bilenky, M.; Leung, D.; Tang, J.X.; Shinkai, Y.; Mager, D.L.; Jones, S.; Hirst, M.; et al. DNA Methylation and SETDB1/H3K9me3 Regulate Predominantly Distinct Sets of Genes, Retroelements, and Chimeric Transcripts in MESCs. Cell Stem Cell 2011, 8, 676–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, A.H.; O’Carroll, D.; Scherthan, H.; Mechtler, K.; Sauer, S.; Schöfer, C.; Weipoltshammer, K.; Pagani, M.; Lachner, M.; Kohlmaier, A.; et al. Loss of the Suv39h Histone Methyltransferases Impairs Mammalian Heterochromatin and Genome Stability. Cell 2001, 107, 323–337. [Google Scholar] [CrossRef] [Green Version]
- Rao, V.K.; Pal, A.; Taneja, R. A Drive in SUVs: From Development to Disease. Epigenetics 2017, 12, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Robbez-Masson, L.; Tie, C.H.C.; Conde, L.; Tunbak, H.; Husovsky, C.; Tchasovnikarova, I.A.; Timms, R.T.; Herrero, J.; Lehner, P.J.; Rowe, H.M. The HUSH Complex Cooperates with TRIM28 to Repress Young Retrotransposons and New Genes. Genome Res. 2018, 28, 836–845. [Google Scholar] [CrossRef] [Green Version]
- Müller, I.; Moroni, A.S.; Shlyueva, D.; Sahadevan, S.; Schoof, E.M.; Radzisheuskaya, A.; Højfeldt, J.W.; Tatar, T.; Koche, R.P.; Huang, C.; et al. MPP8 Is Essential for Sustaining Self-Renewal of Ground-State Pluripotent Stem Cells. Nat. Commun. 2021, 12, 3034. [Google Scholar] [CrossRef]
- Harten, S.K.; Bruxner, T.J.; Bharti, V.; Blewitt, M.; Nguyen, T.-M.-T.; Whitelaw, E.; Epp, T. The First Mouse Mutants of D14Abb1e (Fam208a) Show That It Is Critical for Early Development. Mamm. Genome 2014, 25, 293–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, Q.; Shen, Y.; Wang, Y.; Wang, X.; Francisco, J.C.; Luo, Z.; Lin, C. Striking a Balance: Regulation of Transposable Elements by Zfp281 and Mll2 in Mouse Embryonic Stem Cells. Nucleic Acids Res. 2017, 45, 12301–12310. [Google Scholar] [CrossRef] [Green Version]
- Achwal, C.W.; Iyer, C.A.; Chandra, H.S. Immunochemical Evidence for the Presence of 5mC, 6mA and 7mG in Human, Drosophila and Mealybug DNA. FEBS Lett. 1983, 158, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Ratel, D.; Ravanat, J.-L.; Berger, F.; Wion, D. N6-Methyladenine: The Other Methylated Base of DNA. Bioessays 2006, 28, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA Methylation on N(6)-Adenine in Mammalian Embryonic Stem Cells. Nature 2016, 532, 329–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adhikari, S.; Xiao, W.; Zhao, Y.-L.; Yang, Y.-G. M(6)A: Signaling for MRNA Splicing. RNA Biol. 2016, 13, 756–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Gao, M.; He, J.; Wu, K.; Lin, S.; Jin, L.; Chen, Y.; Liu, H.; Shi, J.; Wang, X.; et al. The RNA M6A Reader YTHDC1 Silences Retrotransposons and Guards ES Cell Identity. Nature 2021, 591, 322–326. [Google Scholar] [CrossRef]
- Chen, C.; Liu, W.; Guo, J.; Liu, Y.; Liu, X.; Liu, J.; Dou, X.; Le, R.; Huang, Y.; Li, C.; et al. Nuclear M6A Reader YTHDC1 Regulates the Scaffold Function of LINE1 RNA in Mouse ESCs and Early Embryos. Protein Cell 2021, 12, 455–474. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xie, W. Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol. 2018, 28, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Matsui, Y.; Okamura, D. Mechanisms of Germ-Cell Specification in Mouse Embryos. Bioessays 2005, 27, 136–143. [Google Scholar] [CrossRef]
- Ginsburg, M.; Snow, M.H.; McLaren, A. Primordial Germ Cells in the Mouse Embryo during Gastrulation. Development 1990, 110, 521–528. [Google Scholar] [CrossRef]
- Sato, M.; Kimura, T.; Kurokawa, K.; Fujita, Y.; Abe, K.; Masuhara, M.; Yasunaga, T.; Ryo, A.; Yamamoto, M.; Nakano, T. Identification of PGC7, a New Gene Expressed Specifically in Preimplantation Embryos and Germ Cells. Mech. Dev. 2002, 113, 91–94. [Google Scholar] [CrossRef]
- Saitou, M.; Barton, S.C.; Surani, M.A. A Molecular Programme for the Specification of Germ Cell Fate in Mice. Nature 2002, 418, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Lou, C.; Goodier, J.L.; Qiang, R. A Potential New Mechanism for Pregnancy Loss: Considering the Role of LINE-1 Retrotransposons in Early Spontaneous Miscarriage. Reprod. Biol. Endocrinol. 2020, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Seisenberger, S.; Andrews, S.; Krueger, F.; Arand, J.; Walter, J.; Santos, F.; Popp, C.; Thienpont, B.; Dean, W.; Reik, W. The Dynamics of Genome-Wide DNA Methylation Reprogramming in Mouse Primordial Germ Cells. Mol. Cell 2012, 48, 849–862. [Google Scholar] [CrossRef] [Green Version]
- Guibert, S.; Forné, T.; Weber, M. Global Profiling of DNA Methylation Erasure in Mouse Primordial Germ Cells. Genome Res. 2012, 22, 633–641. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, H.; Sakurai, T.; Miura, F.; Imai, M.; Mochiduki, K.; Yanagisawa, E.; Sakashita, A.; Wakai, T.; Suzuki, Y.; Ito, T.; et al. High-Resolution DNA Methylome Analysis of Primordial Germ Cells Identifies Gender-Specific Reprogramming in Mice. Genome Res. 2013, 23, 616–627. [Google Scholar] [CrossRef] [Green Version]
- Bourc’his, D.; Bestor, T.H. Meiotic Catastrophe and Retrotransposon Reactivation in Male Germ Cells Lacking Dnmt3L. Nature 2004, 431, 96–99. [Google Scholar] [CrossRef]
- Kato, Y.; Kaneda, M.; Hata, K.; Kumaki, K.; Hisano, M.; Kohara, Y.; Okano, M.; Li, E.; Nozaki, M.; Sasaki, H. Role of the Dnmt3 Family in de Novo Methylation of Imprinted and Repetitive Sequences during Male Germ Cell Development in the Mouse. Hum. Mol. Genet. 2007, 16, 2272–2280. [Google Scholar] [CrossRef]
- Barau, J.; Teissandier, A.; Zamudio, N.; Roy, S.; Nalesso, V.; Hérault, Y.; Guillou, F.; Bourc’his, D. The DNA Methyltransferase DNMT3C Protects Male Germ Cells from Transposon Activity. Science 2016, 354, 909–912. [Google Scholar] [CrossRef]
- Jansz, N. DNA Methylation Dynamics at Transposable Elements in Mammals. Essays Biochem. 2019, 63, 677–689. [Google Scholar] [CrossRef]
- Stewart, K.R.; Veselovska, L.; Kelsey, G. Establishment and Functions of DNA Methylation in the Germline. Epigenomics 2016, 8, 1399–1413. [Google Scholar] [CrossRef] [Green Version]
- Hajkova, P.; Erhardt, S.; Lane, N.; Haaf, T.; El-Maarri, O.; Reik, W.; Walter, J.; Surani, M.A. Epigenetic Reprogramming in Mouse Primordial Germ Cells. Mech. Dev. 2002, 117, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Lees-Murdock, D.J.; De Felici, M.; Walsh, C.P. Methylation Dynamics of Repetitive DNA Elements in the Mouse Germ Cell Lineage. Genomics 2003, 82, 230–237. [Google Scholar] [CrossRef]
- Pezic, D.; Manakov, S.A.; Sachidanandam, R.; Aravin, A.A. PiRNA Pathway Targets Active LINE1 Elements to Establish the Repressive H3K9me3 Mark in Germ Cells. Genes Dev. 2014, 28, 1410–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aravin, A.A.; Sachidanandam, R.; Bourc’his, D.; Schaefer, C.; Pezic, D.; Toth, K.F.; Bestor, T.; Hannon, G.J. A PiRNA Pathway Primed by Individual Transposons Is Linked to de Novo DNA Methylation in Mice. Mol. Cell 2008, 31, 785–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains PiRNA Production in Most Animals. Mol. Cell 2018, 71, 775–790.e5. [Google Scholar] [CrossRef] [Green Version]
- Cox, D.N.; Chao, A.; Baker, J.; Chang, L.; Qiao, D.; Lin, H. A Novel Class of Evolutionarily Conserved Genes Defined by Piwi Are Essential for Stem Cell Self-Renewal. Genes Dev. 1998, 12, 3715–3727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, R.J.; Weiner, M.M.; Lin, H. PIWI Proteins and PIWI-Interacting RNAs in the Soma. Nature 2014, 505, 353–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.W. PIWI Proteins and PiRNAs in the Nervous System. Mol. Cells 2019, 42, 828–835. [Google Scholar] [CrossRef]
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Totoki, Y.; Toyoda, A.; Ikawa, M.; Asada, N.; Kojima, K.; Yamaguchi, Y.; Ijiri, T.W.; et al. DNA Methylation of Retrotransposon Genes Is Regulated by Piwi Family Members MILI and MIWI2 in Murine Fetal Testes. Genes Dev. 2008, 22, 908–917. [Google Scholar] [CrossRef] [Green Version]
- Manakov, S.A.; Pezic, D.; Marinov, G.K.; Pastor, W.A.; Sachidanandam, R.; Aravin, A.A. MIWI2 and MILI Have Differential Effects on PiRNA Biogenesis and DNA Methylation. Cell Rep. 2015, 12, 1234–1243. [Google Scholar] [CrossRef] [Green Version]
- Vagin, V.V.; Wohlschlegel, J.; Qu, J.; Jonsson, Z.; Huang, X.; Chuma, S.; Girard, A.; Sachidanandam, R.; Hannon, G.J.; Aravin, A.A. Proteomic Analysis of Murine Piwi Proteins Reveals a Role for Arginine Methylation in Specifying Interaction with Tudor Family Members. Genes Dev. 2009, 23, 1749–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, Y.; Wilson, A.F.; DeFalco, T.; Meetei, A.R.; Namekawa, S.H.; Pang, Q. FANCD2 Is Required for the Repression of Germline Transposable Elements. Reproduction 2020, 159, 659–668. [Google Scholar] [CrossRef]
