Identification of Chicken Transglutaminase 1 and In Situ Localization of Transglutaminase Activity in Avian Skin and Esophagus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Transglutaminase In Situ Activity Assay
2.3. RNA Preparation and RT-PCR
2.4. Chicken TGM1 Sequence Assembly In Silico
2.5. Molecular Phylogenetics
3. Results
3.1. Identification of Chicken TGM1
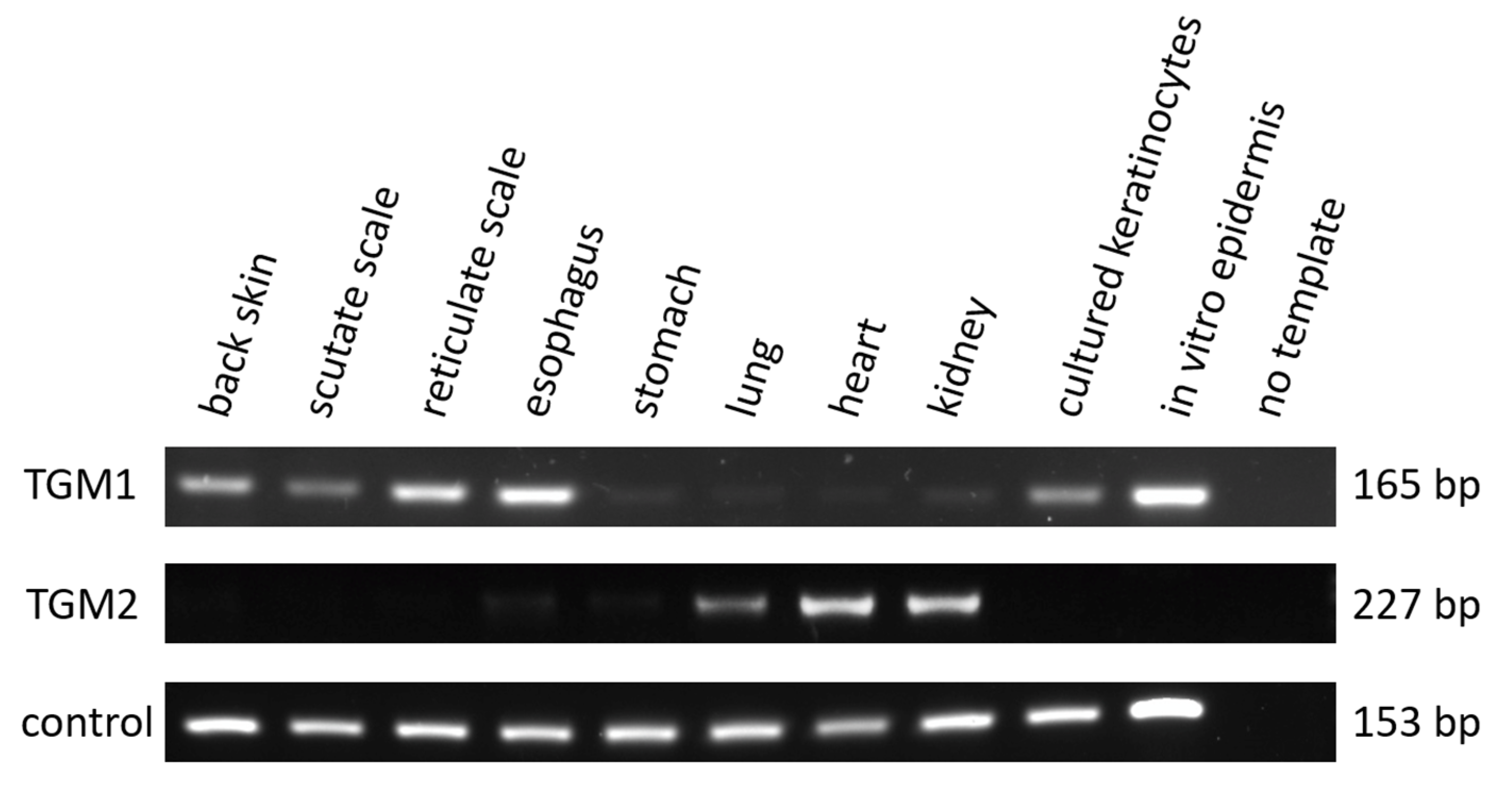
3.2. TGM1 Is Expressed in the Epidermis and Esophagus of the Chicken
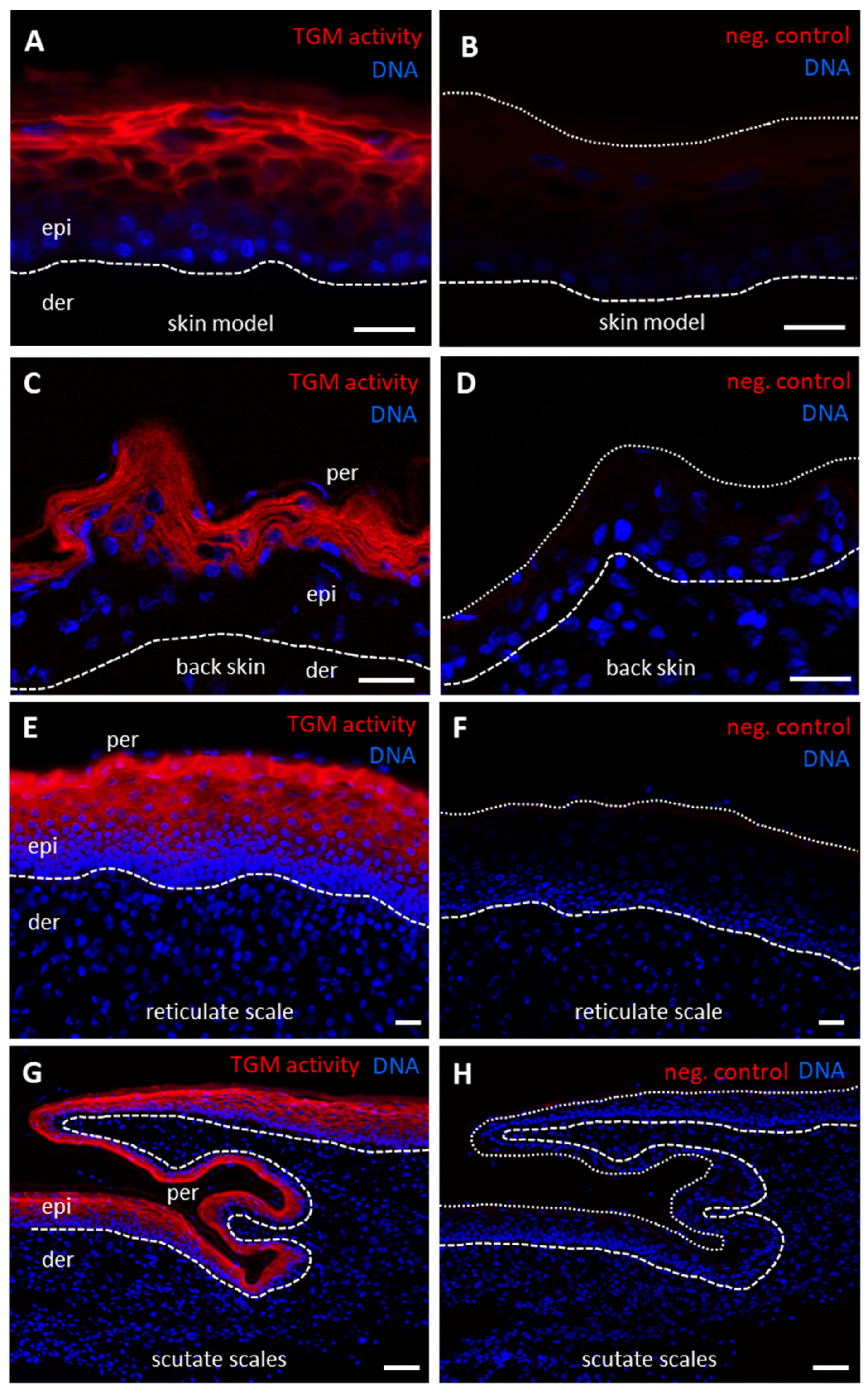
3.3. TGM Activity Is Present in Differentiated Epithelial Cells of Chicken Skin and Esophagus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Candi, E.; Schmidt, R.; Melino, G. The cornified envelope: A model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 2005, 6, 328–340. [Google Scholar] [CrossRef]
- Eckhart, L.; Lippens, S.; Tschachler, E.; Declercq, W. Cell death by cornification. Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1833, 3471–3480. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Amagai, M. Dissecting the formation, structure and barrier function of the stratum corneum. Int. Immunol. 2015, 27, 269–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mischke, D.; Korge, B.P.; Marenholz, I.; Volz, A.; Ziegler, A. Genes Encoding Structural Proteins of Epidermal Cornification and S100 Calcium-Binding Proteins Form a Gene Complex (“Epidermal Differentiation Complex”) on Human Chromosome 1q21. J. Investig. Dermatol. 1996, 106, 989–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, J.; Toulza, E.; Hsu, C.Y.; Pellerin, L.; Balica, S.; Mazereeuw-Hautier, J.; Paul, C.; Serre, G.; Jonca, N.; Simon, M. Update on the epidermal differentiation complex. Front. Biosci. 2012, 17, 1517–1532. [Google Scholar] [CrossRef] [Green Version]
- Kypriotou, M.; Huber, M.; Hohl, D. The human epidermal differentiation complex: Cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp. Dermatol. 2012, 21, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Vanhoutteghem, A.; Djian, P.; Green, H. Ancient origin of the gene encoding involucrin, a precursor of the cross-linked envelope of epidermis and related epithelia. Proc. Natl. Acad. Sci. USA 2008, 105, 15481–15486. [Google Scholar] [CrossRef] [Green Version]
