Genes Associated with Disturbed Cerebral Neurogenesis in the Embryonic Brain of Mouse Models of Down Syndrome
Abstract
1. Introduction
2. Analyses of the Brains of People with DS
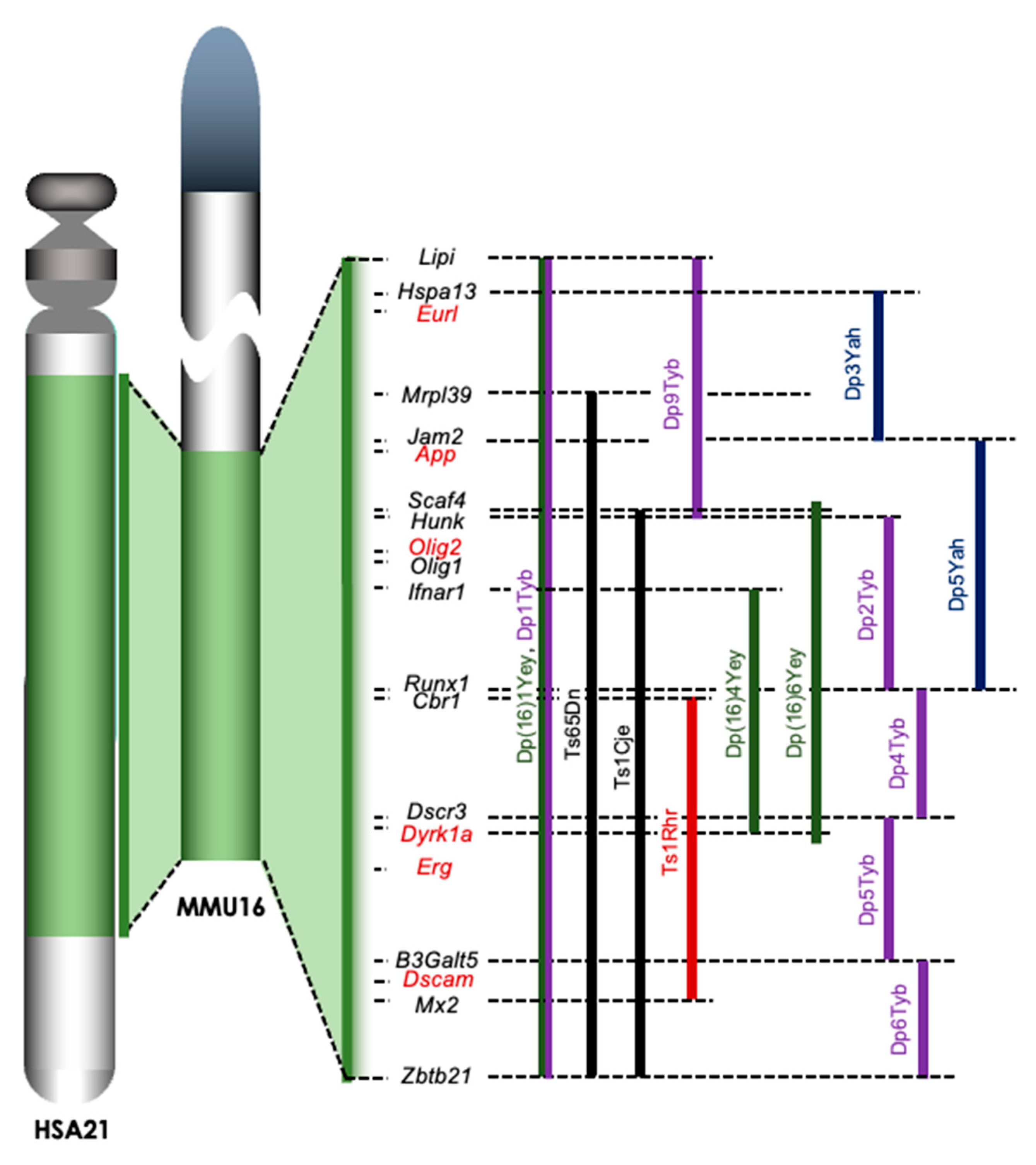
3. Analyses of the Brains of Mouse Models for DS
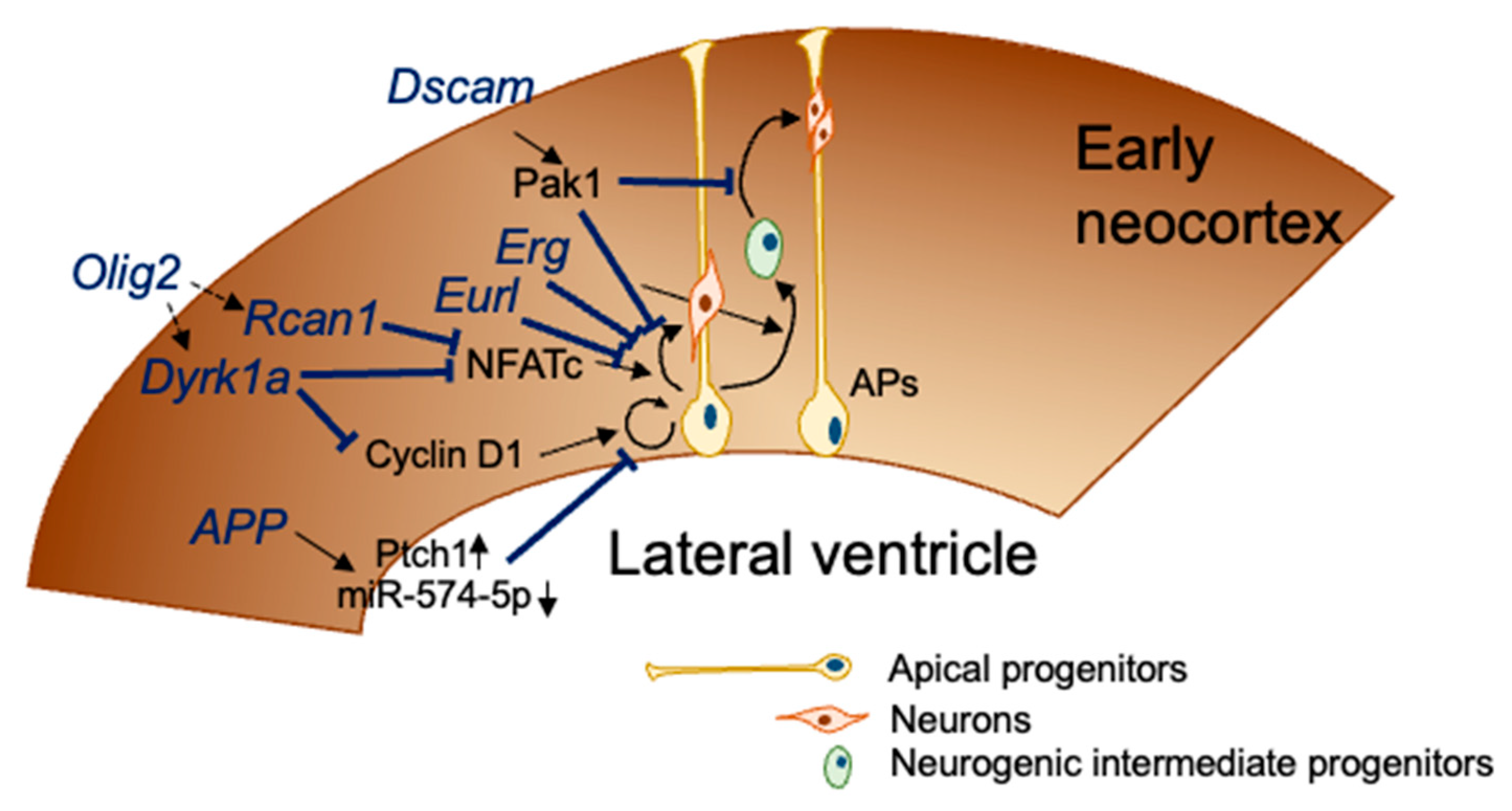
4. Candidate Genes Related to the Impaired Embryonic Neurogenesis in DS
4.1. Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase 1A (Dyrk1a) and Regulator of Calcineurin 1 (Rcan1)
4.2. Amyloid Precursor Protein (App)
4.3. Down Syndrome Cell-Adhesion Molecule (DSCAM)
4.4. Oligodendrocyte Transcription Factor 2 (Olig2)
4.5. Expressed in Undifferentiated Retina and Lens of Chick Embryos (EURL/C21ORF91)
4.6. ETS Transcription Factor ERG
5. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Ross, M.H.; Galaburda, A.M.; Kemper, T.L. Down’s syndrome: Is there a decreased population of neurons? Neurology 1984, 34, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Sidor, B.; Wisniewski, K.E.; Shepard, T.H.; Sersen, E.A. Brain growth in Down syndrome subjects 15 to 22 weeks of gestational age and birth to 60 months. Clin. Neuropathol. 1990, 4, 181–190. [Google Scholar]
- Chakrabarti, L.; Galdzicki, Z.; Haydar, H.F. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J. Neurosci. 2007, 27, 11483–11495. [Google Scholar] [CrossRef]
- Guidi, S.; Bonasoni, P.; Ceccarelli, C.; Santini, D.; Gualtieri, F.; Ciani, E.; Bartesaghi, R. Neurogenesis impairment and increased cell death reduce total neuron number in the hippocampal region of fetuses with Down syndrome. Brain Pathol. 2008, 18, 180–197. [Google Scholar] [CrossRef]
- Ishihara, K.; Amano, K.; Takaki, E.; Shimohata, A.; Sago, H.; Epstein, C.J.; Yamakawa, K. Enlarged brain ventricles and impaired neurogenesis in the Ts1Cje and Ts2Cje mouse models of Down syndrome. Cereb. Cortex 2010, 20, 1131–1143. [Google Scholar] [CrossRef] [PubMed]
- Takashima, S.; Becker, L.E.; Armstrong, D.L.; Chan, F. Abnormal neuronal development in the visual cortex of the human fetus and infant with down’s syndrome. A quantitative and qualitative Golgi study. Brain Res. 1981, 225, 1–21. [Google Scholar] [CrossRef]
- Uguagliati, B.; Al-Absi, A.R.; Stagni, F.; Emili, M.; Giacomini, A.; Guidi, S.; Nyengaard, J.R.; Bartesaghi, R. Early appearance of developmental alterations in the dendritic tree of the hippocampal granule cells in the Ts65Dn model of Down syndrome. Hippcampus 2021, 31, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, M.A.; Kola, I. The “gene dosage effect” hypothesis versus the “amplified developmental instability” hypothesis in Down syndrome. J. Neural. Transm. Suppl. 1999, 57, 293–303. [Google Scholar]