- Lacroix, M.; El Messaoudi, S.; Rodier, G.; Le Cam, A.; Sardet, C.; Fabbrizio, E. The Histone-Binding Protein COPR5 Is Required for Nuclear Functions of the Protein Arginine Methyltransferase PRMT5. EMBO Rep. 2008, 9, 452–458. [Google Scholar] [CrossRef] [Green Version]
- Paul, C.; Delpech, H.; Haouzi, D.; Hamamah, S.; Sardet, C.; Fabbrizio, E. Coprs Inactivation Leads to a Derepression of LINE1 Transposons in Spermatocytes. FEBS Open Bio 2019, 9, 159–168. [Google Scholar] [CrossRef]
- Vourekas, A.; Zheng, Q.; Alexiou, P.; Maragkakis, M.; Kirino, Y.; Gregory, B.D.; Mourelatos, Z. Mili and Miwi Target RNA Repertoire Reveals PiRNA Biogenesis and Function of Miwi in Spermiogenesis. Nat. Struct. Mol. Biol. 2012, 19, 773–781. [Google Scholar] [CrossRef] [Green Version]
- De Fazio, S.; Bartonicek, N.; Di Giacomo, M.; Abreu-Goodger, C.; Sankar, A.; Funaya, C.; Antony, C.; Moreira, P.N.; Enright, A.J.; O’Carroll, D. The Endonuclease Activity of Mili Fuels PiRNA Amplification That Silences LINE1 Elements. Nature 2011, 480, 259–263. [Google Scholar] [CrossRef]
- Zheng, K.; Xiol, J.; Reuter, M.; Eckardt, S.; Leu, N.A.; McLaughlin, K.J.; Stark, A.; Sachidanandam, R.; Pillai, R.S.; Wang, P.J. Mouse MOV10L1 Associates with Piwi Proteins and Is an Essential Component of the Piwi-Interacting RNA (PiRNA) Pathway. Proc. Natl. Acad. Sci. USA 2010, 107, 11841–11846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, M.; Chuma, S.; Tanaka, T.; Franz, T.; Stark, A.; Pillai, R.S. Loss of the Mili-Interacting Tudor Domain-Containing Protein-1 Activates Transposons and Alters the Mili-Associated Small RNA Profile. Nat. Struct. Mol. Biol. 2009, 16, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Saxe, J.P.; Chen, M.; Zhao, H.; Lin, H. Tdrkh Is Essential for Spermatogenesis and Participates in Primary PiRNA Biogenesis in the Germline. EMBO J. 2013, 32, 1869–1885. [Google Scholar] [CrossRef]
- Tan, K.; Kim, M.E.; Song, H.-W.; Skarbrevik, D.; Babajanian, E.; Bedrosian, T.A.; Gage, F.H.; Wilkinson, M.F. The Rhox Gene Cluster Suppresses Germline LINE1 Transposition. Proc. Natl. Acad. Sci. USA 2021, 118, e2024785118. [Google Scholar] [CrossRef] [PubMed]
- Ungewitter, E.K.; Rotgers, E.; Kang, H.S.; Lichti-Kaiser, K.; Li, L.; Grimm, S.A.; Jetten, A.M.; Yao, H.H.-C. Loss of Glis3 Causes Dysregulation of Retrotransposon Silencing and Germ Cell Demise in Fetal Mouse Testis. Sci. Rep. 2018, 8, 9662. [Google Scholar] [CrossRef] [PubMed]
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Takamatsu, K.; Chuma, S.; Kojima-Kita, K.; Shiromoto, Y.; Asada, N.; Toyoda, A.; Fujiyama, A.; et al. MVH in PiRNA Processing and Gene Silencing of Retrotransposons. Genes Dev. 2010, 24, 887–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, R.R.; Tokuzawa, Y.; Yang, Z.; Hayashi, E.; Ichisaka, T.; Kajita, S.; Asano, Y.; Kunieda, T.; Sachidanandam, R.; Chuma, S.; et al. Tudor Domain Containing 12 (TDRD12) Is Essential for Secondary PIWI Interacting RNA Biogenesis in Mice. Proc. Natl. Acad. Sci. USA 2013, 110, 16492–16497. [Google Scholar] [CrossRef] [Green Version]
- Xiol, J.; Cora, E.; Koglgruber, R.; Chuma, S.; Subramanian, S.; Hosokawa, M.; Reuter, M.; Yang, Z.; Berninger, P.; Palencia, A.; et al. A Role for Fkbp6 and the Chaperone Machinery in PiRNA Amplification and Transposon Silencing. Mol. Cell 2012, 47, 970–979. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, T.; Watanabe, T.; Kuramochi-Miyagawa, S.; Takemoto, N.; Shiromoto, Y.; Kudo, A.; Kanai-Azuma, M.; Tashiro, F.; Miyazaki, S.; Katanaya, A.; et al. Mouse GTSF1 Is an Essential Factor for Secondary PiRNA Biogenesis. EMBO Rep. 2018, 19, e42054. [Google Scholar] [CrossRef]
- Pandey, R.R.; Homolka, D.; Olotu, O.; Sachidanandam, R.; Kotaja, N.; Pillai, R.S. Exonuclease Domain-Containing 1 Enhances MIWI2 PiRNA Biogenesis via Its Interaction with TDRD12. Cell Rep. 2018, 24, 3423–3432.e4. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Chen, K.-M.; Pandey, R.R.; Homolka, D.; Reuter, M.; Janeiro, B.K.R.; Sachidanandam, R.; Fauvarque, M.-O.; McCarthy, A.A.; Pillai, R.S. PIWI Slicing and EXD1 Drive Biogenesis of Nuclear PiRNAs from Cytosolic Targets of the Mouse PiRNA Pathway. Mol. Cell 2016, 61, 138–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Lan, Y.; Pandey, R.R.; Homolka, D.; Berger, S.L.; Pillai, R.S.; Bartolomei, M.S.; Wang, P.J. TEX15 Associates with MILI and Silences Transposable Elements in Male Germ Cells. Genes Dev. 2020, 34, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Pastor, W.A.; Stroud, H.; Nee, K.; Liu, W.; Pezic, D.; Manakov, S.; Lee, S.A.; Moissiard, G.; Zamudio, N.; Bourc’his, D.; et al. MORC1 Represses Transposable Elements in the Mouse Male Germline. Nat. Commun. 2014, 5, 5795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Giacomo, M.; Comazzetto, S.; Sampath, S.C.; Sampath, S.C.; O’Carroll, D. G9a Co-Suppresses LINE1 Elements in Spermatogonia. Epigenetics Chromatin 2014, 7, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-Interacting RNAs: Small RNAs with Big Functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Carroll, D.; Scherthan, H.; Peters, A.H.; Opravil, S.; Haynes, A.R.; Laible, G.; Rea, S.; Schmid, M.; Lebersorger, A.; Jerratsch, M.; et al. Isolation and Characterization of Suv39h2, a Second Histone H3 Methyltransferase Gene That Displays Testis-Specific Expression. Mol. Cell. Biol. 2000, 20, 9423–9433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahadevan, I.A.; Kumar, S.; Rao, M.R.S. Linker Histone Variant H1t Is Closely Associated with Repressed Repeat-Element Chromatin Domains in Pachytene Spermatocytes. Epigenetics Chromatin 2020, 13, 9. [Google Scholar] [CrossRef]
- Mishra, L.N.; Shalini, V.; Gupta, N.; Ghosh, K.; Suthar, N.; Bhaduri, U.; Rao, M.R.S. Spermatid-Specific Linker Histone HILS1 Is a Poor Condenser of DNA and Chromatin and Preferentially Associates with LINE-1 Elements. Epigenetics Chromatin 2018, 11, 43. [Google Scholar] [CrossRef]
- Dong, J.; Wang, X.; Cao, C.; Wen, Y.; Sakashita, A.; Chen, S.; Zhang, J.; Zhang, Y.; Zhou, L.; Luo, M.; et al. UHRF1 Suppresses Retrotransposons and Cooperates with PRMT5 and PIWI Proteins in Male Germ Cells. Nat. Commun. 2019, 10, 4705. [Google Scholar] [CrossRef] [Green Version]
- Bostick, M.; Kim, J.K.; Estève, P.-O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 Plays a Role in Maintaining DNA Methylation in Mammalian Cells. Science 2007, 317, 1760–1764. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Zhang, J.; Chen, R.; Wang, L.; Li, B.; Cheng, H.; Duan, X.; Zhu, H.; Wei, W.; Li, J.; et al. Dissecting the Precise Role of H3K9 Methylation in Crosstalk with DNA Maintenance Methylation in Mammals. Nat. Commun. 2016, 7, 12464. [Google Scholar] [CrossRef] [Green Version]
- Sheng, Y.; Wang, H.; Liu, D.; Zhang, C.; Deng, Y.; Yang, F.; Zhang, T.; Zhang, C. Methylation of Tumor Suppressor Gene CDH13 and SHP1 Promoters and Their Epigenetic Regulation by the UHRF1/PRMT5 Complex in Endometrial Carcinoma. Gynecol. Oncol. 2016, 140, 145–151. [Google Scholar] [CrossRef]
- Li, X.; Zhang, J.; Jia, R.; Cheng, V.; Xu, X.; Qiao, W.; Guo, F.; Liang, C.; Cen, S. The MOV10 Helicase Inhibits LINE-1 Mobility. J. Biol. Chem. 2013, 288, 21148–21160. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Guo, R.; Cui, Y.; Zhu, Z.; Zhang, Y.; Wu, H.; Zheng, B.; Yue, Q.; Bai, S.; Zeng, W.; et al. An Essential Role for PNLDC1 in PiRNA 3′ End Trimming and Male Fertility in Mice. Cell Res. 2017, 27, 1392–1396. [Google Scholar] [CrossRef]
- Ding, D.; Liu, J.; Dong, K.; Midic, U.; Hess, R.A.; Xie, H.; Demireva, E.Y.; Chen, C. PNLDC1 Is Essential for PiRNA 3′ End Trimming and Transposon Silencing during Spermatogenesis in Mice. Nat. Commun. 2017, 8, 819. [Google Scholar] [CrossRef] [Green Version]
- Ding, D.; Liu, J.; Dong, K.; Melnick, A.F.; Latham, K.E.; Chen, C. Mitochondrial Membrane-Based Initial Separation of MIWI and MILI Functions during Pachytene PiRNA Biogenesis. Nucleic Acids Res. 2019, 47, 2594–2608. [Google Scholar] [CrossRef] [PubMed]
- Wheldon, L.M.; Abakir, A.; Ferjentsik, Z.; Dudnakova, T.; Strohbuecker, S.; Christie, D.; Dai, N.; Guan, S.; Foster, J.M.; Corrêa, I.R.; et al. Transient Accumulation of 5-Carboxylcytosine Indicates Involvement of Active Demethylation in Lineage Specification of Neural Stem Cells. Cell Rep. 2014, 7, 1353–1361. [Google Scholar] [CrossRef] [PubMed]
