Ectopic Overexpression of Maize Heat Stress Transcription Factor ZmHsf05 Confers Drought Tolerance in Transgenic Rice
Abstract
:1. Introduction
2. Materials and Methods
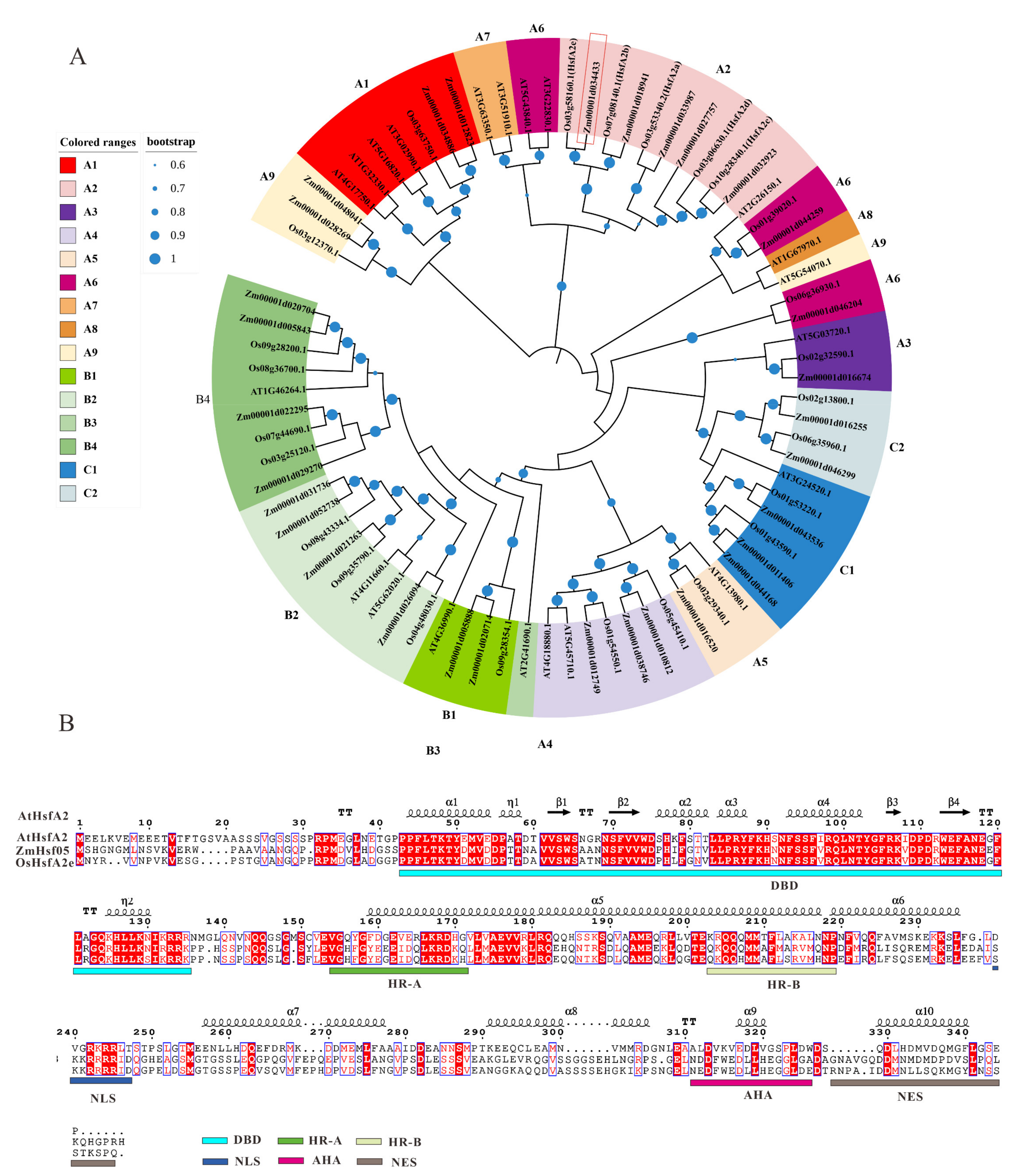
2.1. Gene Structural Features and Protein Sequence Alignments
2.2. Plant Materials, Growth Conditions, and Transformation
2.3. Total RNA Extraction and RT-qPCR Analysis
2.4. Phenotypic Analysis
2.5. Measurement of Physiological Indices
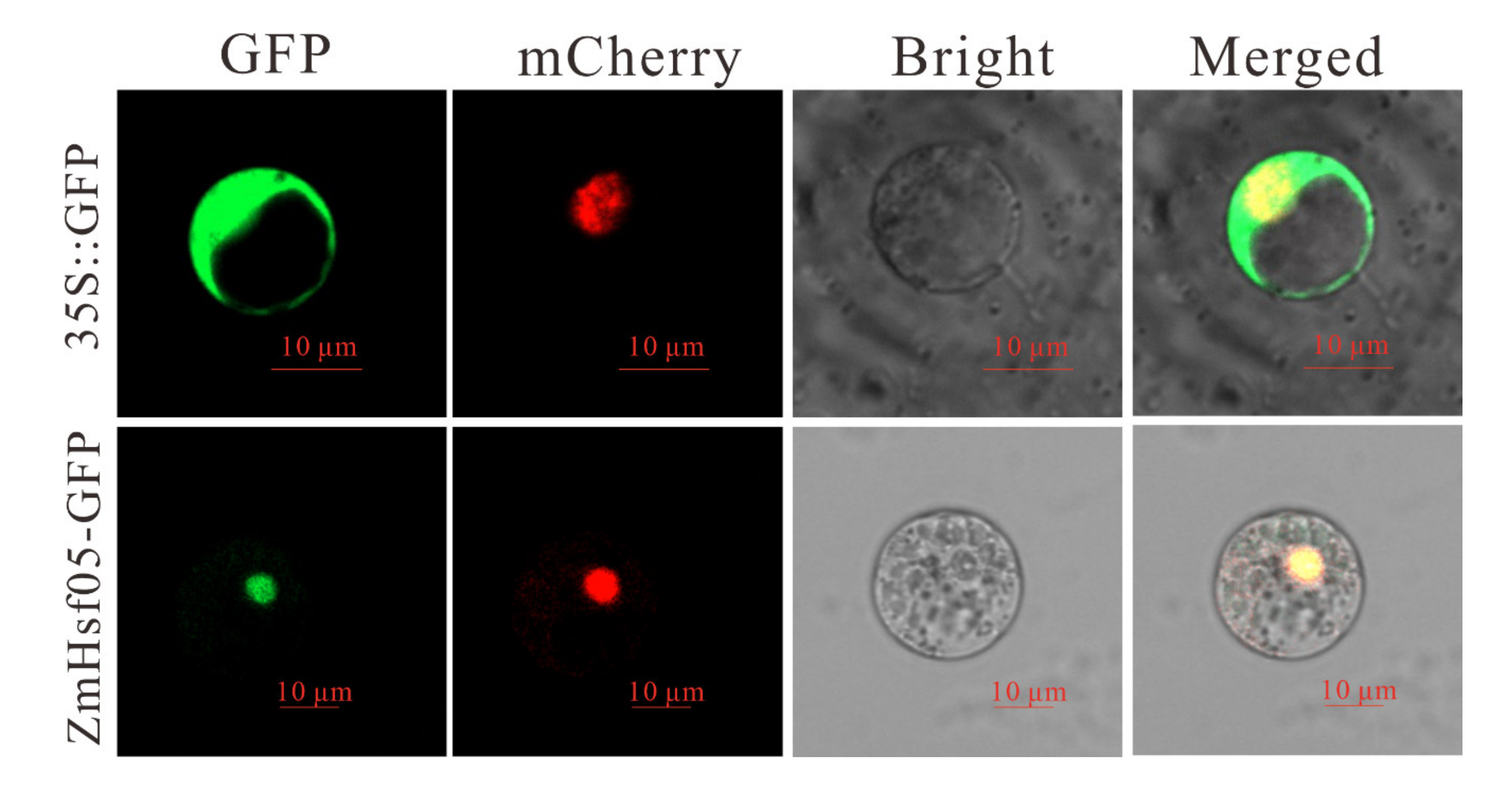
2.6. Subcellular Localization Analysis
2.7. Transactivation Activity Assays
2.8. Statistical Analysis
3. Results
3.1. Molecular Characterization of ZmHsf05 Sequence
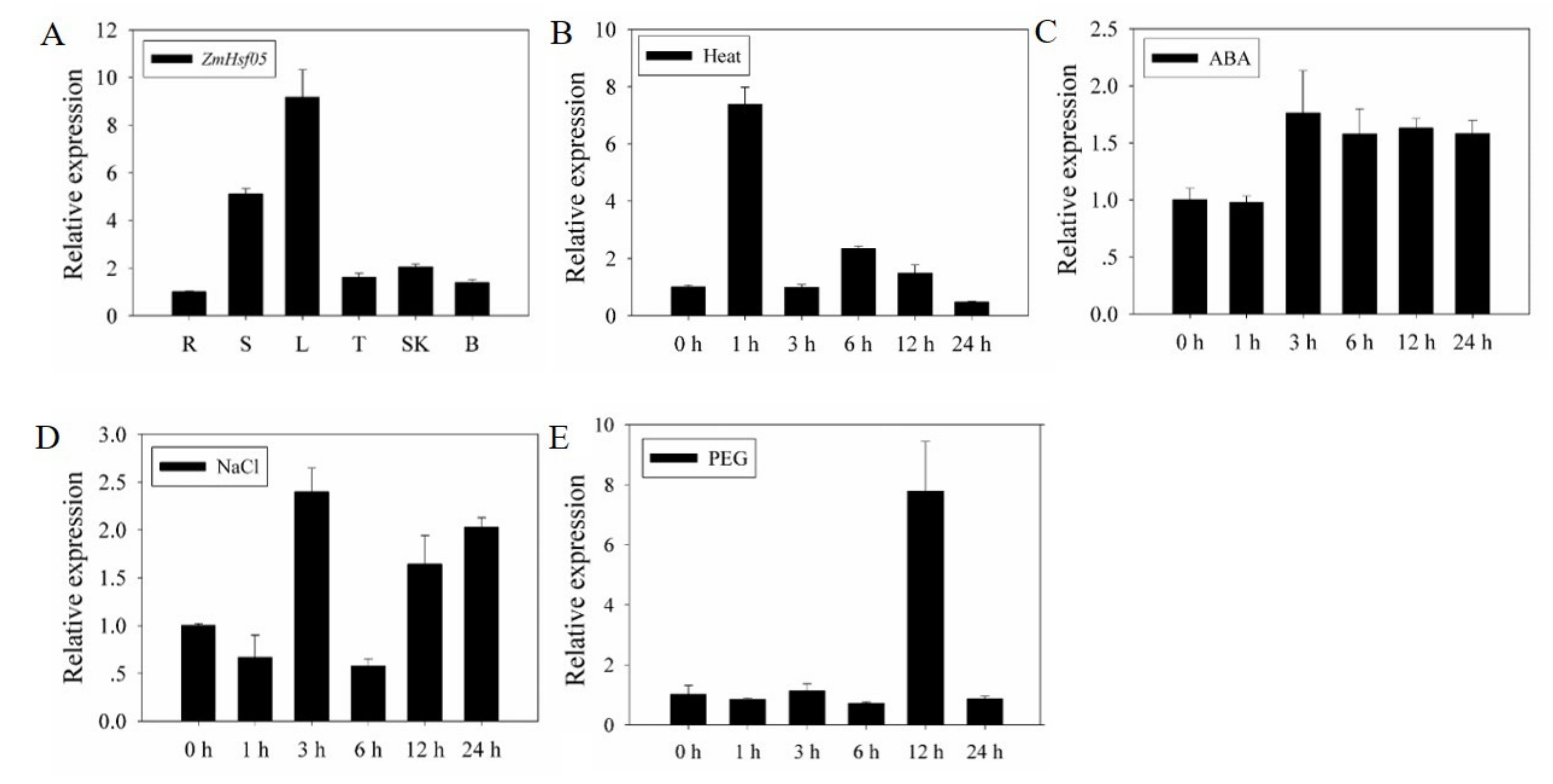
3.2. Tissue-Specific and Stress-Induced Expression Pattern of ZmHsf05
3.3. Subcellular Location Analysis of ZmHsf05
3.4. ZmHsf05 Has Transactivation Activity in Yeast
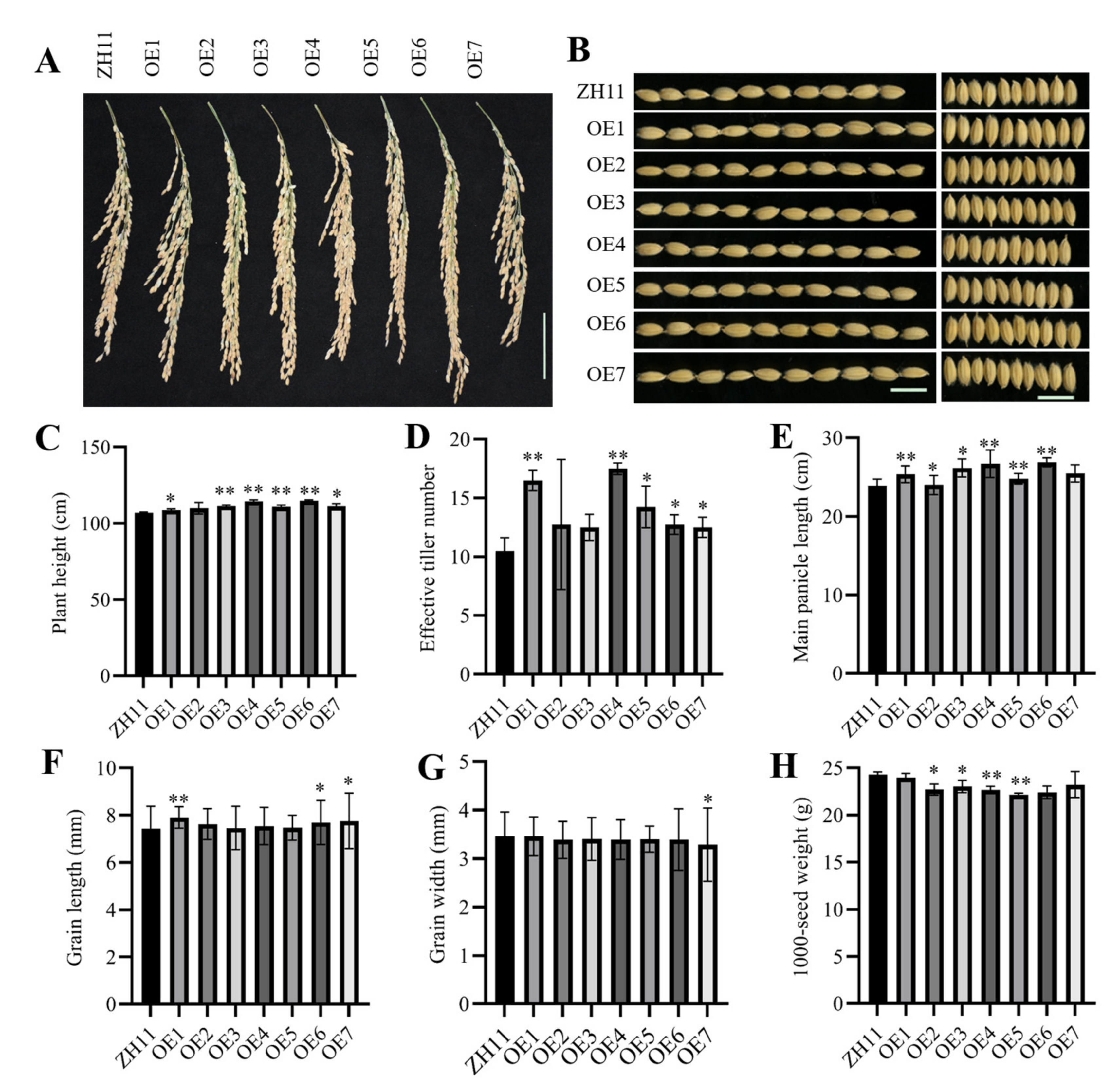
3.5. Overexpression of ZmHsf05 in Rice Alters Agronomical Traits in the Field Trials
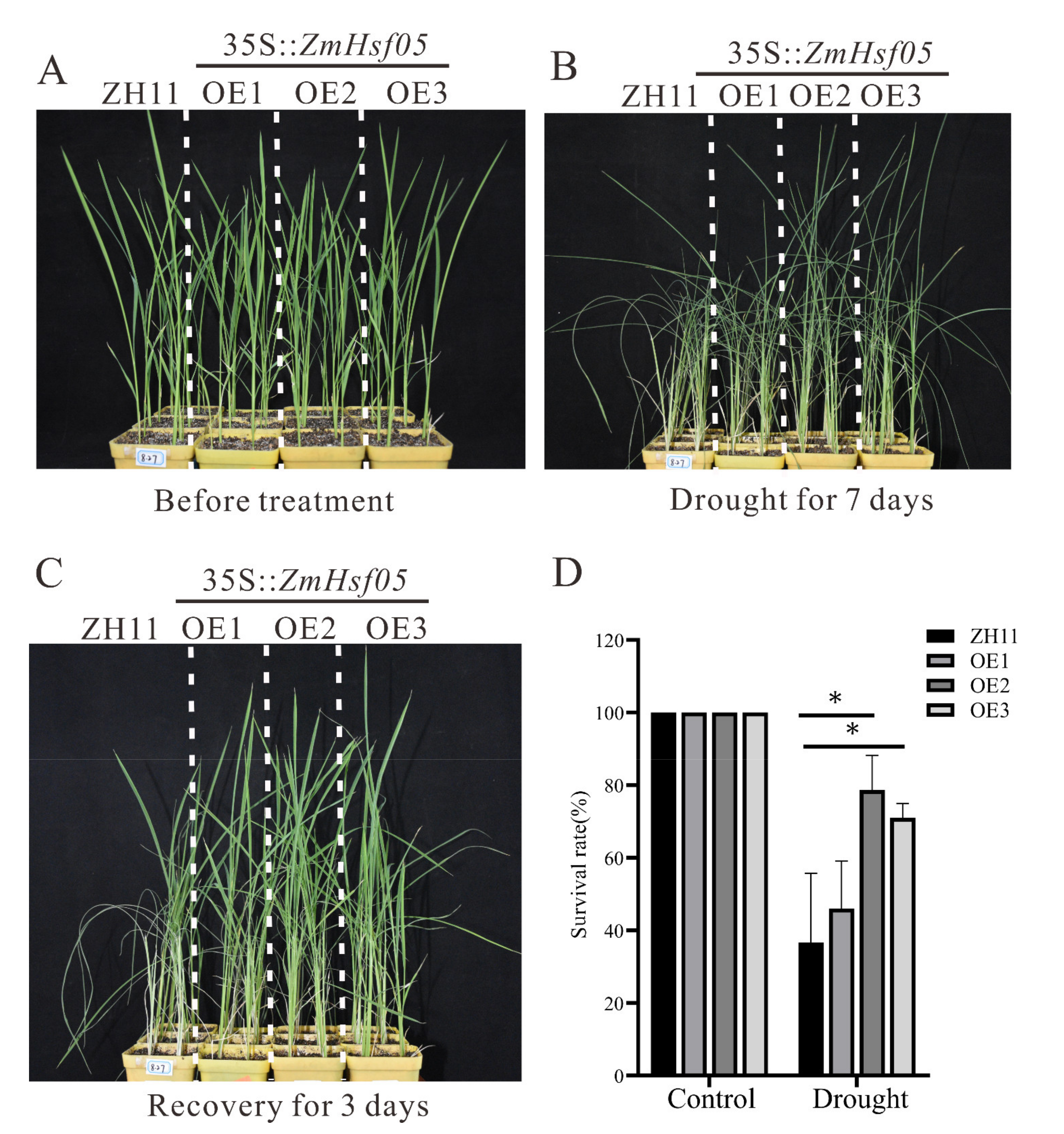
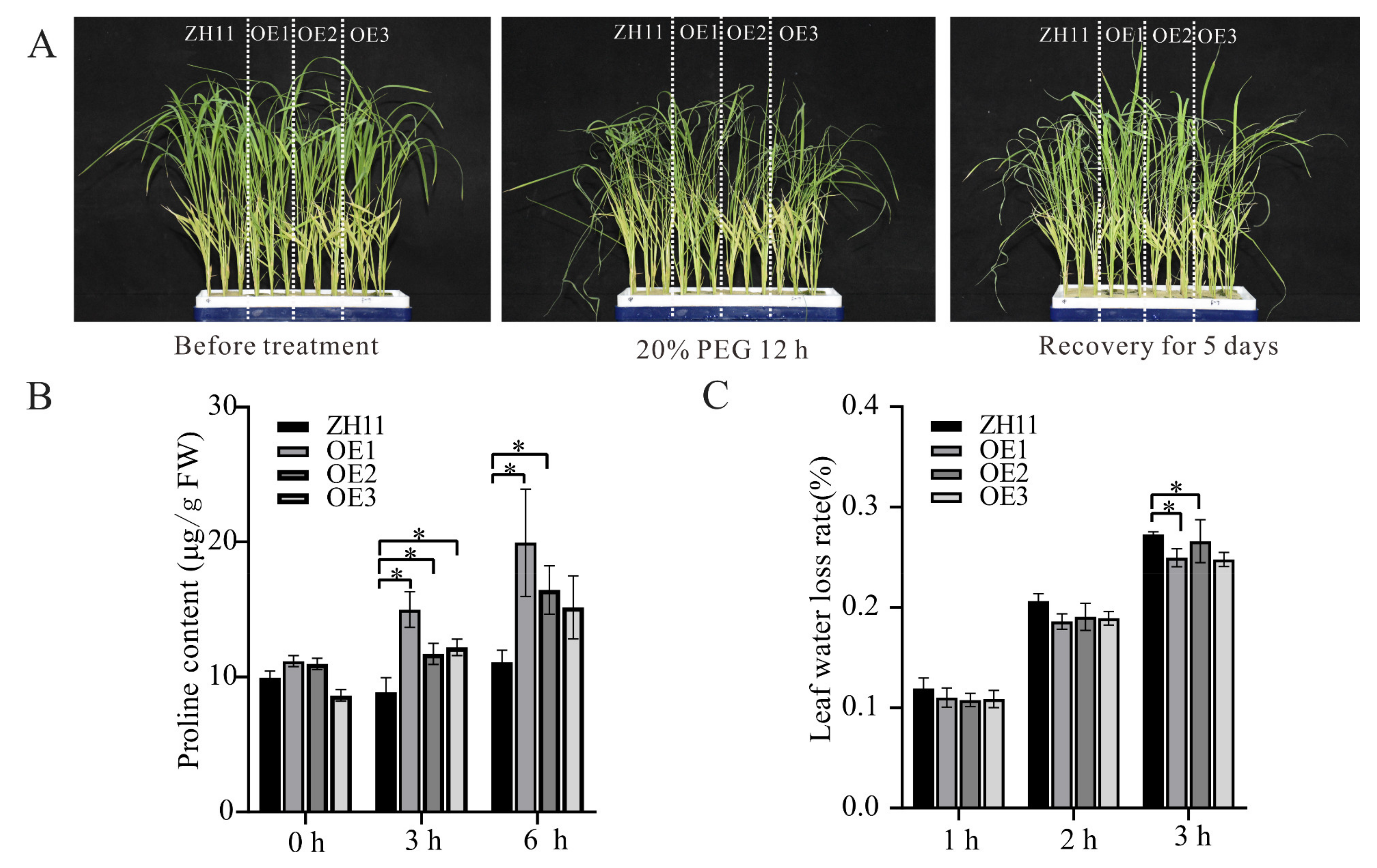
3.6. Overexpression of ZmHsf05 Enhances Drought Tolerance in Rice
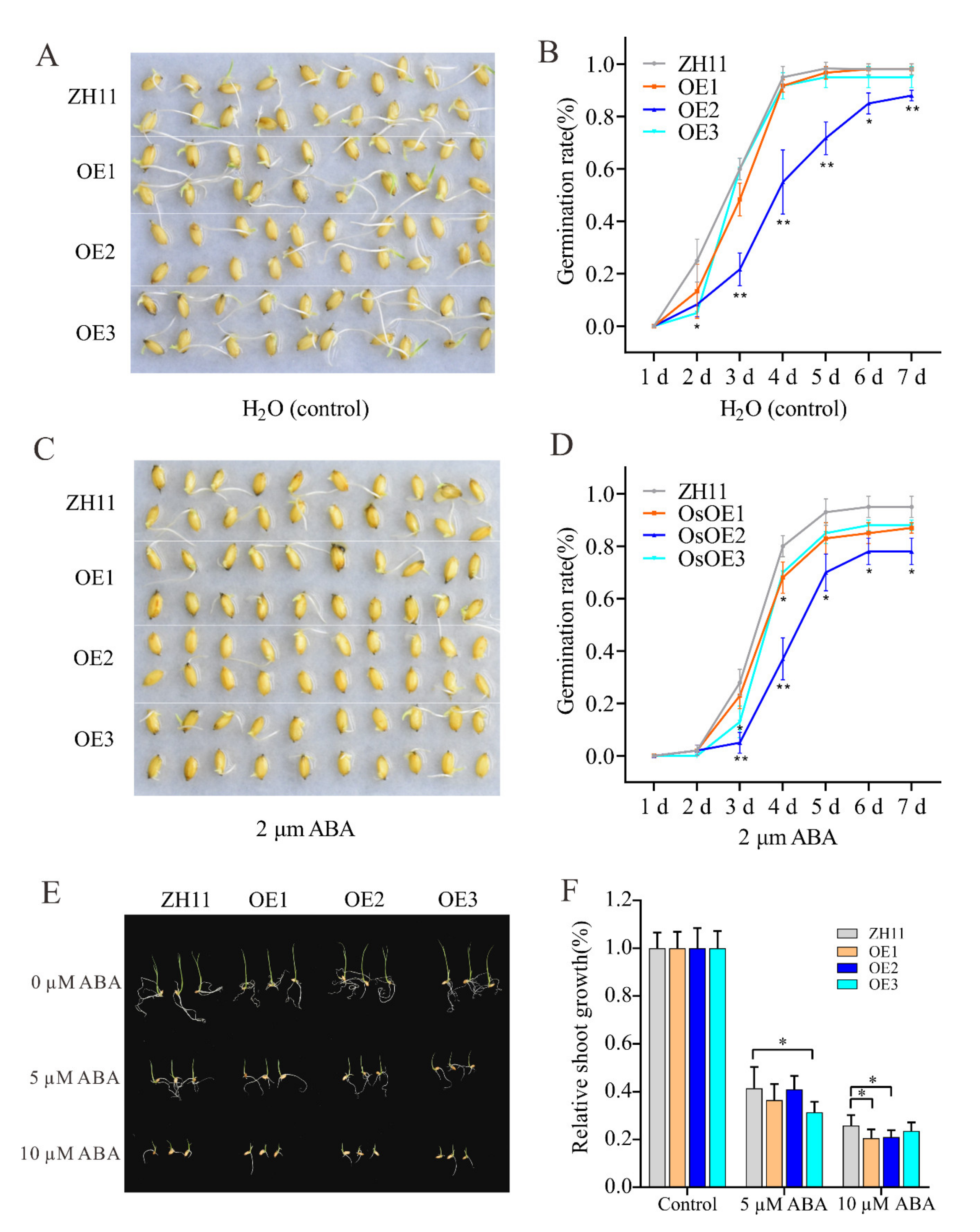
3.7. Overexpression of ZmHsf05 Increases Sensitivity to Exogenous ABA Treatment
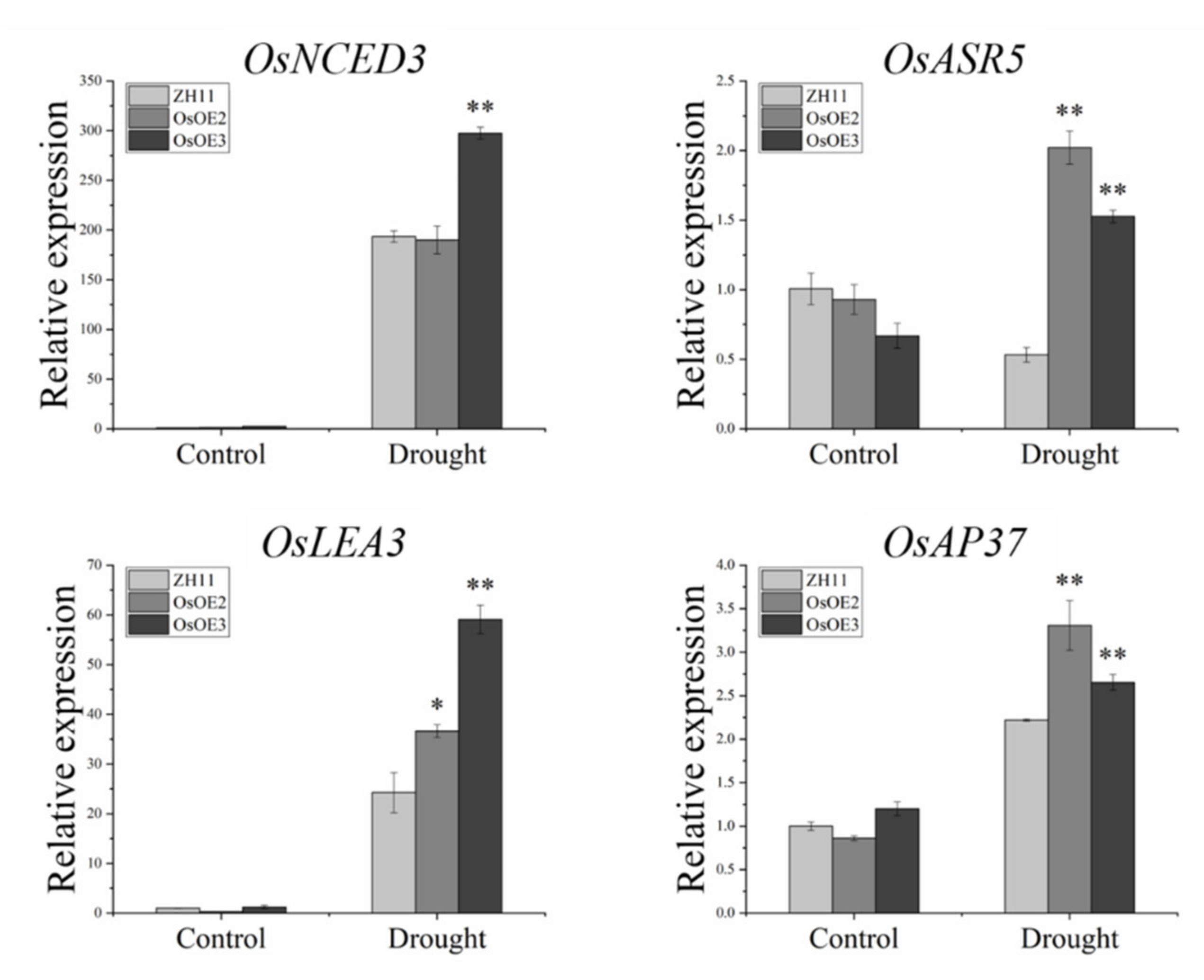
3.8. Expression Analysis of Stress-Related Genes in ZmHsf05 Transgenic Rice
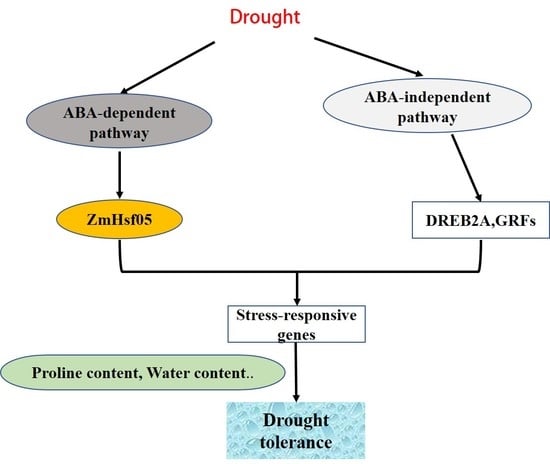
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smertenko, A. Can Peroxisomes Inform Cellular Response to Drought? Trends Plant Sci. 2017, 22, 1005–1007. [Google Scholar] [CrossRef]