- Sturgeon, X.; Gardiner, K.J. Transcript catalogs of human chromosome 21 and orthologous chimpanzee and mouse regions. Mamm. Genome 2011, 22, 261–271. [Google Scholar] [CrossRef]
- Guidi, S.; Ciani, E.; Bonasoni, P.; Santini, D.; Bartesaghi, R. Widespread proliferation impairment and hypocellularity in the cerebellum of fetuses with Down syndrome. Brain Pathol. 2011, 21, 361–373. [Google Scholar] [CrossRef]
- Larsen, K.B.; Laursen, H.; Graem, N.; Samuelsen, G.B.; Bogdanovic, N.; Pakkenberg, B. Reduced cell number in the neocortical part of the human fetal brain in Down syndrome. Ann. Anat. 2008, 190, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Guidi, S.; Giacomini, A.; Stagni, F.; Emili, M.; Uguagliati, B.; Bonasoni, M.P.; Bartesaghi, R. Abnormal development of the inferior temporal region in fetuses with Down syndrome. Brain Pathol. 2018, 28, 986–998. [Google Scholar] [CrossRef]
- Contestabile, A.; Fila, T.; Ceccarelli, C.; Bonasoni, P.; Bonapace, L.; Santini, D.; Bartesaghi, R.; Ciani, E. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with Down syndrome and in Ts65Dn mice. Hippocampus 2007, 17, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Kanaumi, T.; Milenkovic, I.; Adle-Biassette, H.; Aronica, E.; Kovacs, G.G. Non-neuronal cell responses differ between normal and Down syndrome developing brains. Int. J. Dev. Neurosci. 2013, 31, 796–803. [Google Scholar] [CrossRef]
- Golden, J.A.; Hyman, B.T. Development of the superior temporal neocortex is anomalous in trisomy 21. J. Neuropathol. Exp. Neurol. 1994, 53, 513–520. [Google Scholar] [CrossRef]
- Engidawork, E.; Lubec, G. Molecular changes in fetal Down syndrome brain. J. Neurochem. 2003, 84, 895–904. [Google Scholar] [CrossRef]
- Lu, J.; Lian, G.; Zhou, H.; Esposito, G.; Steardo, L.; Delli-Bovi, L.C.; Hecht, J.L.; Lu, Q.R.; Sheen, V. OLIG2 over-expression impairs proliferation of human Down syndrome neural progenitors. Hum. Mol. Genet. 2012, 21, 2330–2340. [Google Scholar] [CrossRef]
- Hibaoui, Y.; Grad, I.; Letourneau, A.; Sailani, M.R.; Dahoun, S.; Santoni, F.A.; Gimelli, S.; Guipponi, M.; Pelte, M.F.; Bena, F.; et al. Modelling and rescuing neurodevelopmental defect of Down syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO Mol. Med. 2014, 6, 259–277. [Google Scholar] [CrossRef]
- Weick, J.P.; Held, D.L.; Bonadurer, G.F., 3rd; Doers, M.E.; Liu, Y.; Maguire, C.; Clark, A.; Knackert, J.A.; Molinarolo, K.; Musser, M.; et al. Deficits in human trisomy 21 iPSCs and neurons. Proc. Natl. Acad. Sci. USA 2013, 110, 9962–9967. [Google Scholar] [CrossRef]
