Telomerase (hTERT) Overexpression Reveals a Promising Prognostic Biomarker and Therapeutical Target in Different Clinical Subtypes of Pediatric Acute Lymphoblastic Leukaemia
Abstract
:1. Introduction
2. Patients and Methods
2.1. Biological Samples
2.2. Extraction of RNA and Reverse Transcription of RNA to cDNA
2.3. Genetic Fusion Identification
2.3.1. Electrophoresis in Agarose Gel
2.3.2. Sequencing of Fusioned Fragments
2.4. Validation of Gene Expression by Real-Time Quantitative Polymerase Chain Reaction (qPCR)
2.5. hTERT Protein-Protein Interaction Network and Functional Enrichment Analysis
2.6. Statistical Analysis
3. Results
3.1. Clinical Features of Acute Lymphoblastic Leukemia Patients
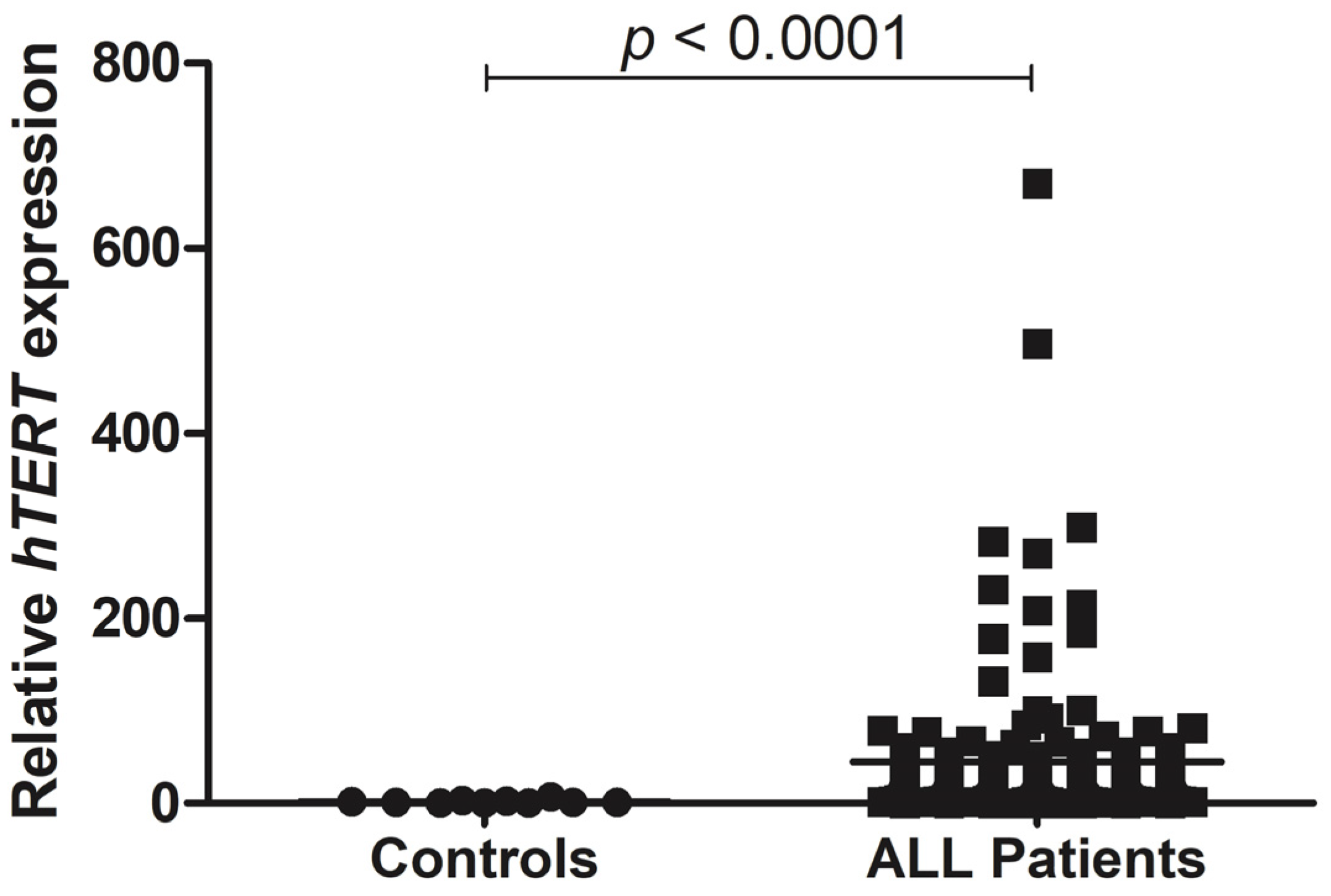
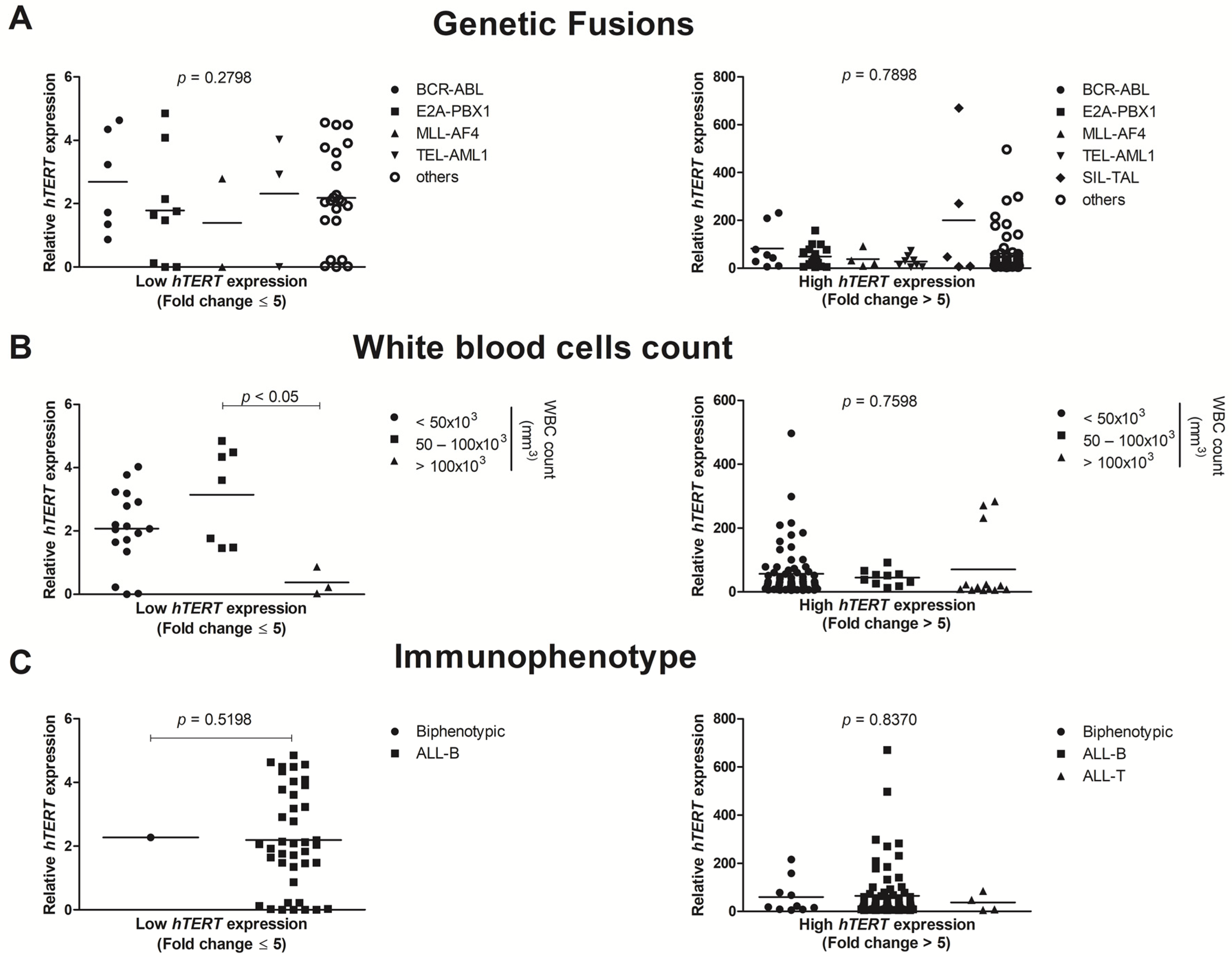
3.2. hTERT Shows Enhanced Gene Expression in ALL Pediatric Patients
3.3. Patient’s Overall Survival
3.4. PPI Network and Biological Functions of hTERT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Inaba, H.; Mullighan, C.G. Pediatric acute lymphoblastic leukemia. Haematologica 2020, 105, 2524–2539. [Google Scholar] [CrossRef]
- Philip, D.G.P.; Pizzo, A. Histologic Classification of Pediatric Gonadal and Extragonadal Tumors. Princ. Pract. Pediatr. Oncol. 2017, 53, 1689–1699. [Google Scholar]