- Blythe, M.J.; Kocer, A.; Rubio-Roldan, A.; Giles, T.; Abakir, A.; Ialy-Radio, C.; Wheldon, L.M.; Bereshchenko, O.; Bruscoli, S.; Kondrashov, A.; et al. LINE-1 Transcription in Round Spermatids Is Associated with Accretion of 5-Carboxylcytosine in Their Open Reading Frames. Commun. Biol. 2021, 4, 691. [Google Scholar] [CrossRef] [PubMed]
- Kuramochi-Miyagawa, S.; Kimura, T.; Ijiri, T.W.; Isobe, T.; Asada, N.; Fujita, Y.; Ikawa, M.; Iwai, N.; Okabe, M.; Deng, W.; et al. Mili, a Mammalian Member of Piwi Family Gene, Is Essential for Spermatogenesis. Development 2004, 131, 839–849. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Lin, H. Miwi, a Murine Homolog of Piwi, Encodes a Cytoplasmic Protein Essential for Spermatogenesis. Dev. Cell 2002, 2, 819–830. [Google Scholar] [CrossRef] [Green Version]
- Carmell, M.A.; Girard, A.; van de Kant, H.J.G.; Bourc’his, D.; Bestor, T.H.; de Rooij, D.G.; Hannon, G.J. MIWI2 Is Essential for Spermatogenesis and Repression of Transposons in the Mouse Male Germline. Dev. Cell 2007, 12, 503–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, P.; Rozhkov, N.V.; Li, F.; Cárdenas, F.L.; Davydenko, O.; Davydenk, O.; Vandivier, L.E.; Gregory, B.D.; Hannon, G.J.; Schultz, R.M. Essential Role for Endogenous SiRNAs during Meiosis in Mouse Oocytes. PLoS Genet. 2015, 11, e1005013. [Google Scholar] [CrossRef] [Green Version]
- Tam, O.H.; Aravin, A.A.; Stein, P.; Girard, A.; Murchison, E.P.; Cheloufi, S.; Hodges, E.; Anger, M.; Sachidanandam, R.; Schultz, R.M.; et al. Pseudogene-Derived Small Interfering RNAs Regulate Gene Expression in Mouse Oocytes. Nature 2008, 453, 534–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taborska, E.; Pasulka, J.; Malik, R.; Horvat, F.; Jenickova, I.; Jelić Matošević, Z.; Svoboda, P. Restricted and Non-Essential Redundancy of RNAi and PiRNA Pathways in Mouse Oocytes. PLoS Genet. 2019, 15, e1008261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.-Q.; Sun, F.; Handel, M.A.; Schimenti, J.C.; Eppig, J.J. Meiosis Arrest Female 1 (MARF1) Has Nuage-like Function in Mammalian Oocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 18653–18660. [Google Scholar] [CrossRef] [Green Version]
- Ollinger, R.; Childs, A.J.; Burgess, H.M.; Speed, R.M.; Lundegaard, P.R.; Reynolds, N.; Gray, N.K.; Cooke, H.J.; Adams, I.R. Deletion of the Pluripotency-Associated Tex19.1 Gene Causes Activation of Endogenous Retroviruses and Defective Spermatogenesis in Mice. PLoS Genet. 2008, 4, e1000199. [Google Scholar] [CrossRef] [Green Version]
- Reichmann, J.; Reddington, J.P.; Best, D.; Read, D.; Ollinger, R.; Meehan, R.R.; Adams, I.R. The Genome-Defence Gene Tex19.1 Suppresses LINE-1 Retrotransposons in the Placenta and Prevents Intra-Uterine Growth Retardation in Mice. Hum. Mol. Genet. 2013, 22, 1791–1806. [Google Scholar] [CrossRef]
- Tarabay, Y.; Achour, M.; Teletin, M.; Ye, T.; Teissandier, A.; Mark, M.; Bourc’his, D.; Viville, S. Tex19 Paralogs Are New Members of the PiRNA Pathway Controlling Retrotransposon Suppression. J. Cell Sci. 2017, 130, 1463–1474. [Google Scholar] [CrossRef] [Green Version]
- MacLennan, M.; García-Cañadas, M.; Reichmann, J.; Khazina, E.; Wagner, G.; Playfoot, C.J.; Salvador-Palomeque, C.; Mann, A.R.; Peressini, P.; Sanchez, L.; et al. Mobilization of LINE-1 Retrotransposons Is Restricted by Tex19.1 in Mouse Embryonic Stem Cells. eLife 2017, 6, e26152. [Google Scholar] [CrossRef]
- Muñoz-López, M.; Vilar, R.; Philippe, C.; Rahbari, R.; Richardson, S.R.; Andres-Anton, M.; Widmann, T.; Cano, D.; Cortés, J.L.; Rubio-Roldán, A.; et al. LINE-1 Retrotransposition Impacts the Genome of Human Pre-Implantation Embryos and Extraembryonic Tissues. bioRxiv 2019. [Google Scholar] [CrossRef]
- He, Z.; Li, J.; Hwa, Y.L.; Brost, B.; Fang, Q.; Jiang, S.-W. Transition of LINE-1 DNA Methylation Status and Altered Expression in First and Third Trimester Placentas. PLoS ONE 2014, 9, e96994. [Google Scholar] [CrossRef] [PubMed]
- Evrony, G.D.; Cai, X.; Lee, E.; Hills, L.B.; Elhosary, P.C.; Lehmann, H.S.; Parker, J.J.; Atabay, K.D.; Gilmore, E.C.; Poduri, A.; et al. Single-Neuron Sequencing Analysis of L1 Retrotransposition and Somatic Mutation in the Human Brain. Cell 2012, 151, 483–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evrony, G.D.; Lee, E.; Mehta, B.K.; Benjamini, Y.; Johnson, R.M.; Cai, X.; Yang, L.; Haseley, P.; Lehmann, H.S.; Park, P.J.; et al. Cell Lineage Analysis in Human Brain Using Endogenous Retroelements. Neuron 2015, 85, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Upton, K.R.; Gerhardt, D.J.; Jesuadian, J.S.; Richardson, S.R.; Sánchez-Luque, F.J.; Bodea, G.O.; Ewing, A.D.; Salvador-Palomeque, C.; van der Knaap, M.S.; Brennan, P.M.; et al. Ubiquitous L1 Mosaicism in Hippocampal Neurons. Cell 2015, 161, 228–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breton-Larrivée, M.; Elder, E.; McGraw, S. DNA Methylation, Environmental Exposures and Early Embryo Development. Anim. Reprod. 2019, 16, 465–474. [Google Scholar] [CrossRef] [Green Version]
- Valencia, A.M.; Kadoch, C. Chromatin Regulatory Mechanisms and Therapeutic Opportunities in Cancer. Nat. Cell Biol. 2019, 21, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Sen, G.L.; Reuter, J.A.; Webster, D.E.; Zhu, L.; Khavari, P.A. DNMT1 Maintains Progenitor Function in Self-Renewing Somatic Tissue. Nature 2010, 463, 563–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jönsson, M.E.; Ludvik Brattås, P.; Gustafsson, C.; Petri, R.; Yudovich, D.; Pircs, K.; Verschuere, S.; Madsen, S.; Hansson, J.; Larsson, J.; et al. Activation of Neuronal Genes via LINE-1 Elements upon Global DNA Demethylation in Human Neural Progenitors. Nat. Commun. 2019, 10, 3182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muotri, A.R.; Marchetto, M.C.N.; Coufal, N.G.; Oefner, R.; Yeo, G.; Nakashima, K.; Gage, F.H. L1 Retrotransposition in Neurons Is Modulated by MeCP2. Nature 2010, 468, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zingler, N.; Schumann, G.; Strätling, W.H. Methyl-CpG-Binding Protein 2 Represses LINE-1 Expression and Retrotransposition but Not Alu Transcription. Nucleic Acids Res. 2001, 29, 4493–4501. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.G.; Swergold, G.D.; Ozato, K.; Thayer, R.E. Binding of the Ubiquitous Nuclear Transcription Factor YY1 to a Cis Regulatory Sequence in the Human LINE-1 Transposable Element. Hum. Mol. Genet. 1993, 2, 1697–1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapushesky, M.; Emam, I.; Holloway, E.; Kurnosov, P.; Zorin, A.; Malone, J.; Rustici, G.; Williams, E.; Parkinson, H.; Brazma, A. Gene Expression Atlas at the European Bioinformatics Institute. Nucleic Acids Res. 2010, 38, D690–D698. [Google Scholar] [CrossRef] [Green Version]
- Wolf, D.; Goff, S.P. TRIM28 Mediates Primer Binding Site-Targeted Silencing of Murine Leukemia Virus in Embryonic Cells. Cell 2007, 131, 46–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, D.; Goff, S.P. Embryonic Stem Cells Use ZFP809 to Silence Retroviral DNAs. Nature 2009, 458, 1201–1204. [Google Scholar] [CrossRef] [Green Version]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J. SETDB1: A Novel KAP-1-Associated Histone H3, Lysine 9-Specific Methyltransferase That Contributes to HP1-Mediated Silencing of Euchromatic Genes by KRAB Zinc-Finger Proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, A.V.; Peng, H.; Yurchenko, V.; Yap, K.L.; Negorev, D.G.; Schultz, D.C.; Psulkowski, E.; Fredericks, W.J.; White, D.E.; Maul, G.G.; et al. PHD Domain-Mediated E3 Ligase Activity Directs Intramolecular Sumoylation of an Adjacent Bromodomain Required for Gene Silencing. Mol. Cell 2007, 28, 823–837. [Google Scholar] [CrossRef] [Green Version]
- Rowe, H.M.; Jakobsson, J.; Mesnard, D.; Rougemont, J.; Reynard, S.; Aktas, T.; Maillard, P.V.; Layard-Liesching, H.; Verp, S.; Marquis, J.; et al. KAP1 Controls Endogenous Retroviruses in Embryonic Stem Cells. Nature 2010, 463, 237–240. [Google Scholar] [CrossRef]
- Tchasovnikarova, I.A.; Timms, R.T.; Matheson, N.J.; Wals, K.; Antrobus, R.; Göttgens, B.; Dougan, G.; Dawson, M.A.; Lehner, P.J. GENE SILENCING. Epigenetic Silencing by the HUSH Complex Mediates Position-Effect Variegation in Human Cells. Science 2015, 348, 1481–1485. [Google Scholar] [CrossRef] [Green Version]
- Tchasovnikarova, I.A.; Timms, R.T.; Douse, C.H.; Roberts, R.C.; Dougan, G.; Kingston, R.E.; Modis, Y.; Lehner, P.J. Hyperactivation of HUSH Complex Function by Charcot-Marie-Tooth Disease Mutation in MORC2. Nat. Genet. 2017, 49, 1035–1044. [Google Scholar] [CrossRef]
- Ren, W.; Fan, H.; Grimm, S.A.; Guo, Y.; Kim, J.J.; Yin, J.; Li, L.; Petell, C.J.; Tan, X.-F.; Zhang, Z.-M.; et al. Direct Readout of Heterochromatic H3K9me3 Regulates DNMT1-Mediated Maintenance DNA Methylation. Proc. Natl. Acad. Sci. USA 2020, 117, 18439–18447. [Google Scholar] [CrossRef]