- Lobell, D.B.; Roberts, M.J.; Schlenker, W.; Braun, N.; Little, B.B.; Rejesus, R.M.; Hammer, G.L. Greater sensitivity to drought accompanies maize yield increase in the U.S. Midwest. Science 2014, 344, 516–569. [Google Scholar] [CrossRef]
- Salekdeh, G.H.; Reynolds, M.; Bennett, J.; Boyer, J. Conceptual framework for drought phenotyping during molecular breeding. Trends Plant Sci. 2009, 14, 488–496. [Google Scholar] [CrossRef]
- Rueda, M.; Godoy, O.; Hawkins, B.A. Spatial and evolutionary parallelism between shade and drought tolerance explains the distributions of conifers in the conterminous United States. Glob. Ecol. Biogeogr. 2017, 26, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Ripullone, F.; Borghetti, M.; Raddi, S.; Vicinelli, E.; Baraldi, R.; Guerrieri, M.R.; Nole, A.; Magnani, F. Physiological and structural changes in response to altered precipitation regimes in a Mediterranean macchia ecosystem. Trees 2009, 23, 823–834. [Google Scholar] [CrossRef]
- Dong, X.J.; Zhang, X.S. Some observations of the adaptations of sandy shrubs to the arid environment in the Mu Us Sandland: Leaf water relations and anatomic features. J. Arid Environ. 2001, 48, 41–48. [Google Scholar] [CrossRef]
- Fraser, L.H.; Greenall, A.; Carlyle, C.; Turkington, R.; Friedman, C.R. Adaptive phenotypic plasticity of Pseudoroegneria spicata: Response of stomatal density, leaf area and biomass to changes in water supply and increased temperature. Ann. Bot. 2009, 103, 769–775. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Zhou, G. Responses of leaf stomatal density to water status and its relationship with photosynthesis in a grass. J. Exp. Bot. 2008, 59, 3317–3325. [Google Scholar] [CrossRef] [Green Version]
- Lopes, M.S.; Reynolds, M.P. Partitioning of assimilates to deeper roots is associated with cooler canopies and increased yield under drought in wheat. Funct. Plant Biol. 2010, 37, 147–156. [Google Scholar] [CrossRef]
- De Smet, I.; Signora, L.; Beeckman, T.; Inze, D.; Foyer, C.H.; Zhang, H. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. Cell Mol. Biol. 2003, 33, 543–555. [Google Scholar] [CrossRef] [Green Version]
- Mukarram, M.; Choudhary, S.; Kurjak, D.; Petek, A.; Khan, M.M.A. Drought: Sensing, signalling, effects and tolerance in higher plants. Physiol. Plant 2021, 172, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Abscisic Acid and abiotic stress signaling. Plant Signal Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, Y.; Yin, Z.; Jiang, J.; Zhang, M.; Guo, X.; Ye, Z.; Zhao, Y.; Xiong, H.; Zhang, Z.; et al. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H(2) O(2) signalling in rice. Plant Biotechnol. J. 2017, 15, 183–196. [Google Scholar] [CrossRef]
- Yan, H.; Jia, H.; Chen, X.; Hao, L.; An, H.; Guo, X. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014, 55, 2060–2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, H.; Takasaki, H.; Takahashi, F.; Suzuki, T.; Iuchi, S.; Mitsuda, N.; Ohme-Takagi, M.; Ikeda, M.; Seo, M.; Yamaguchi-Shinozaki, K.; et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11178–E11187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, S.