- Tang, X.Y.; Xu, L.; Wang, J.; Hong, Y.; Wang, Y.; Zhu, Q.; Wang, D.; Zhang, X.Y.; Liu, C.Y.; Fang, K.H.; et al. DSCAM/PAK1 pathway suppression reverses neurogenesis deficits in iPSC-derived cerebral organoids from patients with Down syndrome. J. Clin. Investig. 2021, 131, e135763. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, P.D.; Bradburn, M.; Campbell, M.J.; Cooper, C.L.; Graham, R.; Jarvis, D.; Kilby, M.D.; Mason, G.; Mooney, C.; Robson, S.C.; et al. Use of MRI in the diagnosis of fetal brain abnormalities in utero (MERIDIAN): A multicentre, prospective cohort study. Lancet 2017, 389, 538–546. [Google Scholar] [CrossRef]
- Tarui, T.; Im, K.; Madan, N.; Madankumar, R.; Skotko, B.G.; Schwartz, A.; Sharr, C.; Ralston, S.J.; Kitano, R.; Akiyama, S.; et al. Quantitative MRI analyses of regional brain growth in living fetuses with Down syndrome. Cereb. Cortex 2020, 30, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Davisson, M.T.; Schmidt, C.; Akeson, E.C. Segmental trisomy of murine chromosome 16: A new model system for studying Down syndrome. In Molecular Genetics of Chromosome 21 and Down Syndrome; Patterson, D., Epstein, C.J., Eds.; Wiley-Liss: New York, NY, USA, 1990; pp. 263–280. [Google Scholar]
- Haydar, T.F.; Reeves, R.H. Trisomy and early brain development. Trends Neurosci. 2012, 35, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Sago, H.; Carlson, E.J.; Smith, D.J.; Kilbridge, J.; Rubin, E.M.; Mobley, W.C.; Epstein, C.J.; Huang, T.T. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc. Natl. Acad. Sci. USA 1998, 95, 6256–6261. [Google Scholar] [CrossRef]
- Sago, H.; Carlson, E.J.; Smith, D.J.; Rubin, E.M.; Crnic, L.S.; Huang, T.T.; Epstein, C.J. Genetic dissection of region associated with behavioral abnormalities in mouse models for Down syndrome. Pediatr. Res. 2000, 48, 606–613. [Google Scholar] [CrossRef]
- Nakano-Kobayashi, A.; Awaya, T.; Kii, I.; Sumida, Y.; Okuno, Y.; Yoshida, S.; Sumida, T.; Inoue, H.; Hosoya, T.; Hagiwara, M. Prenatal neurogenesis induction therapy normalizes brain structure and function in Down syndrome mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10268–10273. [Google Scholar] [CrossRef]
- Kurabayashi, N.; Sanada, K. Increased dosage of DYRK1A and DSCR1 delays neuronal differentiation in neocortical progenitor cells. Genes Dev. 2013, 27, 2708–2721. [Google Scholar] [CrossRef]
- Aziz, N.M.; Guedj, F.; Pennings, J.L.A.; Olmos-Serrano, J.L.; Siegel, A.; Haydar, T.F.; Bianchi, D.W. Lifespan analysis of brain development, gene expression and behavioral phenotypes in the Ts1Cje, Ts65Dn and Dp(16)1/Yey mouse models of Down syndrome. Dis. Model. Mech. 2018, 11, dmm031013. [Google Scholar] [CrossRef]
- Herault, Y.; Delabar, J.M.; Fisher, E.M.C.; Tybulewicz, V.L.J.; Yu, E.; Brault, V. Rodent models in Down syndrome research: Impact and future opportunities. Dis. Model. Mech. 2017, 10, 1165–1186. [Google Scholar] [CrossRef]