- Ren, W.; Fan, H.; Grimm, S.A.; Kim, J.J.; Li, L.; Guo, Y.; Petell, C.J.; Tan, X.-F.; Zhang, Z.-M.; Coan, J.P.; et al. DNMT1 Reads Heterochromatic H4K20me3 to Reinforce LINE-1 DNA Methylation. Nat. Commun. 2021, 12, 2490. [Google Scholar] [CrossRef]
- Lee, E.J.; Banerjee, S.; Zhou, H.; Jammalamadaka, A.; Arcila, M.; Manjunath, B.S.; Kosik, K.S. Identification of PiRNAs in the Central Nervous System. RNA 2011, 17, 1090–1099. [Google Scholar] [CrossRef] [Green Version]
- Nandi, S.; Chandramohan, D.; Fioriti, L.; Melnick, A.M.; Hébert, J.M.; Mason, C.E.; Rajasethupathy, P.; Kandel, E.R. Roles for Small Noncoding RNAs in Silencing of Retrotransposons in the Mammalian Brain. Proc. Natl. Acad. Sci. USA 2016, 113, 12697–12702. [Google Scholar] [CrossRef] [Green Version]
- Leighton, L.J.; Wei, W.; Marshall, P.R.; Ratnu, V.S.; Li, X.; Zajaczkowski, E.L.; Spadaro, P.A.; Khandelwal, N.; Kumar, A.; Bredy, T.W. Disrupting the Hippocampal Piwi Pathway Enhances Contextual Fear Memory in Mice. Neurobiol. Learn Mem. 2019, 161, 202–209. [Google Scholar] [CrossRef]
- Burns, K.H. Transposable Elements in Cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef]
- Rodic, N. LINE-1 Activity and Regulation in Cancer. Front. Biosci. (Landmark Ed.) 2018, 23, 1680–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleh, A.; Macia, A.; Muotri, A.R. Transposable Elements, Inflammation, and Neurological Disease. Front. Neurol. 2019, 10, 894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misiak, B.; Ricceri, L.; Sąsiadek, M.M. Transposable Elements and Their Epigenetic Regulation in Mental Disorders: Current Evidence in the Field. Front. Genet. 2019, 10, 580. [Google Scholar] [CrossRef] [PubMed]
- Lapp, H.E.; Hunter, R.G. Early Life Exposures, Neurodevelopmental Disorders, and Transposable Elements. Neurobiol. Stress 2019, 11, 100174. [Google Scholar] [CrossRef]
- Rohrback, S.; Siddoway, B.; Liu, C.S.; Chun, J. Genomic Mosaicism in the Developing and Adult Brain. Dev. Neurobiol. 2018, 78, 1026–1048. [Google Scholar] [CrossRef] [PubMed]
- Suarez, N.A.; Macia, A.; Muotri, A.R. LINE-1 Retrotransposons in Healthy and Diseased Human Brain. Dev. Neurobiol. 2018, 78, 434–455. [Google Scholar] [CrossRef]
- Basil, P.; Li, Q.; Sham, P.-C.; McAlonan, G.M. LINE1 and Mecp2 Methylation of the Adult Striatum and Prefrontal Cortex Exposed to Prenatal Immune Activation. Data Brief 2019, 25, 104003. [Google Scholar] [CrossRef] [PubMed]
- Caliri, A.W.; Caceres, A.; Tommasi, S.; Besaratinia, A. Hypomethylation of LINE-1 Repeat Elements and Global Loss of DNA Hydroxymethylation in Vapers and Smokers. Epigenetics 2020, 15, 816–829. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Barchitta, M.; Fallico, M.; Castellino, N.; Reibaldi, M.; Agodi, A. Characterization of SIRT1/DNMTs Functions and LINE-1 Methylation in Patients with Age-Related Macular Degeneration. J. Clin. Med. 2019, 8, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Meter, M.; Kashyap, M.; Rezazadeh, S.; Geneva, A.J.; Morello, T.D.; Seluanov, A.; Gorbunova, V. SIRT6 Represses LINE1 Retrotransposons by Ribosylating KAP1 but This Repression Fails with Stress and Age. Nat. Commun. 2014, 5, 5011. [Google Scholar] [CrossRef]
- Vazquez, B.N.; Thackray, J.K.; Simonet, N.G.; Chahar, S.; Kane-Goldsmith, N.; Newkirk, S.J.; Lee, S.; Xing, J.; Verzi, M.P.; An, W.; et al. SIRT7 Mediates L1 Elements Transcriptional Repression and Their Association with the Nuclear Lamina. Nucleic Acids Res. 2019, 47, 7870–7885. [Google Scholar] [CrossRef] [Green Version]
- Montoya-Durango, D.E.; Ramos, K.A.; Bojang, P.; Ruiz, L.; Ramos, I.N.; Ramos, K.S. LINE-1 Silencing by Retinoblastoma Proteins Is Effected through the Nucleosomal and Remodeling Deacetylase Multiprotein Complex. BMC Cancer 2016, 16, 38. [Google Scholar] [CrossRef] [Green Version]
- Sahnane, N.; Ottini, G.; Turri-Zanoni, M.; Furlan, D.; Battaglia, P.; Karligkiotis, A.; Albeni, C.; Cerutti, R.; Mura, E.; Chiaravalli, A.M.; et al. Comprehensive Analysis of HPV Infection, EGFR Exon 20 Mutations and LINE1 Hypomethylation as Risk Factors for Malignant Transformation of Sinonasal-Inverted Papilloma to Squamous Cell Carcinoma. Int. J. Cancer 2019, 144, 1313–1320. [Google Scholar] [CrossRef]
- Tahara, S.; Tahara, T.; Horiguchi, N.; Okubo, M.; Terada, T.; Yoshida, D.; Funasaka, K.; Nakagawa, Y.; Shibata, T.; Tsukamoto, T.; et al. Lower LINE-1 Methylation Is Associated with Promoter Hypermethylation and Distinct Molecular Features in Gastric Cancer. Epigenomics 2019, 11, 1651–1659. [Google Scholar] [CrossRef]
- Ponomarev, I.; Wang, S.; Zhang, L.; Harris, R.A.; Mayfield, R.D. Gene Coexpression Networks in Human Brain Identify Epigenetic Modifications in Alcohol Dependence. J. Neurosci. 2012, 32, 1884–1897. [Google Scholar] [CrossRef]
- Okudaira, N.; Ishizaka, Y.; Nishio, H. Retrotransposition of Long Interspersed Element 1 Induced by Methamphetamine or Cocaine. J. Biol. Chem. 2014, 289, 25476–25485. [Google Scholar] [CrossRef] [Green Version]
- Moszczynska, A.; Flack, A.; Qiu, P.; Muotri, A.R.; Killinger, B.A. Neurotoxic Methamphetamine Doses Increase LINE-1 Expression in the Neurogenic Zones of the Adult Rat Brain. Sci. Rep. 2015, 5, 14356. [Google Scholar] [CrossRef] [Green Version]
- Kale, S.P.; Moore, L.; Deininger, P.L.; Roy-Engel, A.M. Heavy Metals Stimulate Human LINE-1 Retrotransposition. Int. J. Environ. Res. Public Health 2005, 2, 14–23. [Google Scholar] [CrossRef] [Green Version]
- Kalayasiri, R.; Kraijak, K.; Maes, M.; Mutirangura, A. Methamphetamine (MA) Use Induces Specific Changes in LINE-1 Partial Methylation Patterns, Which Are Associated with MA-Induced Paranoia: A Multivariate and Neuronal Network Study. Mol. Neurobiol. 2019, 56, 4258–4272. [Google Scholar] [CrossRef]
- Del Re, B.; Giorgi, G. Long INterspersed Element-1 Mobility as a Sensor of Environmental Stresses. Environ. Mol. Mutagen. 2020, 61, 465–493. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Hernandez, A.; Kuo, C.-C.; Rentero-Garrido, P.; Tang, W.-Y.; Redon, J.; Ordovas, J.M.; Navas-Acien, A.; Tellez-Plaza, M. Environmental Chemicals and DNA Methylation in Adults: A Systematic Review of the Epidemiologic Evidence. Clin. Epigenetics 2015, 7, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardelli, M. The Epigenetic Alterations of Endogenous Retroelements in Aging. Mech. Ageing Dev. 2018, 174, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Salameh, Y.; Bejaoui, Y.; El Hajj, N. DNA Methylation Biomarkers in Aging and Age-Related Diseases. Front. Genet. 2020, 11, 171. [Google Scholar] [CrossRef]
- Basil, P.; Li, Q.; Dempster, E.L.; Mill, J.; Sham, P.-C.; Wong, C.C.Y.; McAlonan, G.M. Prenatal Maternal Immune Activation Causes Epigenetic Differences in Adolescent Mouse Brain. Transl. Psychiatry 2014, 4, e434. [Google Scholar] [CrossRef] [PubMed]
- Basil, P.; Li, Q.; Gui, H.; Hui, T.C.K.; Ling, V.H.M.; Wong, C.C.Y.; Mill, J.; McAlonan, G.M.; Sham, P.-C. Prenatal Immune Activation Alters the Adult Neural Epigenome but Can Be Partly Stabilised by a N-3 Polyunsaturated Fatty Acid Diet. Transl. Psychiatry 2018, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Conway, F.; Brown, A.S. Maternal Immune Activation and Related Factors in the Risk of Offspring Psychiatric Disorders. Front. Psychiatry 2019, 10, 430. [Google Scholar] [CrossRef] [PubMed]
- Amodeo, D.A.; Lai, C.-Y.; Hassan, O.; Mukamel, E.A.; Behrens, M.M.; Powell, S.B. Maternal Immune Activation Impairs Cognitive Flexibility and Alters Transcription in Frontal Cortex. Neurobiol. Dis. 2019, 125, 211–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.-H.; Lee, J.-H.; Lee, H.-Y.; Min, K.-J. Sirtuin Signaling in Cellular Senescence and Aging. BMB Rep. 2019, 52, 24–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagunas-Rangel, F.A. Current Role of Mammalian Sirtuins in DNA Repair. DNA Repair. 2019, 80, 85–92. [Google Scholar] [CrossRef]
- Mendes, K.L.; de Farias Lelis, D.; Santos, S.H.S. Nuclear Sirtuins and Inflammatory Signaling Pathways. Cytokine Growth Factor Rev. 2017, 38, 98–105. [Google Scholar] [CrossRef]
- Zhou, S.; Tang, X.; Chen, H.-Z. Sirtuins and Insulin Resistance. Front. Endocrinol. 2018, 9, 748. [Google Scholar] [CrossRef] [Green Version]