-G.; Chen, H.-C.; Huang, W.-Y.; Chu, Y.-C.; Shii, C.-T.; Cheng, W.-H. Ectopic expression of rice OsNCED3 in Arabidopsis increases ABA level and alters leaf morphology. Plant Sci. 2010, 178, 12–22. [Google Scholar] [CrossRef]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plantarum 2021, 172, 847–868. [Google Scholar] [CrossRef]
- Long, L.; Gao, W.; Xu, L.; Liu, M.; Luo, X.; He, X.; Yang, X.; Zhang, X.; Zhu, L. GbMPK3, a mitogen-activated protein kinase from cotton, enhances drought and oxidative stress tolerance in tobacco. Plant Cell Tissue Organ Cult. 2014, 116, 153–162. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Meng, H.; Wen, H.; Fan, Y.; Zhao, J. Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 2011, 75, 365–378. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xiao, S.; Li, W.; Feng, W.; Li, J.; Wu, Z.; Gao, X.; Liu, F.; Shao, M. Overexpression of a Harpin-encoding gene hrf1 in rice enhances drought tolerance. J. Exp. Bot. 2011, 62, 4229–4238. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Sun, S.J.; Xu, D.Q.; Yang, X.; Bao, Y.M.; Wang, Z.F.; Tang, H.J.; Zhang, H. Increased tolerance of rice to cold, drought and oxidative stresses mediated by the overexpression of a gene that encodes the zinc finger protein ZFP245. Biochem. Biophys. Res. Commun. 2009, 389, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Tyerman, S.D.; Niemietz, C.M.; Bramley, H. Plant aquaporins: Multifunctional water and solute channels with expanding roles. Plant Cell Environ. 