- Strasser, B.; Mlitz, V.; Hermann, M.; Rice, R.H.; Eigenheer, R.A.; Alibardi, L.; Tschachler, E.; Eckhart, L. Evolutionary Origin and Diversification of Epidermal Barrier Proteins in Amniotes. Mol. Biol. Evol. 2014, 31, 3194–3205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holthaus, K.B.; Strasser, B.; Sipos, W.; Schmidt, H.; Mlitz, V.; Sukseree, S.; Weissenbacher, A.; Tschachler, E.; Alibardi, L.; Eckhart, L. Comparative Genomics Identifies Epidermal Proteins Associated with the Evolution of the Turtle Shell. Mol. Biol. Evol. 2015, 33, 726–737. [Google Scholar] [CrossRef]
- Holthaus, K.B.; Mlitz, V.; Strasser, B.; Tschachler, E.; Alibardi, L.; Eckhart, L. Identification and comparative analysis of the epidermal differentiation complex in snakes. Sci. Rep. 2017, 7, 45338. [Google Scholar] [CrossRef] [Green Version]
- Holthaus, K.B.; Strasser, B.; Lachner, J.; Sukseree, S.; Sipos, W.; Weissenbacher, A.; Tschachler, E.; Alibardi, L.; Eckhart, L. Comparative Analysis of Epidermal Differentiation Genes of Crocodilians Suggests New Models for the Evolutionary Origin of Avian Feather Proteins. Genome Biol. Evol. 2018, 10, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Holthaus, K.B.; Alibardi, L.; Tschachler, E.; Eckhart, L. Identification of epidermal differentiation genes of the tuatara pro-vides insights into the early evolution of lepidosaurian skin. Sci. Rep. 2020, 10, 12844. [Google Scholar] [CrossRef]
- Alibardi, L. Vertebrate keratinization evolved into cornification mainly due to transglutaminase and sulfhydryl oxidase activities on epidermal proteins: An immunohistochemical survey. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2021. [Google Scholar] [CrossRef]
- Eckert, R.L.; Kaartinen, M.T.; Nurminskaya, M.; Belkin, A.M.; Colak, G.; Johnson, G.V.; Mehta, K. Transglutaminase regu-lation of cell function. Physiol. Rev. 2014, 94, 383–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassidy, A.; van Steensel, M.; Steijlen, P.M.; van Geel, M.; van der Velden, J.; Morley, S.M.; Terrinoni, A.; Melino, G.; Candi, E.; McLean, W.I. A Homozygous Missense Mutation in TGM5 Abolishes Epidermal Transglutaminase 5 Activity and Causes Acral Peeling Skin Syndrome. Am. J. Hum. Genet. 2005, 77, 909–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chermnykh, E.S.; Alpeeva, E.V.; Vorotelyak, E.A. Transglutaminase 3: The Involvement in Epithelial Differentiation and Cancer. Cells 2020, 9, 1996. [Google Scholar] [CrossRef] [PubMed]