- Li, Z.; Yu, T.; Morishima, M.; Pao, A.; LaDuca, J.; Conroy, J.; Nowak, N.; Matsui, S.; Shiraishi, I.; Yu, Y. Duplication of the entire 22.9-Mb human chromosome 21 syntenic region on mouse chromosome 16 causes cardiovascular and gastrointestinal abnormalities. Hum. Mol. Genet. 2007, 16, 1359–1366. [Google Scholar] [CrossRef]
- Goodliffe, J.W.; Olmos-Serrano, J.L.; Aziz, N.M.; Pennings, J.L.A.; Guedj, F.; Bianchi, D.W.; Haydar, T.F. Absence of prenatal forebrain defects in the Dp(16)1Yey/+ mouse model of Down syndrome. J. Neurosci. 2016, 36, 2926–2944. [Google Scholar] [CrossRef]
- Guimera, J.; Casas, C.; Pucharcos, C.; Solans, A.; Domenech, A.; Planas, A.M.; Ashley, J.; Lovett, M.; Estivill, X.; Pritchard, M.A. A human homologue of drosophila minibrain (Mnb) is expressed in the neuronal regions affected in Down syndrome and maps to the critical region. Hum. Mol. Genet. 1996, 5, 1305–1310. [Google Scholar] [CrossRef]
- Tejedor, F.J.; Hämmerle, B. MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J. 2011, 278, 223–235. [Google Scholar] [CrossRef]
- Dehay, C.; Kennedy, H. Cell-cycle control and cortical development. Nat. Rev. Neurosci. 2007, 8, 438–450. [Google Scholar] [CrossRef]
- Gotz, M.; Huttner, W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 777–788. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Lin, J.-R.; Tsai, F.-C.; Meyer, T. Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle. Mol. Cell 2013, 52, 87–100. [Google Scholar] [CrossRef]
- Najas, S.; Arranz, J.; Lochhead, P.A.; Ashford, A.L.; Oxley, D.; Delabar, J.M.; Cook, S.J.; Barallobre, M.J.; Arbones, M.L. DYRK1A-mediated Cyclin D1 degradation in neural stem cells contributes to the neurogenic cortical defects in Down syndrome. EBioMedicine 2015, 2, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Arron, J.R.; Winslow, M.M.; Polleri, A.; Chang, C.P.; Wu, H.; Gao, X.; Neilson, J.R.; Chen, L.; Heit, J.J.; Kim, S.K.; et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 2006, 441, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wu, Y.; Herculano, B.; Song, W. RCAN1 overexpression exacerbates calcium overloading-induced neuronal apoptosis. PLoS ONE 2014, 9, e95471. [Google Scholar]
- Khacho, M.; Clark, A.; Svoboda, D.S.; MacLaurin, J.G.; Lagace, D.C.; Park, D.S.; Slack, R.S. Mitochondrial dysfunction underlies cognitive defects as a result of neural stem cell depletion and impaired neurogenesis. Hum. Mol. Genet. 2017, 26, 3327–3341. [Google Scholar] [CrossRef]
- Khacho, M.; Harris, R.; Slack, R.S. Mitochondria as central regulators of neural stem cell fate and cognitive function. Nat. Rev. Neurosci. 2019, 20, 34–48. [Google Scholar] [CrossRef]
- Peiris, H.; Dubach, D.; Jessup, C.F.; Unterweger, P.; Raghupathi, R.; Muyderman, H.; Zanin, M.P.; Mackenzie, K.; Pritchard, M.A.; Keating, D.J. RCAN1 regulates mitochondrial function and increases susceptibility to oxidative stress in mammalian cells. Oxid. Med. Cell Longev. 2014, 2014, 520316. [Google Scholar] [CrossRef]