2002, 25, 173–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Nover, L.; Bharti, K.; Doring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperon 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Jia, H.X.; Li, J.B.; Huang, J.; Lu, M.Z.; Hu, J.J. The heat shock factor gene family in Salix suchowensis: A genome-wide survey and expression profiling during development and abiotic stresses. Front. Plant Sci. 2015, 6, 748. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.C.; Niu, C.Y.; Yang, C.R.; Jinn, T.L. The Heat Stress Factor HSFA6b Connects ABA Signaling and ABA-Mediated Heat Responses. Plant Physiol. 2016, 172, 1182–1199. [Google Scholar] [CrossRef]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; von Koskull-Doring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. Cell Mol. Biol. 2004, 39, 98–112. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [Green Version]
- Doring, P.; Treuter, E.; Kistner, C.; Lyck, R.; Chen, A.; Nover, L. The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 2000, 12, 265–278. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Hwang, S.M.; Kim, D.W.; Woo, M.S.; Jeong, H.S.; Son, Y.S.; Akhter, S.; Choi, G.J.; Bahk, J.D. Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant Cell Environ. 2014, 37, 1202–1222. [Google Scholar] [CrossRef] [PubMed]
- Personat, J.M.; Tejedor-Cano, J.; Prieto-Dapena, P.; Almoguera, C.; Jordano, J. Co-overexpression of two Heat Shock Factors results in enhanced seed longevity and in synergistic effects on seedling tolerance to severe dehydration and oxidative stress. BMC Plant Biol. 2014, 14, 56. [Google Scholar] [CrossRef] [Green Version]
- Bi, H.; Zhao, Y.; Li, H.; Liu, W. Wheat Heat Shock Factor TaHsfA6f Increases ABA Levels and Enhances Tolerance to Multiple Abiotic Stresses in Transgenic Plants. Int. J. Mol. Sci. 2020, 21, 3121. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.D.; Zhang, M.; Gao, D.J.; Zhou, K.; Tang, S.J.; Zhou, B.; Lv, Y.M. Rice OsHSFA3 Gene Improves Drought Tolerance by Modulating Polyamine Biosynthesis Depending on Abscisic Acid and ROS Levels. Int. J. Mol. Sci. 2020, 21, 1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Liu, W.; Yu, L.; Guo, Z.; Chen, Z.; Jiang, S.; Xu, H.; Fang, H.; Wang, Y.; Zhang, Z.; et al. Heat Shock Factor A8a Modulates Flavonoid Synthesis and Drought Tolerance. Plant Physiol. 2020, 184, 1273–1290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, G.; Fu, C.; Duan, S.; Hu, D.; Guo, X. Genome-wide identification, transcriptome analysis and alternative splicing events of Hsf family genes in maize. Sci. Rep. 2020, 10, 8073. [Google Scholar] [CrossRef]
- Jiang, Y.; Zheng, Q.; Chen, L.; Liang, Y.; Wu, J. Ectopic overexpression of maize heat shock transcription factor gene ZmHsf04 confers increased thermo and salt-stress tolerance in transgenic Arabidopsis. Acta Physiol. Plant. 2018, 40, 1–12. [Google Scholar]