- Grenard, P.; Bates, M.K.; Aeschlimann, D. Evolution of Transglutaminase Genes: Identification of a Transglutaminase Gene Cluster on Human Chromosome 15q15. J. Biol. Chem. 2001, 276, 33066–33078. [Google Scholar] [CrossRef] [Green Version]
- Russell, L.J.; DiGiovanna, J.J.; Rogers, G.R.; Steinert, P.M.; Hashem, N.; Compton, J.G.; Bale, S.J. Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat. Genet. 1995, 9, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, M.; Yamashita, F.; Ishida-Yamamoto, A.; Yamada, K.; Kinoshita, C.; Fushiki, S.; Ueda, E.; Morishima, Y.; Tabata, K.; Yasuno, H.; et al. Defective stratum corneum and early neonatal death in mice lacking the gene for transglutaminase 1 (keratinocyte transglutaminase). Proc. Natl. Acad. Sci. USA 1998, 95, 1044–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuramoto, N.; Takizawa, T.; Takizawa, T.; Matsuki, M.; Morioka, H.; Robinson, J.M.; Yamanishi, K. Development of ich-thyosiform skin compensates for defective permeability barrier function in mice lacking transglutaminase 1. J. Clin. Investig. 2002, 109, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Michel, S.; Bernerd, F.; Jetten, A.M.; E Floyd, E.; Shroot, B.; Reichert, U. Expression of Keratinocyte Transglutamine mRNA Revealed by In Situ Hybridization. J. Investig. Dermatol. 1992, 98, 364–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contzler, R.; Favre, B.; Huber, M.; Hohl, D. Cornulin, a new member of the “fused gene” family, is expressed during epi-dermal differentiation. J. Investig. Dermatol. 2005, 124, 990–997. [Google Scholar] [CrossRef] [Green Version]
- Heimann, R.; Rice, R.H. Rat esophageal and epidermal keratinocytes: Intrinsic differences in culture and derivation of con-tinuous lines. J. Cell Physiol. 1983, 117, 362–367. [Google Scholar] [CrossRef] [Green Version]
- Cruz, S.I.R.; Phillips, M.A.; Kültz, D.; Rice, R.H. Tgm1-like transglutaminases in tilapia (Oreochromis mossambicus). PLoS ONE 2017, 12, e0177016. [Google Scholar] [CrossRef]
- Kikuta, A.; Furukawa, E.; Ogawa, R.; Suganuma, N.; Saitoh, M.; Nishimaki, T.; Katsumura, T.; Oota, H.; Kawamoto, T.; Tatsukawa, H.; et al. Biochemical Characterization of Medaka (Oryzias latipes) Transglutaminases, OlTGK1 and OlTGK2, as Orthologues of Human Keratinocyte-Type Transglutaminase. PLoS ONE 2015, 10, e0144194. [Google Scholar] [CrossRef] [Green Version]