- Shukkur, E.A.; Shimohata, A.; Akagi, T.; Yu, W.; Yamaguchi, M.; Murayama, M.; Chui, D.; Takeuchi, T.; Amano, K.; Subramhanya, K.H.; et al. Mitochondrial dysfunction and tau hyperphosphorylation in Ts1Cje, a mouse model for down syndrome. Hum. Mol. Genet. 2006, 15, 2752–2762. [Google Scholar] [CrossRef] [PubMed]
- Trazzi, S.; Mitrugno, V.M.; Valli, E.; Fuchs, C.; Rizzi, S.; Guidi, S.; Perini, G.; Bartesaghi, R.; Ciani, E. APP-dependent up-regulation of Ptch1 underlies proliferation impairment of neural precursors in Down syndrome. Hum. Mol. Genet. 2011, 20, 1560–1573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Thevapriya, S.; Kim, P.J.; Yu, W.-P.; Je, H.S.; Tan, E.K.; Zeng, L. Amyloid precursor protein regulates neurogenesis by antagonizing miR-574-5p in the developing cerebral cortex. Nat. Commun. 2014, 5, 3330. [Google Scholar] [CrossRef][Green Version]
- Chen, B.E.; Kondo, M.; Garnier, A.; Watson, F.L.; Puettmann-Holgado, R.; Lamar, D.R.; Schmucker, D. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell 2006, 125, 607–620. [Google Scholar] [CrossRef]
- Fuerst, P.G.; Koizumi, A.; Masland, R.H.; Burgess, R.W. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature 2008, 451, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Fuerst, P.G.; Bruce, F.; Tian, M.; Wei, W.; Elstrott, J.; Feller, M.B.; Erskine, L.; Singer, J.H.; Burgess, R.W. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron 2009, 64, 484–497. [Google Scholar] [CrossRef]
- Li, H.-L.; Huang, B.S.; Vishwasrao, H.; Sutedja, N.; Chen, W.; Jin, I.; Hawkins, R.D.; Bailey, C.H.; Kandel, E.R. Dscam mediates remodeling of glutamate receptors in aplysia during de novo and learning-related synapse formation. Neuron 2009, 61, 527–540. [Google Scholar] [CrossRef]
- Liu, G.; Li, W.; Wang, L.; Kar, A.; Guan, K.-L.; Rao, Y.; Wu, J.Y. DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc. Natl. Acad. Sci. USA 2009, 106, 2951–2956. [Google Scholar] [CrossRef] [PubMed]
- Maynard, K.R.; Stein, E. DSCAM contributes to dendrite arborization and spine formation in the developing cerebral cortex. J. Neurosci. 2012, 32, 16637–16650. [Google Scholar] [CrossRef] [PubMed]
- Thiry, L.; Lemieux, M.D.; Laflamme, O.; Bretzner, F. Role of DSCAM in the evelopment of the spinal locomotor and sensorimotor circuits. J. Neurophysiol. 2016, 115, 1338–1354. [Google Scholar] [CrossRef] [PubMed]
- Laflamme, O.D.; Lemieux, M.; Thiry, L.; Bretzner, F. DSCAM mutation impairs motor cortex network dynamic and voluntary motor functions. Cereb. Cortex 2019, 29, 2313–2330. [Google Scholar] [CrossRef] [PubMed]