- Zhang, H.; Li, G.; Hu, D.; Zhang, Y.; Zhang, Y.; Shao, H.; Zhao, L.; Yang, R.; Guo, X. Functional characterization of maize heat shock transcription factor gene ZmHsf01 in thermotolerance. PeerJ 2020, 8, e8926. [Google Scholar]
- Bateman, A.; Birney, E.; Durbin, R.; Eddy, S.R.; Finn, R.D.; Sonnhammer, E.L. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999, 27, 260–262. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, K.; Zhang, J.; Li, X.; Xu, K.; Zhang, Y.; Qi, J.; Yu, D.; Wang, J.; Li, C. JcDREB2, a Physic Nut AP2/ERF Gene, Alters Plant Growth and Salinity Stress Responses in Transgenic Rice. Front. Plant Sci. 2017, 8, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, S.W.; Cai, C.Y.; Ren, H.B.; Wang, W.J.; Xiang, M.Q.; Tang, X.S.; Zhu, C.P.; Yin, T.F.; Zhang, L.; Zhu, Q. An Efficient Plant Regeneration and Transformation System of Ma Bamboo (Dendrocalamus latiflorus Munro) Started from Young Shoot as Explant. Front. Plant Sci. 2017, 8, 1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, S.; Hu, W.; Deng, X.; Zhang, Y.; Liu, X.; Zhao, X.; Luo, Q.; Jin, Z.; Li, Y.; Zhou, S.; et al. A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol. 2014, 14, 133. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.M.; Yao, D.M.; Lin, F.; Jiang, M.Y. PEG-mediated transient gene expression and silencing system in maize mesophyll protoplasts: A valuable tool for signal transduction study in maize. Acta Physiol. Plant. 2014, 36, 1271–1281. [Google Scholar] [CrossRef]
- Tang, Q.Y.; Zhang, C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef]