- Obinata, A.; Endo, H.; Obinata, H.E.A. Induction of epidermal transglutaminase by hydrocortisone in chick embryonic skin. Nature 1977, 270, 440–441. [Google Scholar] [CrossRef] [PubMed]
- Obinata, A.; Endo, H. Induction of chick epidermal transglutaminase by hydrocortisone in ovo and in vitro with reference to the differentiation of epidermal cells. J. Biol. Chem. 1979, 254, 8487–8490. [Google Scholar] [CrossRef]
- Vanhoutteghem, A.; Londero, T.; Ghinea, N.; Djian, P. Serial cultivation of chicken keratinocytes, a composite cell type that accumulates lipids and synthesizes a novel beta-keratin. Differentiation 2004, 72, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Greenwold, M.J.; Sawyer, R.H. Genomic organization and molecular phylogenies of the beta (beta) keratin multigene family in the chicken (Gallus gallus) and zebra finch (Taeniopygia guttata), implications for feather evolution. BMC Evol. Biol. 2010, 10, 148. [Google Scholar] [CrossRef] [Green Version]
- Rice, R.H.; Winters, B.R.; Durbin-Johnson, B.P.; Rocke, D. Chicken Corneocyte Cross-Linked Proteome. J. Proteome Res. 2013, 12, 771–776. [Google Scholar] [CrossRef]
- Alibardi, L.; Holthaus, K.B.; Sukseree, S.; Hermann, M.; Tschachler, E.; Eckhart, L. Immunolocalization of a Histidine-Rich Epidermal Differentiation Protein in the Chicken Supports the Hypothesis of an Evolutionary Developmental Link between the Embryonic Subperiderm and Feather Barbs and Barbules. PLoS ONE 2016, 11, e0167789. [Google Scholar] [CrossRef]
- Davis, A.C.; Greenwold, M.J.; Sawyer, R.H. Complex Gene Loss and Duplication Events Have Facilitated the Evolution of Multiple Loricrin Genes in Diverse Bird Species. Genome Biol. Evol. 2019, 11, 984–1001. [Google Scholar] [CrossRef] [PubMed]
- Lachner, J.; Ehrlich, F.; Mlitz, V.; Hermann, M.; Alibardi, L.; Tschachler, E.; Eckhart, L. Immunolocalization and phylogenetic profiling of the feather protein with the highest cysteine content. Protoplasma 2019, 256, 1257–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alibardi, L.; Eckhart, L. Immunolocalization of epidermal differentiation complex proteins reveals distinct molecular com-positions of cells that control structure and mechanical properties of avian skin appendages. J. Morphol. 2021, 282, 917–933. [Google Scholar] [CrossRef]