- De Céu Teixeira, M.; Sanchez-Lopez, E.; Espina, M.; Garcia, M.L.; Durazzo, A.; Lucarini, M.; Novellino, E.; Souto, S.B.; Santini, A.; Souto, E.B. Sirtuins and SIRT6 in Carcinogenesis and in Diet. Int. J. Mol. Sci. 2019, 20, 4945. [Google Scholar] [CrossRef] [Green Version]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D.; et al. L1 Drives IFN in Senescent Cells and Promotes Age-Associated Inflammation. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Van Meter, M.; Ablaeva, J.; Ke, Z.; Gonzalez, R.S.; Taguchi, T.; De Cecco, M.; Leonova, K.I.; Kogan, V.; Helfand, S.L.; et al. LINE1 Derepression in Aged Wild-Type and SIRT6-Deficient Mice Drives Inflammation. Cell Metab. 2019, 29, 871–885.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumari, P.; Popescu, D.; Yue, S.; Bober, E.; Ianni, A.; Braun, T. Sirt7 Inhibits Sirt1-Mediated Activation of Suv39h1. Cell Cycle 2018, 17, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Min, B.; Jeon, K.; Park, J.S.; Kang, Y.-K. Demethylation and Derepression of Genomic Retroelements in the Skeletal Muscles of Aged Mice. Aging Cell 2019, 18, e13042. [Google Scholar] [CrossRef] [Green Version]
- Fujita, N.; Jaye, D.L.; Geigerman, C.; Akyildiz, A.; Mooney, M.R.; Boss, J.M.; Wade, P.A. MTA3 and the Mi-2/NuRD Complex Regulate Cell Fate during B Lymphocyte Differentiation. Cell 2004, 119, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.-L.; Jin, S.-G.; Pfeifer, G.P. MBD3L1 Is a Transcriptional Repressor That Interacts with Methyl-CpG-Binding Protein 2 (MBD2) and Components of the NuRD Complex. J. Biol. Chem. 2004, 279, 52456–52464. [Google Scholar] [CrossRef] [Green Version]
- Ramírez, J.; Dege, C.; Kutateladze, T.G.; Hagman, J. MBD2 and Multiple Domains of CHD4 Are Required for Transcriptional Repression by Mi-2/NuRD Complexes. Mol. Cell. Biol. 2012, 32, 5078–5088. [Google Scholar] [CrossRef] [Green Version]
- Bojang, P.; Ramos, K.S. Epigenetic Reactivation of LINE-1 Retrotransposon Disrupts NuRD Corepressor Functions and Induces Oncogenic Transformation in Human Bronchial Epithelial Cells. Mol. Oncol. 2018, 12, 1342–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montoya-Durango, D.E.; Liu, Y.; Teneng, I.; Kalbfleisch, T.; Lacy, M.E.; Steffen, M.C.; Ramos, K.S. Epigenetic Control of Mammalian LINE-1 Retrotransposon by Retinoblastoma Proteins. Mutat. Res. 2009, 665, 20–28. [Google Scholar] [CrossRef] [Green Version]
- Ishak, C.A.; Marshall, A.E.; Passos, D.T.; White, C.R.; Kim, S.J.; Cecchini, M.J.; Ferwati, S.; MacDonald, W.A.; Howlett, C.J.; Welch, I.D.; et al. An RB-EZH2 Complex Mediates Silencing of Repetitive DNA Sequences. Mol. Cell 2016, 64, 1074–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamdorf, M.; Idica, A.; Zisoulis, D.G.; Gamelin, L.; Martin, C.; Sanders, K.J.; Pedersen, I.M. MiR-128 Represses L1 Retrotransposition by Binding Directly to L1 RNA. Nat. Struct. Mol. Biol. 2015, 22, 824–831. [Google Scholar] [CrossRef]
- Idica, A.; Sevrioukov, E.A.; Zisoulis, D.G.; Hamdorf, M.; Daugaard, I.; Kadandale, P.; Pedersen, I.M. MicroRNA MiR-128 Represses LINE-1 (L1) Retrotransposition by down-Regulating the Nuclear Import Factor TNPO1. J. Biol. Chem. 2017, 292, 20494–20508. [Google Scholar] [CrossRef] [Green Version]
- Fung, L.; Guzman, H.; Sevrioukov, E.; Idica, A.; Park, E.; Bochnakian, A.; Daugaard, I.; Jury, D.; Mortazavi, A.; Zisoulis, D.G.; et al. MiR-128 Restriction of LINE-1 (L1) Retrotransposition Is Dependent on Targeting HnRNPA1 MRNA. Int. J. Mol. Sci. 2019, 20, 1955. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M.; Kose, S.; Okumura, N.; Imai, K.; Furuta, M.; Sakiyama, N.; Tomii, K.; Horton, P.; Takao, T.; Imamoto, N. Identification of Cargo Proteins Specific for the Nucleocytoplasmic Transport Carrier Transportin by Combination of an in Vitro Transport System and Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC)-Based Quantitative Proteomics. Mol. Cell Proteom. 2013, 12, 145–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Twyffels, L.; Gueydan, C.; Kruys, V. Transportin-1 and Transportin-2: Protein Nuclear Import and Beyond. FEBS Lett. 2014, 588, 1857–1868. [Google Scholar] [CrossRef] [PubMed]
- Reed, R.; Hurt, E. A Conserved MRNA Export Machinery Coupled to Pre-MRNA Splicing. Cell 2002, 108, 523–531. [Google Scholar] [CrossRef] [Green Version]
- Li, P.W.-L.; Li, J.; Timmerman, S.L.; Krushel, L.A.; Martin, S.L. The Dicistronic RNA from the Mouse LINE-1 Retrotransposon Contains an Internal Ribosome Entry Site Upstream of Each ORF: Implications for Retrotransposition. Nucleic Acids Res. 2006, 34, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Peddigari, S.; Li, P.W.-L.; Rabe, J.L.; Martin, S.L. HnRNPL and Nucleolin Bind LINE-1 RNA and Function as Host Factors to Modulate Retrotransposition. Nucleic Acids Res. 2013, 41, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of Primary MicroRNAs by the Microprocessor Complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor Complex Mediates the Genesis of MicroRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 Complex in Primary MicroRNA Processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [Green Version]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of Small RNAs in Animals. Nat. Rev. Mol. Cell. Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Heras, S.R.; Macias, S.; Plass, M.; Fernandez, N.; Cano, D.; Eyras, E.; Garcia-Perez, J.L.; Cáceres, J.F. The Microprocessor Controls the Activity of Mammalian Retrotransposons. Nat. Struct. Mol. Biol. 2013, 20, 1173–1181. [Google Scholar] [CrossRef]
- Ikeda, T.; Shimoda, M.; Ebrahimi, D.; VandeBerg, J.L.; Harris, R.S.; Koito, A.; Maeda, K. Opossum APOBEC1 Is a DNA Mutator with Retrovirus and Retroelement Restriction Activity. Sci. Rep. 2017, 7, 46719. [Google Scholar] [CrossRef] [Green Version]
- Lahouassa, H.; Daddacha, W.; Hofmann, H.; Ayinde, D.; Logue, E.C.; Dragin, L.; Bloch, N.; Maudet, C.; Bertrand, M.; Gramberg, T.; et al. SAMHD1 Restricts the Replication of Human Immunodeficiency Virus Type 1 by Depleting the Intracellular Pool of Deoxynucleoside Triphosphates. Nat. Immunol. 2012, 13, 223–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, K.; Du, J.; Han, X.; Goodier, J.L.; Li, P.; Zhou, X.; Wei, W.; Evans, S.L.; Li, L.; Zhang, W.; et al. Modulation of LINE-1 and Alu/SVA Retrotransposition by Aicardi-Goutières Syndrome-Related SAMHD1. Cell Rep. 2013, 4, 1108–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renner, T.M.; Bélanger, K.; Goodwin, L.R.; Campbell, M.; Langlois, M.-A. Characterization of Molecular Attributes That Influence LINE-1 Restriction by All Seven Human APOBEC3 Proteins. Virology 2018, 520, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Du, J.; Peng, Y.; Li, P.; Wang, S.; Wang, Y.; Hou, J.; Kang, J.; Zheng, W.; Hua, S.; et al. LINE1 Contributes to Autoimmunity through Both RIG-I- and MDA5-Mediated RNA Sensing Pathways. J. Autoimmun. 2018, 90, 105–115. [Google Scholar] [CrossRef]
- Tunbak, H.; Enriquez-Gasca, R.; Tie, C.H.C.; Gould, P.A.; Mlcochova, P.; Gupta, R.K.; Fernandes, L.; Holt, J.; van der Veen, A.G.; Giampazolias, E.; et al. The HUSH Complex Is a Gatekeeper of Type I Interferon through Epigenetic Regulation of LINE-1s. Nat. Commun. 2020, 11, 5387. [Google Scholar] [CrossRef]
- Rostami, M.R.; Bradic, M. The Derepression of Transposable Elements in Lung Cells Is Associated with the Inflammatory Response and Gene Activation in Idiopathic Pulmonary Fibrosis. Mob. DNA 2021, 12, 14. [Google Scholar] [CrossRef]
- Kawano, K.; Doucet, A.J.; Ueno, M.; Kariya, R.; An, W.; Marzetta, F.; Kuroki, M.; Turelli, P.; Sukegawa, S.; Okada, S.; et al. HIV-1 Vpr and P21 Restrict LINE-1 Mobility. Nucleic Acids Res. 2018, 46, 8454–8470. [Google Scholar] [CrossRef] [Green Version]
- Schöbel, A.; Nguyen-Dinh, V.; Schumann, G.G.; Herker, E. Hepatitis C Virus Infection Restricts Human LINE-1 Retrotransposition in Hepatoma Cells. PLoS Pathog. 2021, 17, e1009496. [Google Scholar] [CrossRef]
- Crow, Y.J.; Chase, D.S.; Lowenstein Schmidt, J.; Szynkiewicz, M.; Forte, G.M.A.; Gornall, H.L.; Oojageer, A.; Anderson, B.; Pizzino, A.; Helman, G.; et al. Characterization of Human Disease Phenotypes Associated with Mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet. A 2015, 167A, 296–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.; Dong, B.; Doucet, A.J.; Moldovan, J.B.; Moran, J.V.; Silverman, R.H. RNase L Restricts the Mobility of Engineered Retrotransposons in Cultured Human Cells. Nucleic Acids Res. 2014, 42, 3803–3820. [Google Scholar] [CrossRef] [PubMed]
- Malathi, K.; Dong, B.; Gale, M.; Silverman, R.H. Small Self-RNA Generated by RNase L Amplifies Antiviral Innate Immunity. Nature 2007, 448, 816–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reikine, S.; Nguyen, J.B.; Modis, Y. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front. Immunol. 2014, 5, 342. [Google Scholar] [CrossRef] [Green Version]