- Lin, Y.X.; Jiang, H.Y.; Chu, Z.X.; Tang, X.L.; Zhu, S.W.; Cheng, B.J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12, 76. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.Z. OsLEA3, a late embryogenesis abundant protein gene from rice, confers tolerance to water deficit and salt stress to transgenic rice. Russ. J. Plant Physiol. 2008, 55, 530–537. [Google Scholar] [CrossRef]
- Mao, C.; Ding, J.; Zhang, B.; Xi, D.; Ming, F. OsNAC2 positively affects salt-induced cell death and binds to the OsAP37 and OsCOX11 promoters. Plant J. Cell Mol. Biol. 2018, 94, 454–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 2008, 227, 957–967. [Google Scholar] [CrossRef]
- Jiang, D.; Zhou, L.; Chen, W.; Ye, N.; Xia, J.; Zhuang, C. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways. Rice 2019, 12, 76. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Si, W.; Liang, Q.; Chen, L.; Song, F.; Chen, Y.; Jiang, H. Ectopic Overexpression of Maize Heat Stress Transcription Factor ZmHsf05 Confers Drought Tolerance in Transgenic Rice. Genes 2021, 12, 1568. https://doi.org/10.3390/genes12101568
Si W, Liang Q, Chen L, Song F, Chen Y, Jiang H. Ectopic Overexpression of Maize Heat Stress Transcription Factor ZmHsf05 Confers Drought Tolerance in Transgenic Rice. Genes. 2021; 12(10):1568. https://doi.org/10.3390/genes12101568
Chicago/Turabian StyleSi, Weina, Qizhi Liang, Li Chen, Feiyang Song, You Chen, and Haiyang Jiang. 2021. "Ectopic Overexpression of Maize Heat Stress Transcription Factor ZmHsf05 Confers Drought Tolerance in Transgenic Rice" Genes 12, no. 10: 1568. https://doi.org/10.3390/genes12101568