- Mlitz, V.; Strasser, B.; Jaeger, K.; Hermann, M.; Ghannadan, M.; Buchberger, M.; Alibardi, L.; Tschachler, E.; Eckhart, L. Trichohyalin-Like Proteins Have Evolutionarily Conserved Roles in the Morphogenesis of Skin Appendages. J. Investig. Dermatol. 2014, 134, 2685–2692. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.; Greenwold, M. Evolution of an Epidermal Differentiation Complex (EDC) Gene Family in Birds. Genes 2021, 12, 767. [Google Scholar] [CrossRef]
- Mlitz, V.; Hermann, M.; Buchberger, M.; Tschachler, E.; Eckhart, L. The Trichohyalin-Like Protein Scaffoldin Is Expressed in the Multilayered Periderm during Development of Avian Beak and Egg Tooth. Genes 2021, 12, 248. [Google Scholar] [CrossRef]
- Feng, S.; Stiller, J.; Deng, Y.; Armstrong, J.; Fang, Q.; Reeve, A.H.; Xie, D.; Chen, G.; Guo, C.; Faircloth, B.C.; et al. Dense sampling of bird diversity increases power of comparative genomics. Nature 2020, 587, 252–257. [Google Scholar] [CrossRef]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef]
- Lachner, J.; Derdak, S.; Mlitz, V.; Wagner, T.; Holthaus, K.B.; Ehrlich, F.; Mildner, M.; Tschachler, E.; Eckhart, L. An in vitro model of avian skin reveals evolutionarily conserved transcriptional regulation of epidermal barrier formation. J. Investig. Dermatol. 2021. [Google Scholar] [CrossRef]
- Raghunath, M.; Hennies, H.C.; Velten, F.; Wiebe, V.; Steinert, P.M.; Reis, A.; Traupe, H. A novel in situ method for the detection of deficient transglutaminase activity in the skin. Arch. Dermatol. Res. 1998, 290, 621–627. [Google Scholar] [CrossRef]
- Hohl, D.; Aeschlimann, D.; Huber, M. In vitro and rapid in situ transglutaminase assays for congenital ichthyoses—A com-parative study. J. Investig. Dermatol. 1998, 110, 268–271. [Google Scholar] [CrossRef] [Green Version]
- Cau, L.; Pendaries, V.; Lhuillier, E.; Thompson, P.R.; Serre, G.; Takahara, H.; Méchin, M.-C.; Simon, M. Lowering relative humidity level increases epidermal protein deimination and drives human filaggrin breakdown. J. Dermatol. Sci. 2017, 86, 106–113. [Google Scholar] [CrossRef] [Green Version]
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques 1993, 15, 532–536. [Google Scholar]
- Lachner, J.; Mlitz, V.; Tschachler, E.; Eckhart, L. Epidermal cornification is preceded by the expression of a keratino-cyte-specific set of pyroptosis-related genes. Sci. Rep. 2017, 7, 17446. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akaike, H. Information Theory and an Extension of the Maximum Likelihood Principle. In Selected Papers of Hirotugu Akaike; Parzen, E., Tanabe, K., Kitagawa, G., Eds.; Springer Series in Statistics (Perspectives in Statistics): New York, NY, USA, 1998; pp. 199–213. [Google Scholar]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarova, K.S.; Aravind, L.; Koonin, E.V. A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases. Protein Sci. 1999, 8, 1714–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawyer, R.H.; Knapp, L.W.; O’Guin, M.W. The Skin of Birds. Epidermis, dermis and appendages. In Biology of the Integument; Bereither-Hahn, J., Matoltsy, A.G., Sylvia-Richards, R., Eds.; Springer: Berlin, Germany, 1986; Volume 2 Ver-tebrates, pp. 194–238. [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.-C.; Wu, P.; Lin, G.-W.; Chen, C.-K.; Yeh, C.-Y.; Tsai, S.; Yan, J.; Jiang, T.-X.; Lai, Y.-C.; Huang, D.; et al. Folding Keratin Gene Clusters during Skin Regional Specification. Dev. Cell 2020, 53, 561–576.e9. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Y.; Lin, K.-R.; Chen, Y.-J.; Chiang, Y.-J.; Ho, K.-C.; Shen, L.-F.; Song, I.-W.; Liu, K.-M.; Yang-Yen, H.-F.; Chen, Y.-T.; et al. Palmitoyl Acyltransferase Activity of ZDHHC13 Regulates Skin Barrier Development Partly by Controlling PADi3 and TGM1 Protein Stability. J. Investig. Dermatol. 2020, 140, 959–970.e3. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sachslehner, A.P.; Surbek, M.; Lachner, J.; Paudel, S.; Eckhart, L. Identification of Chicken Transglutaminase 1 and In Situ Localization of Transglutaminase Activity in Avian Skin and Esophagus. Genes 2021, 12, 1565. https://doi.org/10.3390/genes12101565
Sachslehner AP, Surbek M, Lachner J, Paudel S, Eckhart L. Identification of Chicken Transglutaminase 1 and In Situ Localization of Transglutaminase Activity in Avian Skin and Esophagus. Genes. 2021; 12(10):1565. https://doi.org/10.3390/genes12101565
Chicago/Turabian StyleSachslehner, Attila Placido, Marta Surbek, Julia Lachner, Surya Paudel, and Leopold Eckhart. 2021. "Identification of Chicken Transglutaminase 1 and In Situ Localization of Transglutaminase Activity in Avian Skin and Esophagus" Genes 12, no. 10: 1565. https://doi.org/10.3390/genes12101565
APA StyleSachslehner, A. P., Surbek, M., Lachner, J., Paudel, S., & Eckhart, L. (2021). Identification of Chicken Transglutaminase 1 and In Situ Localization of Transglutaminase Activity in Avian Skin and Esophagus. Genes, 12(10), 1565. https://doi.org/10.3390/genes12101565