- Sachse, S.M.; Lievens, S.; Ribeiro, L.F.; Dascenco, D.; Masschaele, D.; Horré, K.; Misbaer, A.; Vanderroost, N.; De Smet, A.S.; Salta, E.; et al. Nuclear import of the DSCAM-cytoplasmic domain drives signaling capable of inhibiting synapse formation. EMBO J. 2019, 38, e99669. [Google Scholar] [CrossRef]
- Arimura, N.; Okada, M.; Taya, S.; Dewa, K.I.; Tsuzuki, A.; Uetake, H.; Miyashita, S.; Hashizume, K.; Shimaoka, K.; Egusa, S.; et al. SCAM regulates delamination of neurons in the developing midbrain. Sci. Adv. 2020, 6, eaba1693. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, Y.; Chen, J.-Y.; Ding, Y.-Q.; Song, N.-N. DSCAM and DSCAML1 regulate the radial migration and callosal projection in developing cerebral cortex. Brain Res. 2015, 1594, 61–70. [Google Scholar] [CrossRef]
- Perez-Nunez, R.; Barraza, N.; Gonzalez-Jamett, A.; Cardenas, A.M.; Barnier, J.V.; Caviedes, P. Overexpressed Down Syndrome cell adhesion molecule (DSCAM) deregulates P21-activated Kinase (PAK) activity in an in vitro neuronal model of Down syndrome: Consequences on cell process formation and extension. Neurotox. Res. 2016, 30, 76–87. [Google Scholar] [CrossRef]
- Pan, X.; Chang, X.; Leung, C.; Zhou, Z.; Cao, F.; Xie, W.; Jia, Z. PAK1 regulates cortical development via promoting neuronal migration and progenitor cell proliferation. Mol. Brain 2015, 8, 36. [Google Scholar] [CrossRef]
- Lu, Q.R.; Sun, T.; Zhu, Z.; Ma, N.; Garcia, M.; Stiles, C.D.; Rowitch, D.H. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 2002, 109, 75–86. [Google Scholar] [CrossRef]
- Takebayashi, H.; Nabeshima, Y.; Yoshida, S.; Chisaka, O.; Ikenaka, K.; Nabeshima, Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr. Biol. 2002, 12, 1157–1163. [Google Scholar] [CrossRef]
- Zhou, Q.; Anderson, D.J. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 2002, 109, 61–73. [Google Scholar] [CrossRef]
- Takebayashi, H.; Yoshida, S.; Sugimori, M.; Kosako, H.; Kominami, R.; Nakafuku, M.; Nabeshima, Y. Dynamic expression of basic helix-loop-helix Olig family members: Implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech. Dev. 2000, 99, 143–148. [Google Scholar] [CrossRef]
- Ono, K.; Takebayashi, H.; Ikeda, K.; Furusho, M.; Nishizawa, T.; Watanabe, K.; Ikenaka, K. Regional-and temporal-dependent changes in the differentiation of Olig2 progenitors in the forebrain, and the impact on astrocyte development in the dorsal pallium. Dev. Biol. 2008, 320, 456–468. [Google Scholar] [CrossRef]
- Miyoshi, G.; Butt, S.J.; Takebayashi, H.; Fishell, G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J. Neurosci. 2007, 27, 7786–7798. [Google Scholar] [CrossRef]
- Chakrabarti, L.; Best, T.K.; Cramer, N.P.; Carney, R.S.E.; Isaac, J.T.R.; Galdzicki, Z.; Haydar, T.F. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat. Neurosci. 2010, 13, 927–934. [Google Scholar] [CrossRef]