- Oshiumi, H.; Kouwaki, T.; Seya, T. Accessory Factors of Cytoplasmic Viral RNA Sensors Required for Antiviral Innate Immune Response. Front. Immunol. 2016, 7, 200. [Google Scholar] [CrossRef]
- Arjan-Odedra, S.; Swanson, C.M.; Sherer, N.M.; Wolinsky, S.M.; Malim, M.H. Endogenous MOV10 Inhibits the Retrotransposition of Endogenous Retroelements but Not the Replication of Exogenous Retroviruses. Retrovirology 2012, 9, 53. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Hwang, S.-Y.; Ahn, K. Interplay between RNASEH2 and MOV10 Controls LINE-1 Retrotransposition. Nucleic Acids Res. 2018, 46, 1912–1926. [Google Scholar] [CrossRef]
- Goodier, J.L.; Cheung, L.E.; Kazazian, H.H. MOV10 RNA Helicase Is a Potent Inhibitor of Retrotransposition in Cells. PLoS Genet. 2012, 8, e1002941. [Google Scholar] [CrossRef] [Green Version]
- Skariah, G.; Seimetz, J.; Norsworthy, M.; Lannom, M.C.; Kenny, P.J.; Elrakhawy, M.; Forsthoefel, C.; Drnevich, J.; Kalsotra, A.; Ceman, S. Mov10 Suppresses Retroelements and Regulates Neuronal Development and Function in the Developing Brain. BMC Biol. 2017, 15, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moldovan, J.B.; Moran, J.V. The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLoS Genet. 2015, 11, e1005121. [Google Scholar] [CrossRef] [PubMed]
- Benitez-Guijarro, M.; Lopez-Ruiz, C.; Tarnauskaitė, Ž.; Murina, O.; Mian Mohammad, M.; Williams, T.C.; Fluteau, A.; Sanchez, L.; Vilar-Astasio, R.; Garcia-Canadas, M.; et al. RNase H2, Mutated in Aicardi-Goutières Syndrome, Promotes LINE-1 Retrotransposition. EMBO J. 2018, 37, e98506. [Google Scholar] [CrossRef]
- Warkocki, Z.; Krawczyk, P.S.; Adamska, D.; Bijata, K.; Garcia-Perez, J.L.; Dziembowski, A. Uridylation by TUT4/7 Restricts Retrotransposition of Human LINE-1s. Cell 2018, 174, 1537–1548.e29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampath, P.; Mazumder, B.; Seshadri, V.; Gerber, C.A.; Chavatte, L.; Kinter, M.; Ting, S.M.; Dignam, J.D.; Kim, S.; Driscoll, D.M.; et al. Noncanonical Function of Glutamyl-Prolyl-TRNA Synthetase: Gene-Specific Silencing of Translation. Cell 2004, 119, 195–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapasi, P.; Chaudhuri, S.; Vyas, K.; Baus, D.; Komar, A.A.; Fox, P.L.; Merrick, W.C.; Mazumder, B. L13a Blocks 48S Assembly: Role of a General Initiation Factor in MRNA-Specific Translational Control. Mol. Cell 2007, 25, 113–126. [Google Scholar] [CrossRef] [Green Version]
- Ward, J.R.; Vasu, K.; Deutschman, E.; Halawani, D.; Larson, P.A.; Zhang, D.; Willard, B.; Fox, P.L.; Moran, J.V.; Longworth, M.S. Condensin II and GAIT Complexes Cooperate to Restrict LINE-1 Retrotransposition in Epithelial Cells. PLoS Genet. 2017, 13, e1007051. [Google Scholar] [CrossRef] [Green Version]
- Mukhopadhyay, R.; Jia, J.; Arif, A.; Ray, P.S.; Fox, P.L. The GAIT System: A Gatekeeper of Inflammatory Gene Expression. Trends Biochem. Sci. 2009, 34, 324–331. [Google Scholar] [CrossRef] [Green Version]
- Stetson, D.B. Endogenous Retroelements and Autoimmune Disease. Curr. Opin. Immunol. 2012, 24, 692–697. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Du, J.; Goodier, J.L.; Hou, J.; Kang, J.; Kazazian, H.H.; Zhao, K.; Yu, X.-F. Aicardi-Goutières Syndrome Protein TREX1 Suppresses L1 and Maintains Genome Integrity through Exonuclease-Independent ORF1p Depletion. Nucleic Acids Res. 2017, 45, 4619–4631. [Google Scholar] [CrossRef] [Green Version]
- Stetson, D.B.; Ko, J.S.; Heidmann, T.; Medzhitov, R. Trex1 Prevents Cell-Intrinsic Initiation of Autoimmunity. Cell 2008, 134, 587–598. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, D.; Beresford, P.J.; Zhu, P.; Zhang, D.; Sung, J.-S.; Demple, B.; Perrino, F.W.; Lieberman, J. The Exonuclease TREX1 Is in the SET Complex and Acts in Concert with NM23-H1 to Degrade DNA during Granzyme A-Mediated Cell Death. Mol. Cell 2006, 23, 133–142. [Google Scholar] [CrossRef]
- Vembar, S.S.; Brodsky, J.L. One Step at a Time: Endoplasmic Reticulum-Associated Degradation. Nat. Rev. Mol. Cell. Biol. 2008, 9, 944–957. [Google Scholar] [CrossRef]
- Deutschmann, J.; Gramberg, T. SAMHD1 … and Viral Ways around It. Viruses 2021, 13, 395. [Google Scholar] [CrossRef]
- Chen, S.; Bonifati, S.; Qin, Z.; St Gelais, C.; Wu, L. SAMHD1 Suppression of Antiviral Immune Responses. Trends Microbiol. 2019, 27, 254–267. [Google Scholar] [CrossRef]
- Rice, G.I.; Bond, J.; Asipu, A.; Brunette, R.L.; Manfield, I.W.; Carr, I.M.; Fuller, J.C.; Jackson, R.M.; Lamb, T.; Briggs, T.A.; et al. Mutations Involved in Aicardi-Goutières Syndrome Implicate SAMHD1 as Regulator of the Innate Immune Response. Nat. Genet. 2009, 41, 829–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Li, J.; Xu, F.; Mei, S.; Le Duff, Y.; Yin, L.; Pang, X.; Cen, S.; Jin, Q.; Liang, C.; et al. SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation. PLoS Genet. 2015, 11, e1005367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.E.; Brandariz-Nuñez, A.; Han, K.; Sawyer, S.L.; Kim, B.; Diaz-Griffero, F. Modulation of LINE-1 Retrotransposition by a Human SAMHD1 Polymorphism. Virol. Rep. 2016, 6, 53–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; Peng, Y.; Wang, S.; Hou, J.; Wang, Y.; Sun, T.; Zhao, K. Nucleocytoplasmic Shuttling of SAMHD1 Is Important for LINE-1 Suppression. Biochem. Biophys. Res. Commun. 2019, 510, 551–557. [Google Scholar] [CrossRef]
- Rose, K.M.; Spada, S.J.; Broeckel, R.; McNally, K.L.; Hirsch, V.M.; Best, S.M.; Bouamr, F. From Capsids to Complexes: Expanding the Role of TRIM5α in the Restriction of Divergent RNA Viruses and Elements. Viruses 2021, 13, 446. [Google Scholar] [CrossRef]
- Ganser-Pornillos, B.K.; Chandrasekaran, V.; Pornillos, O.; Sodroski, J.G.; Sundquist, W.I.; Yeager, M. Hexagonal Assembly of a Restricting TRIM5alpha Protein. Proc. Natl. Acad. Sci. USA 2011, 108, 534–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkmann, B.; Wittmann, S.; Lagisquet, J.; Deutschmann, J.; Eissmann, K.; Ross, J.J.; Biesinger, B.; Gramberg, T. Human TRIM5α Senses and Restricts LINE-1 Elements. Proc. Natl. Acad. Sci. USA 2020, 117, 17965–17976. [Google Scholar] [CrossRef] [PubMed]
- Goodier, J.L. Restricting Retrotransposons: A Review. Mob. DNA 2016, 7, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silvas, T.V.; Schiffer, C.A. APOBEC3s: DNA-Editing Human Cytidine Deaminases. Protein Sci. 2019, 28, 1552–1566. [Google Scholar] [CrossRef] [PubMed]
- Bogerd, H.P.; Wiegand, H.L.; Hulme, A.E.; Garcia-Perez, J.L.; O’Shea, K.S.; Moran, J.V.; Cullen, B.R. Cellular Inhibitors of Long Interspersed Element 1 and Alu Retrotransposition. Proc. Natl. Acad. Sci. USA 2006, 103, 8780–8785. [Google Scholar] [CrossRef] [Green Version]
- Muckenfuss, H.; Hamdorf, M.; Held, U.; Perković, M.; Löwer, J.; Cichutek, K.; Flory, E.; Schumann, G.G.; Münk, C. APOBEC3 Proteins Inhibit Human LINE-1 Retrotransposition. J. Biol. Chem. 2006, 281, 22161–22172. [Google Scholar] [CrossRef] [Green Version]
- Stenglein, M.D.; Harris, R.S. APOBEC3B and APOBEC3F Inhibit L1 Retrotransposition by a DNA Deamination-Independent Mechanism. J. Biol. Chem. 2006, 281, 16837–16841. [Google Scholar] [CrossRef] [Green Version]
- Kinomoto, M.; Kanno, T.; Shimura, M.; Ishizaka, Y.; Kojima, A.; Kurata, T.; Sata, T.; Tokunaga, K. All APOBEC3 Family Proteins Differentially Inhibit LINE-1 Retrotransposition. Nucleic Acids Res. 2007, 35, 2955–2964. [Google Scholar] [CrossRef]
- Niewiadomska, A.M.; Tian, C.; Tan, L.; Wang, T.; Sarkis, P.T.N.; Yu, X.-F. Differential Inhibition of Long Interspersed Element 1 by APOBEC3 Does Not Correlate with High-Molecular-Mass-Complex Formation or P-Body Association. J. Virol. 2007, 81, 9577–9583. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Goubran, M.H.; Follack, T.B.; Chelico, L. Deamination-Independent Restriction of LINE-1 Retrotransposition by APOBEC3H. Sci. Rep. 2017, 7, 10881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, S.R.; Narvaiza, I.; Planegger, R.A.; Weitzman, M.D.; Moran, J.V. APOBEC3A Deaminates Transiently Exposed Single-Strand DNA during LINE-1 Retrotransposition. eLife 2014, 3, e02008. [Google Scholar] [CrossRef]
- Horn, A.V.; Klawitter, S.; Held, U.; Berger, A.; Vasudevan, A.A.J.; Bock, A.; Hofmann, H.; Hanschmann, K.-M.O.; Trösemeier, J.-H.; Flory, E.; et al. Human LINE-1 Restriction by APOBEC3C Is Deaminase Independent and Mediated by an ORF1p Interaction That Affects LINE Reverse Transcriptase Activity. Nucleic Acids Res. 2014, 42, 396–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, W.; Xu, J.; Yuan, W.; Song, X.; Zhang, J.; Wei, W.; Yu, X.-F.; Yang, Y. APOBEC3DE Inhibits LINE-1 Retrotransposition by Interacting with ORF1p and Influencing LINE Reverse Transcriptase Activity. PLoS ONE 2016, 11, e0157220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pak, V.; Heidecker, G.; Pathak, V.K.; Derse, D. The Role of Amino-Terminal Sequences in Cellular Localization and Antiviral Activity of APOBEC3B. J. Virol. 2011, 85, 8538–8547. [Google Scholar] [CrossRef] [Green Version]