- Ishihara, K.; Kanai, S.; Sago, H.; Yamakawa, K.; Akiba, S. Comparative proteomic profiling reveals aberrant cell proliferation in the brain of embryonic Ts1Cje, a mouse model of Down syndrome. Neuroscience 2014, 281, 1–15. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, H.; Liu, L.; Zhao, C.; Deng, Y.; Chen, L. Disruption of neurogenesis and cortical development in transgenic mice misexpressing Olig2, a gene in the Down syndrome critical region. Neurobiol. Dis. 2015, 77, 106–116. [Google Scholar] [CrossRef]
- Xu, R.; Brawner, A.T.; Li, S.; Liu, J.-J.; Kim, H.; Xue, H.; Pang, Z.P.; Ki, W.Y.; Hart, R.P.; Liu, Y.; et al. OLIG2 drives abnormal neurodevelopmental phenotypes in human iPSC-based organoid and chimeric mouse models of down syndrome. Cell Stem Cell 2019, 24, 908–926. [Google Scholar] [CrossRef]
- Li, S.S.; Qu, Z.D.; Haas, M.; Ngo, L.; Heo, Y.J.; Kang, H.J.; Britto, J.M.; Cullen, H.D.; Vanyai, H.K.; Tan, S.S.; et al. The HSA21 gene EURL/C21ORF91 controls neurogenesis within the cerebral cortex and is implicated in the pathogenesis of Down syndrome. Sci. Rep. 2016, 6, 29514. [Google Scholar] [CrossRef]
- Ait Yahya-Graison, E.; Aubert, J.; Dauphinot, L.; Rivals, I.; Prieur, M.; Golfier, G.; Rossier, J.; Personnaz, L.; Creau, N.; Blehaut, H.; et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: Impact on disease phenotypes. Am. J. Hum. Genet. 2007, 81, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Prandini, P.; Deutsch, S.; Lyle, R.; Gagnebin, M.; Delucinge Vivier, C.; Delorenzi, M.; Gehrig, C.; Descombes, P.; Sherman, S.; Dagna Bricarelli, F.; et al. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. Am. J. Hum. Genet. 2007, 81, 252–263. [Google Scholar] [CrossRef]
- Rost, I.; Fiegler, H.; Fauth, C.; Carr, P.; Bettecken, T.; Kraus, J.; Meyer, C.; Enders, A.; Wirtz, A.; Meitinger, T.; et al. Tetrasomy 21pter --> q21.2 in a male infant without typical Down’s syndrome dysmorphic features but moderate mental retardation. J. Med. Genet. 2004, 41, e26. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Slavotinek, A.M.; Chen, X.N.; Jackson, A.; Gaunt, L.; Campbell, A.; Clayton-Smith, J.; Korenberg, J.R. Partial tetrasomy 21 in a male infant. J. Med. Genet. 2000, 37, E30. [Google Scholar] [CrossRef]
- Emery, B. Regulation of oligodendrocyte differentiation and myelination. Science 2010, 330, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Reiche, L.; Gottle, P.; Lane, L.; Duek, P.; Park, M.; Azim, K.; Schutte, J.; Manousi, A.; Schira-Heinen, J.; Kury, P. C21orf91 Regulates Oligodendroglial Precursor Cell Fate-A Switch in the Glial Lineage? Front. Cell Neurosci. 2021, 16, 653075. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Shimizu, R.; Takata, K.; Kawashita, E.; Amano, K.; Shimohata, A.; Low, D.; Nabe, T.; Sago, H.; Alexander, W.S.; et al. Perturbation of the immune cells and prenatal neurogenesis by the triplication of the Erg gene in mouse models of Down syndrome. Brain Pathol. 2020, 30, 75–91. [Google Scholar] [CrossRef]