- Salamango, D.J.; McCann, J.L.; Demir, Ö.; Brown, W.L.; Amaro, R.E.; Harris, R.S. APOBEC3B Nuclear Localization Requires Two Distinct N-terminal Domain Surfaces. J. Mol. Biol. 2018, 430, 2695–2708. [Google Scholar] [CrossRef]
- MacDuff, D.A.; Demorest, Z.L.; Harris, R.S. AID Can Restrict L1 Retrotransposition Suggesting a Dual Role in Innate and Adaptive Immunity. Nucleic Acids Res. 2009, 37, 1854–1867. [Google Scholar] [CrossRef] [Green Version]
- Orecchini, E.; Doria, M.; Antonioni, A.; Galardi, S.; Ciafrè, S.A.; Frassinelli, L.; Mancone, C.; Montaldo, C.; Tripodi, M.; Michienzi, A. ADAR1 Restricts LINE-1 Retrotransposition. Nucleic Acids Res. 2017, 45, 155–168. [Google Scholar] [CrossRef] [Green Version]
- Frassinelli, L.; Orecchini, E.; Al-Wardat, S.; Tripodi, M.; Mancone, C.; Doria, M.; Galardi, S.; Ciafrè, S.A.; Michienzi, A. The RNA Editing Enzyme ADAR2 Restricts L1 Mobility. RNA Biol. 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Schumann, G.G.; Gogvadze, E.V.; Osanai-Futahashi, M.; Kuroki, A.; Münk, C.; Fujiwara, H.; Ivics, Z.; Buzdin, A.A. Unique Functions of Repetitive Transcriptomes. Int. Rev. Cell Mol. Biol. 2010, 285, 115–188. [Google Scholar] [CrossRef]
- Coin, F.; Marinoni, J.C.; Rodolfo, C.; Fribourg, S.; Pedrini, A.M.; Egly, J.M. Mutations in the XPD Helicase Gene Result in XP and TTD Phenotypes, Preventing Interaction between XPD and the P44 Subunit of TFIIH. Nat. Genet. 1998, 20, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, A.R.; McGibbon, D.; Stefanini, M. Xeroderma Pigmentosum. Orphanet. J. Rare Dis. 2011, 6, 70. [Google Scholar] [CrossRef] [Green Version]
- Laugel, V. Cockayne Syndrome: The Expanding Clinical and Mutational Spectrum. Mech. Ageing Dev. 2013, 134, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Shiloh, Y. ATM (Ataxia Telangiectasia Mutated): Expanding Roles in the DNA Damage Response and Cellular Homeostasis. Biochem. Soc. Trans. 2001, 29, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Stracker, T.H.; Roig, I.; Knobel, P.A.; Marjanović, M. The ATM Signaling Network in Development and Disease. Front. Genet. 2013, 4, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceccaldi, R.; Sarangi, P.; D’Andrea, A.D. The Fanconi Anaemia Pathway: New Players and New Functions. Nat. Rev. Mol. Cell. Biol. 2016, 17, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Brégnard, C.; Guerra, J.; Déjardin, S.; Passalacqua, F.; Benkirane, M.; Laguette, N. Upregulated LINE-1 Activity in the Fanconi Anemia Cancer Susceptibility Syndrome Leads to Spontaneous Pro-Inflammatory Cytokine Production. EBioMedicine 2016, 8, 184–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariumi, Y.; Kawano, K.; Yasuda-Inoue, M.; Kuroki, M.; Fukuda, H.; Siddiqui, R.; Turelli, P.; Tateishi, S. DNA Repair Protein Rad18 Restricts LINE-1 Mobility. Sci. Rep. 2018, 8, 15894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, K.; Tateishi, S.; Kawasuji, M.; Tsurimoto, T.; Inoue, H.; Yamaizumi, M. Rad18 Guides Poleta to Replication Stalling Sites through Physical Interaction and PCNA Monoubiquitination. EMBO J. 2004, 23, 3886–3896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulder, L.C.F.; Chakrabarti, L.A.; Muesing, M.A. Interaction of HIV-1 Integrase with DNA Repair Protein HRad18. J. Biol. Chem. 2002, 277, 27489–27493. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, A.G.; Tateishi, S.; Bieniasz, P.D.; Muesing, M.A.; Yamaizumi, M.; Mulder, L.C.F. Effect of DNA Repair Protein Rad18 on Viral Infection. PLoS Pathog. 2006, 2, e40. [Google Scholar] [CrossRef] [Green Version]
- Harris, C.R.; Dewan, A.; Zupnick, A.; Normart, R.; Gabriel, A.; Prives, C.; Levine, A.J.; Hoh, J. P53 Responsive Elements in Human Retrotransposons. Oncogene 2009, 28, 3857–3865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wylie, A.; Jones, A.E.; D’Brot, A.; Lu, W.-J.; Kurtz, P.; Moran, J.V.; Rakheja, D.; Chen, K.S.; Hammer, R.E.; Comerford, S.A.; et al. P53 Genes Function to Restrain Mobile Elements. Genes Dev. 2016, 30, 64–77. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, B.; Jones, A.E.; Caillet, C.J.; Das, S.; Royer, S.K.; Abrams, J.M. P53 Directly Represses Human LINE1 Transposons. Genes Dev. 2020, 34, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Zaytseva, O.; Kim, N.-H.; Quinn, L.M. MYC in Brain Development and Cancer. Int. J. Mol. Sci. 2020, 21, 7742. [Google Scholar] [CrossRef]
- Sun, X.; Wang, X.; Tang, Z.; Grivainis, M.; Kahler, D.; Yun, C.; Mita, P.; Fenyö, D.; Boeke, J.D. Transcription Factor Profiling Reveals Molecular Choreography and Key Regulators of Human Retrotransposon Expression. Proc. Natl. Acad. Sci. USA 2018, 115, E5526–E5535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briggs, E.M.; Mita, P.; Sun, X.; Ha, S.; Vasilyev, N.; Leopold, Z.R.; Nudler, E.; Boeke, J.D.; Logan, S.K. Unbiased Proteomic Mapping of the LINE-1 Promoter Using CRISPR Cas9. Mob. DNA 2021, 12, 21. [Google Scholar] [CrossRef]
- Yang, N.; Zhang, L.; Zhang, Y.; Kazazian, H.H. An Important Role for RUNX3 in Human L1 Transcription and Retrotransposition. Nucleic Acids Res. 2003, 31, 4929–4940. [Google Scholar] [CrossRef]
- Tchénio, T.; Casella, J.F.; Heidmann, T. Members of the SRY Family Regulate the Human LINE Retrotransposons. Nucleic Acids Res. 2000, 28, 411–415. [Google Scholar] [CrossRef]
- Orqueda, A.J.; Gatti, C.R.; Ogara, M.F.; Falzone, T.L. SOX-11 Regulates LINE-1 Retrotransposon Activity during Neuronal Differentiation. FEBS Lett. 2018, 592, 3708–3719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuwabara, T.; Hsieh, J.; Muotri, A.; Yeo, G.; Warashina, M.; Lie, D.C.; Moore, L.; Nakashima, K.; Asashima, M.; Gage, F.H. Wnt-Mediated Activation of NeuroD1 and Retro-Elements during Adult Neurogenesis. Nat. Neurosci. 2009, 12, 1097–1105. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Yan, K.; Zheng, Q.; Ke, H.; Cheng, J.; Xiong, W.; Shi, X.; Wei, L.; Zhao, M.; Yang, F.; et al. Histone Demethylase KDM4B Promotes DNA Damage by Activating Long Interspersed Nuclear Element-1. Cancer Res. 2019, 79, 86–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.; Chen, L.; Yang, J.; Ye, T.; Chen, Y.; Fang, J. HIF-1α-Induced Histone Demethylase JMJD2B Contributes to the Malignant Phenotype of Colorectal Cancer Cells via an Epigenetic Mechanism. Carcinogenesis 2012, 33, 1664–1673. [Google Scholar] [CrossRef] [Green Version]
- Wilson, C.; Qiu, L.; Hong, Y.; Karnik, T.; Tadros, G.; Mau, B.; Ma, T.; Mu, Y.; New, J.; Louie, R.J.; et al. The Histone Demethylase KDM4B Regulates Peritoneal Seeding of Ovarian Cancer. Oncogene 2017, 36, 2565–2576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhao, L.; Zang, W.; Liu, Z.; Chen, L.; Liu, T.; Xu, D.; Jia, J. Histone Demethylase JMJD2B Is Required for Tumor Cell Proliferation and Survival and Is Overexpressed in Gastric Cancer. Biochem. Biophys. Res. Commun. 2011, 416, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, G.M.; Keri, R.A. FOXA1: A Transcription Factor with Parallel Functions in Development and Cancer. Biosci. Rep. 2012, 32, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Coppé, J.-P.; Desprez, P.-Y.; Krtolica, A.; Campisi, J. The Senescence-Associated Secretory Phenotype: The Dark Side of Tumor Suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Yan, Y.; Deb, S.; Rangasamy, D.; Germann, M.; Malaterre, J.; Eder, N.C.; Ward, R.L.; Hawkins, N.J.; Tothill, R.W.; et al. Cohesin Rad21 Mediates Loss of Heterozygosity and Is Upregulated via Wnt Promoting Transcriptional Dysregulation in Gastrointestinal Tumors. Cell Rep. 2014, 9, 1781–1797. [Google Scholar] [CrossRef] [Green Version]
- Ramos, K.S.; Moore, S.; Runge, I.; Tavera-Garcia, M.A.; Cascone, I.; Courty, J.; Reyes-Reyes, E.M. The Nucleolin Antagonist N6L Inhibits LINE1 Retrotransposon Activity in Non-Small Cell Lung Carcinoma Cells. J. Cancer 2020, 11, 733–740. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.-C.; Rothnagel, J.A.; Upton, K.R. Widespread Exaptation of L1 Transposons for Transcription Factor Binding in Breast Cancer. Int. J. Mol. Sci. 2021, 22, 5625. [Google Scholar] [CrossRef]
- Jacob-Hirsch, J.; Eyal, E.; Knisbacher, B.A.; Roth, J.; Cesarkas, K.; Dor, C.; Farage-Barhom, S.; Kunik, V.; Simon, A.J.; Gal, M.; et al. Whole-Genome Sequencing Reveals Principles of Brain Retrotransposition in Neurodevelopmental Disorders. Cell Res. 2018, 28, 187–203. [Google Scholar] [CrossRef] [Green Version]