- Loughran, S.J.; Kruse, E.A.; Hacking, D.F.; de Graaf, C.A.; Hyland, C.D.; Willson, T.A.; Henley, K.J.; Ellis, S.; Voss, A.K.; Metcalf, D.; et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat. Immunol. 2008, 9, 810–819. [Google Scholar] [CrossRef] [PubMed]
- Kruse, E.A.; Loughran, S.J.; Baldwin, T.M.; Josefsson, E.C.; Ellis, S.; Watson, D.K.; Nurden, P.; Metcalf, D.; Hilton, D.J.; Alexander, W.S.; et al. Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proc. Natl. Acad. Sci. USA 2009, 106, 13814–13819. [Google Scholar] [CrossRef] [PubMed]
- Birdsey, G.M.; Dryden, N.H.; Amsellem, V.; Gebhardt, F.; Sahnan, K.; Haskard, D.O.; Dejana, E.; Mason, J.C.; Randi, A.M. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood 2008, 111, 3498–3506. [Google Scholar] [CrossRef]
- Birdsey, G.M.; Dryden, N.H.; Shah, A.V.; Hannah, R.; Hall, M.D.; Haskard, D.O.; Parsons, M.; Mason, J.C.; Zvelebil, M.; Gottgens, B.; et al. The transcription factor Erg regulates expression of histone deacetylase 6 and multiple pathways involved in endothelial cell migration and angiogenesis. Blood 2012, 119, 894–903. [Google Scholar] [CrossRef]
- Sperone, A.; Dryden, N.H.; Birdsey, G.M.; Madden, L.; Johns, M.; Evans, P.C.; Mason, J.C.; Haskard, D.O.; Boyle, J.J.; Paleolog, E.M.; et al. The transcription factor Erg inhibits vascular inflammation by repressing NF-kappaB activation and proinflammatory gene expression in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Marteau, L.; Pacary, E.; Valable, S.; Bernaudin, M.; Guillemot, F.; Petit, E. Angiopoietin-2 regulates cortical neurogenesis in the developing telencephalon. Cereb. Cortex 2011, 21, 1695–1702. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishihara, K. Genes Associated with Disturbed Cerebral Neurogenesis in the Embryonic Brain of Mouse Models of Down Syndrome. Genes 2021, 12, 1598. https://doi.org/10.3390/genes12101598
Ishihara K. Genes Associated with Disturbed Cerebral Neurogenesis in the Embryonic Brain of Mouse Models of Down Syndrome. Genes. 2021; 12(10):1598. https://doi.org/10.3390/genes12101598
Chicago/Turabian StyleIshihara, Keiichi. 2021. "Genes Associated with Disturbed Cerebral Neurogenesis in the Embryonic Brain of Mouse Models of Down Syndrome" Genes 12, no. 10: 1598. https://doi.org/10.3390/genes12101598
APA StyleIshihara, K. (2021). Genes Associated with Disturbed Cerebral Neurogenesis in the Embryonic Brain of Mouse Models of Down Syndrome. Genes, 12(10), 1598. https://doi.org/10.3390/genes12101598