- Bundo, M.; Toyoshima, M.; Okada, Y.; Akamatsu, W.; Ueda, J.; Nemoto-Miyauchi, T.; Sunaga, F.; Toritsuka, M.; Ikawa, D.; Kakita, A.; et al. Increased L1 Retrotransposition in the Neuronal Genome in Schizophrenia. Neuron 2014, 81, 306–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, G.A.; Crist, R.C.; Karatas, E.T.; Hammond, M.J.; Ewing, A.D.; Ferraro, T.N.; Hahn, C.-G.; Berrettini, W.H. Analysis of LINE-1 Elements in DNA from Postmortem Brains of Individuals with Schizophrenia. Neuropsychopharmacology 2017, 42, 2602–2611. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Shimada, T.; Otowa, T.; Wu, Y.-Y.; Kawamura, Y.; Tochigi, M.; Iwata, Y.; Umekage, T.; Toyota, T.; Maekawa, M.; et al. Genome-Wide Association Study of Autism Spectrum Disorder in the East Asian Populations. Autism. Res. 2016, 9, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.B.; Lee, V.M.-Y.; Trojanowski, J.Q. Gains or Losses: Molecular Mechanisms of TDP43-Mediated Neurodegeneration. Nat. Rev. Neurosci. 2011, 13, 38–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amador-Ortiz, C.; Lin, W.-L.; Ahmed, Z.; Personett, D.; Davies, P.; Duara, R.; Graff-Radford, N.R.; Hutton, M.L.; Dickson, D.W. TDP-43 Immunoreactivity in Hippocampal Sclerosis and Alzheimer’s Disease. Ann. Neurol. 2007, 61, 435–445. [Google Scholar] [CrossRef] [Green Version]
- Nakashima-Yasuda, H.; Uryu, K.; Robinson, J.; Xie, S.X.; Hurtig, H.; Duda, J.E.; Arnold, S.E.; Siderowf, A.; Grossman, M.; Leverenz, J.B.; et al. Co-Morbidity of TDP-43 Proteinopathy in Lewy Body Related Diseases. Acta Neuropathol. 2007, 114, 221–229. [Google Scholar] [CrossRef]
- Schwab, C.; Arai, T.; Hasegawa, M.; Yu, S.; McGeer, P.L. Colocalization of Transactivation-Responsive DNA-Binding Protein 43 and Huntingtin in Inclusions of Huntington Disease. J. Neuropathol. Exp. Neurol. 2008, 67, 1159–1165. [Google Scholar] [CrossRef] [Green Version]
- Weihl, C.C.; Temiz, P.; Miller, S.E.; Watts, G.; Smith, C.; Forman, M.; Hanson, P.I.; Kimonis, V.; Pestronk, A. TDP-43 Accumulation in Inclusion Body Myopathy Muscle Suggests a Common Pathogenic Mechanism with Frontotemporal Dementia. J. Neurol. Neurosurg. Psychiatry 2008, 79, 1186–1189. [Google Scholar] [CrossRef] [Green Version]
- Bose, J.K.; Huang, C.-C.; Shen, C.-K.J. Regulation of Autophagy by Neuropathological Protein TDP-43. J. Biol. Chem. 2011, 286, 44441–44448. [Google Scholar] [CrossRef] [Green Version]
- Mitra, J.; Guerrero, E.N.; Hegde, P.M.; Liachko, N.F.; Wang, H.; Vasquez, V.; Gao, J.; Pandey, A.; Taylor, J.P.; Kraemer, B.C.; et al. Motor Neuron Disease-Associated Loss of Nuclear TDP-43 Is Linked to DNA Double-Strand Break Repair Defects. Proc. Natl. Acad. Sci. USA 2019, 116, 4696–4705. [Google Scholar] [CrossRef] [Green Version]
- Liu, E.Y.; Russ, J.; Cali, C.P.; Phan, J.M.; Amlie-Wolf, A.; Lee, E.B. Loss of Nuclear TDP-43 Is Associated with Decondensation of LINE Retrotransposons. Cell Rep. 2019, 27, 1409–1421.e6. [Google Scholar] [CrossRef] [Green Version]
- Morera, A.A.; Ahmed, N.S.; Schwartz, J.C. TDP-43 Regulates Transcription at Protein-Coding Genes and Alu Retrotransposons. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194434. [Google Scholar] [CrossRef] [PubMed]
- Pereira, G.C.; Sanchez, L.; Schaughency, P.M.; Rubio-Roldán, A.; Choi, J.A.; Planet, E.; Batra, R.; Turelli, P.; Trono, D.; Ostrow, L.W.; et al. Properties of LINE-1 Proteins and Repeat Element Expression in the Context of Amyotrophic Lateral Sclerosis. Mob. DNA 2018, 9, 35. [Google Scholar] [CrossRef]
- Hadlock, K.G.; Miller, R.G.; Jin, X.; Yu, S.; Reis, J.; Mass, J.; Gelinas, D.; Zhang, J.; McGrath, M.S. Elevated Rates of Antibody Reactivity to HML-2/HERV-K but Not Other Endogenous Retroviruses in ALS. Neurology 2004, 5, A37–A38. [Google Scholar]
- Douville, R.; Liu, J.; Rothstein, J.; Nath, A. Identification of Active Loci of a Human Endogenous Retrovirus in Neurons of Patients with Amyotrophic Lateral Sclerosis. Ann. Neurol. 2011, 69, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Samimi, H.; Gamez, M.; Zare, H.; Frost, B. Pathogenic Tau-Induced PiRNA Depletion Promotes Neuronal Death through Transposable Element Dysregulation in Neurodegenerative Tauopathies. Nat. Neurosci. 2018, 21, 1038–1048. [Google Scholar] [CrossRef]
- Guo, C.; Jeong, H.-H.; Hsieh, Y.-C.; Klein, H.-U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018, 23, 2874–2880. [Google Scholar] [CrossRef] [PubMed]
- Ittner, L.M.; Götz, J. Amyloid-β and Tau--a Toxic Pas de Deux in Alzheimer’s Disease. Nat. Rev. Neurosci. 2011, 12, 65–72. [Google Scholar] [CrossRef]
- Rüb, U.; Stratmann, K.; Heinsen, H.; Seidel, K.; Bouzrou, M.; Korf, H.-W. Alzheimer’s Disease: Characterization of the Brain Sites of the Initial Tau Cytoskeletal Pathology Will Improve the Success of Novel Immunological Anti-Tau Treatment Approaches. J. Alzheimers Dis. 2017, 57, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Weingarten, M.D.; Lockwood, A.H.; Hwo, S.Y.; Kirschner, M.W. A Protein Factor Essential for Microtubule Assembly. Proc. Natl. Acad. Sci. USA 1975, 72, 1858–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nizynski, B.; Dzwolak, W.; Nieznanski, K. Amyloidogenesis of Tau Protein. Protein Sci. 2017, 26, 2126–2150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ittner, A.; Ittner, L.M. Dendritic Tau in Alzheimer’s Disease. Neuron 2018, 99, 13–27. [Google Scholar] [CrossRef] [Green Version]
- Hayflick, S.J.; Kurian, M.A.; Hogarth, P. Neurodegeneration with Brain Iron Accumulation. Handb. Clin. Neurol. 2018, 147, 293–305. [Google Scholar] [CrossRef]
- Stanga, S.; Caretto, A.; Boido, M.; Vercelli, A. Mitochondrial Dysfunctions: A Red Thread across Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 3719. [Google Scholar] [CrossRef]
- La Morgia, C.; Maresca, A.; Caporali, L.; Valentino, M.L.; Carelli, V. Mitochondrial Diseases in Adults. J. Intern Med. 2020, 287, 592–608. [Google Scholar] [CrossRef]
- Missiroli, S.; Genovese, I.; Perrone, M.; Vezzani, B.; Vitto, V.A.M.; Giorgi, C. The Role of Mitochondria in Inflammation: From Cancer to Neurodegenerative Disorders. J. Clin. Med. 2020, 9, 740. [Google Scholar] [CrossRef] [Green Version]
- Baeken, M.W.; Moosmann, B.; Hajieva, P. Retrotransposon Activation by Distressed Mitochondria in Neurons. Biochem. Biophys. Res. Commun. 2020, 525, 570–575. [Google Scholar] [CrossRef]
- Giorgi, G.; Marcantonio, P.; Del Re, B. LINE-1 Retrotransposition in Human Neuroblastoma Cells Is Affected by Oxidative Stress. Cell Tissue Res. 2011, 346, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Whongsiri, P.; Pimratana, C.; Wijitsettakul, U.; Sanpavat, A.; Jindatip, D.; Hoffmann, M.J.; Goering, W.; Schulz, W.A.; Boonla, C. Oxidative Stress and LINE-1 Reactivation in Bladder Cancer Are Epigenetically Linked through Active Chromatin Formation. Free Radic Biol. Med. 2019, 134, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.-C.; Wu, P.-C.; Chung, I.-F.; Jiang, J.-H.; Fann, M.-J.; Kao, L.-S. Cell Death Caused by the Synergistic Effects of Zinc and Dopamine Is Mediated by a Stress Sensor Gene Gadd45b—Implication in the Pathogenesis of Parkinson’s Disease. J. Neurochem. 2016, 139, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Ravel-Godreuil, C.; Massiani-Beaudoin, O.; Mailly, P.; Prochiantz, A.; Joshi, R.L.; Fuchs, J. Perturbed DNA Methylation by Gadd45b Induces Chromatin Disorganization, DNA Strand Breaks and Dopaminergic Neuron Death. iScience 2021, 24, 102756. [Google Scholar] [CrossRef] [PubMed]
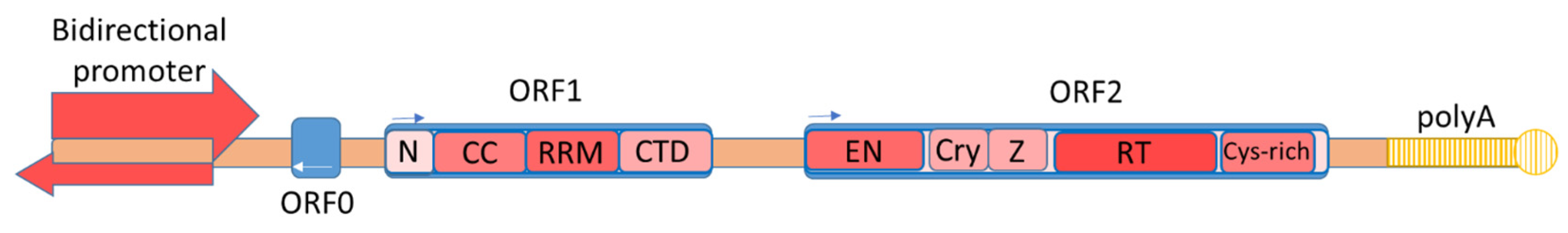
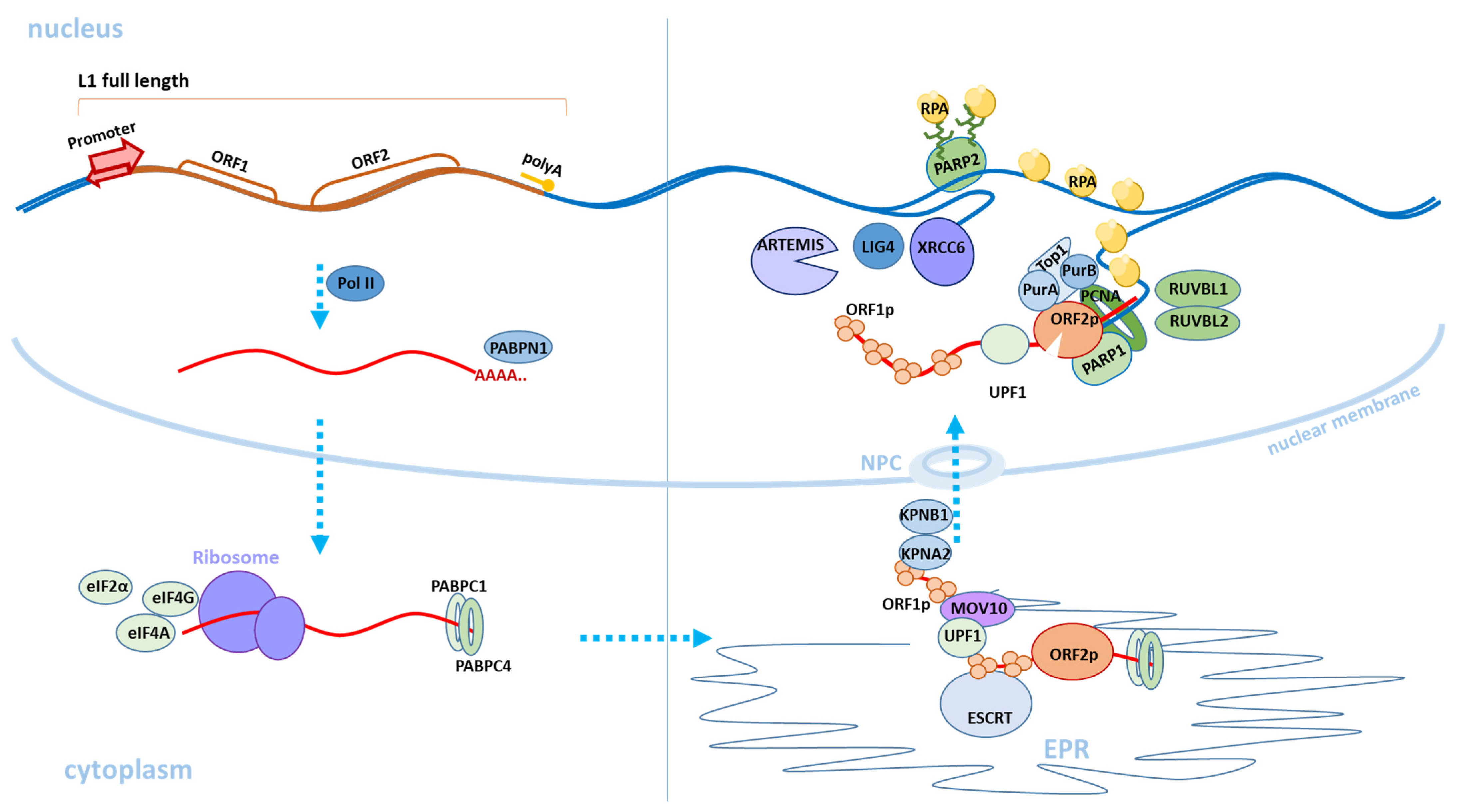
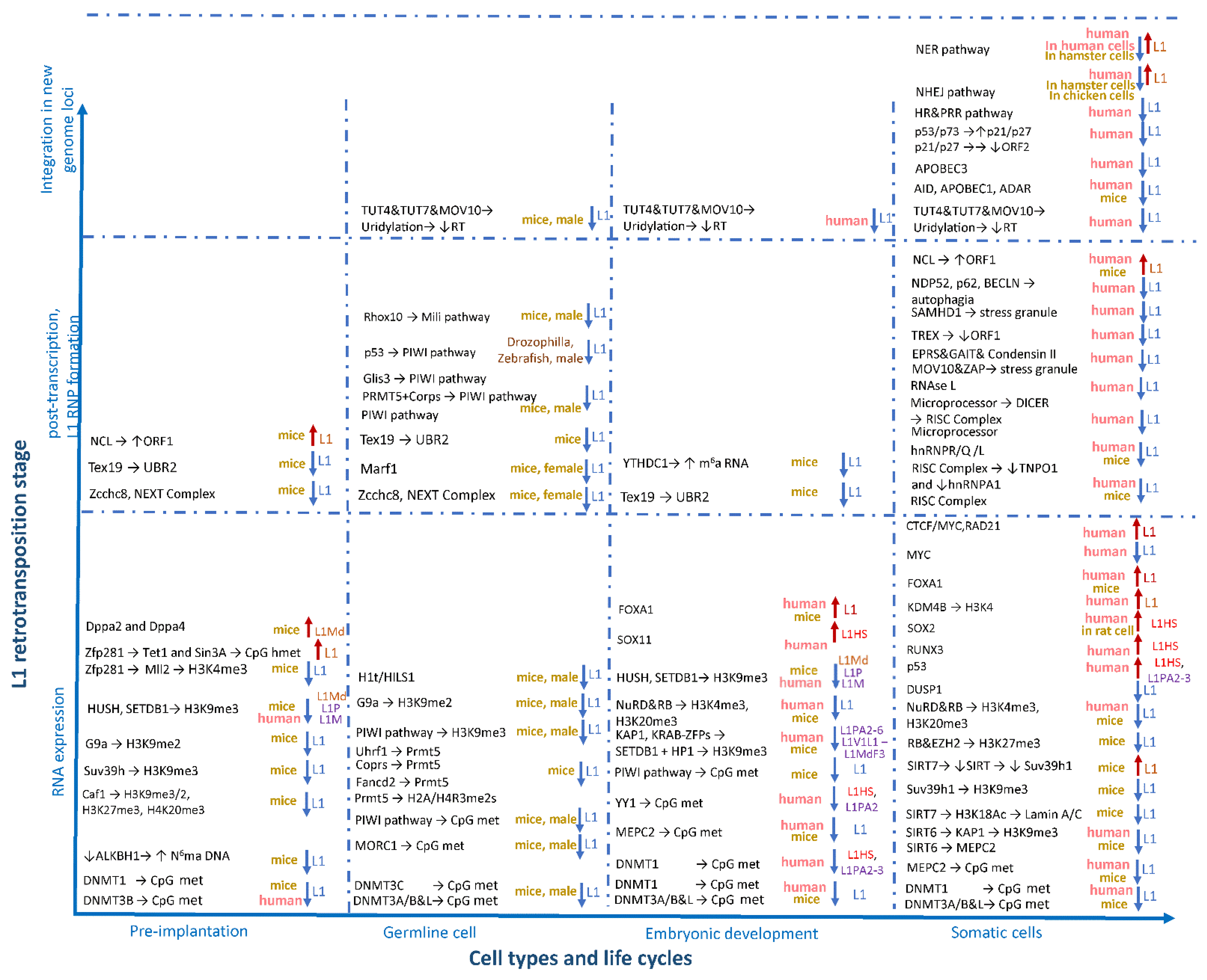
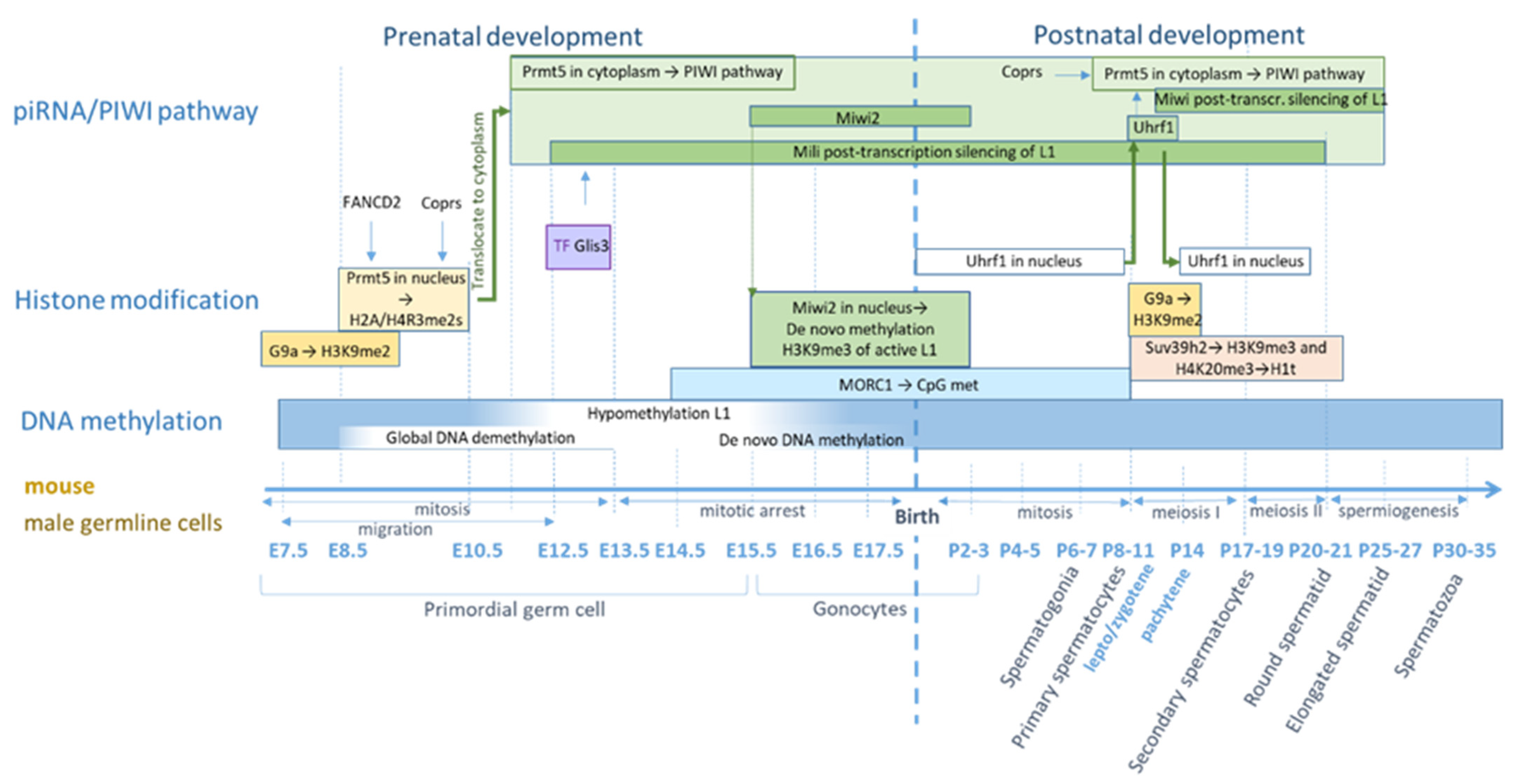
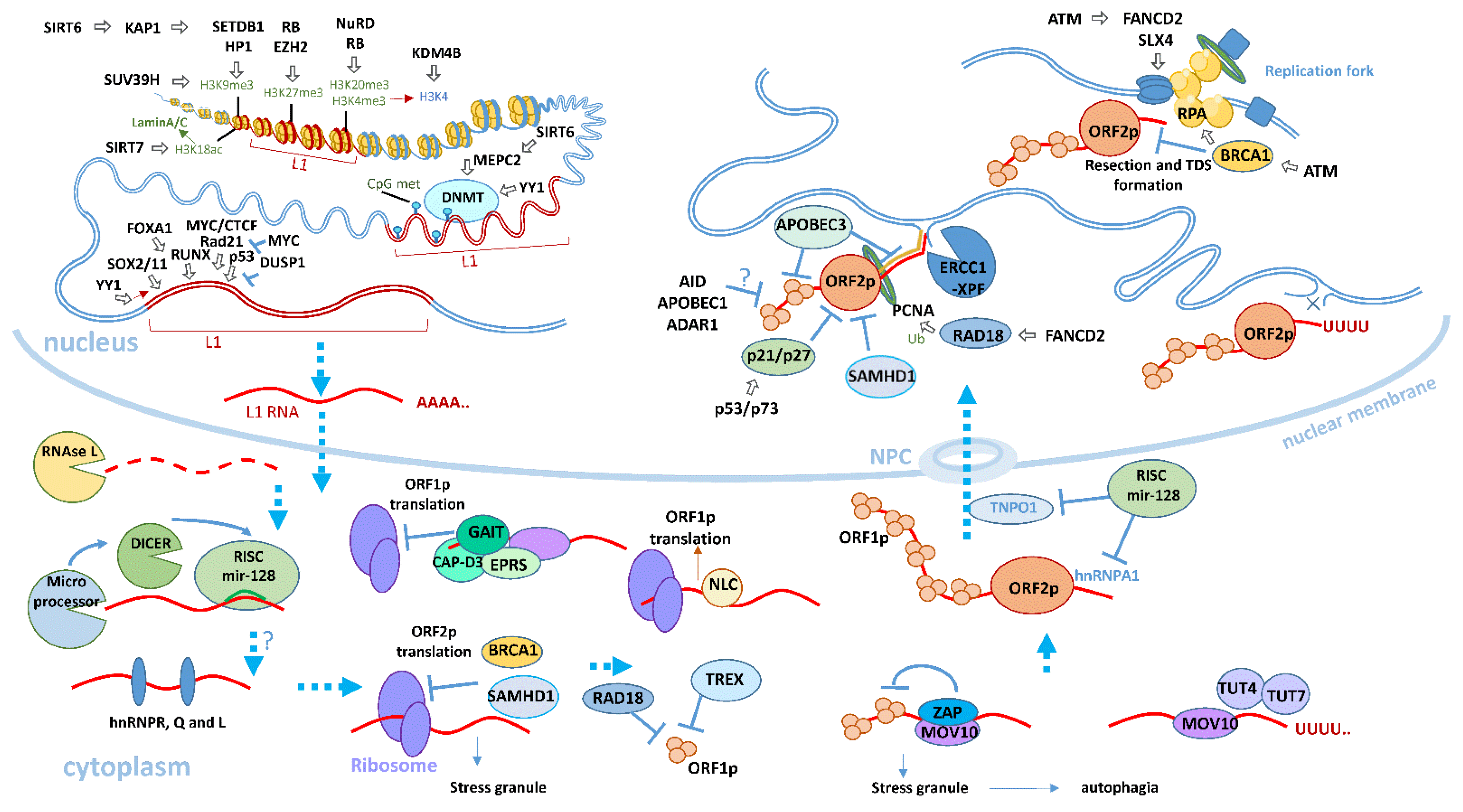
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Protasova, M.S.; Andreeva, T.V.; Rogaev, E.I. Factors Regulating the Activity of LINE1 Retrotransposons. Genes 2021, 12, 1562. https://doi.org/10.3390/genes12101562
Protasova MS, Andreeva TV, Rogaev EI. Factors Regulating the Activity of LINE1 Retrotransposons. Genes. 2021; 12(10):1562. https://doi.org/10.3390/genes12101562
Chicago/Turabian StyleProtasova, Maria Sergeevna, Tatiana Vladimirovna Andreeva, and Evgeny Ivanovich Rogaev. 2021. "Factors Regulating the Activity of LINE1 Retrotransposons" Genes 12, no. 10: 1562. https://doi.org/10.3390/genes12101562
APA StyleProtasova, M. S., Andreeva, T. V., & Rogaev, E. I. (2021). Factors Regulating the Activity of LINE1 Retrotransposons. Genes, 12(10), 1562. https://doi.org/10.3390/genes12101562