- Dasgupta, R.K.; Marini, B.L.; Rudoni, J.; Perissinotti, A.J. A review of CD19-targeted immunotherapies for relapsed or refractory acute lymphoblastic leukemia. J. Oncol. Pharm. Pract. 2018, 24, 453–467. [Google Scholar] [CrossRef]
- Scharff, B.F.S.S.; Modvig, S.; Marquart, H.V.; Christensen, C. Integrin-Mediated Adhesion and Chemoresistance of Acute Lymphoblastic Leukemia Cells Residing in the Bone Marrow or the Central Nervous System. Front. Oncol. 2020, 10, 775. [Google Scholar] [CrossRef] [PubMed]
- Mullighan, C.G. The molecular genetic makeup of acute lymphoblastic leukemia. Hematol. Am. Soc. Hematol. Educ. Program. 2012, 2012, 389–396. [Google Scholar] [CrossRef] [Green Version]
- McNeer, J.L.; Bleyer, A. Acute lymphoblastic leukemia and lymphoblastic lymphoma in adolescents and young adults. Pediatr. Blood Cancer 2018, 65, e26989. [Google Scholar] [CrossRef] [PubMed]
- Greaves, M. A causal mechanism for childhood acute lymphoblastic leukaemia. Nat. Rev. Cancer 2018, 18, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Sanjuan-Pla, A.; Bueno, C.; Prieto, C.; Acha, P.; Stam, R.W.; Marschalek, R.; Menéndez, P. Revisiting the biology of infant t(4;11)/MLL-AF4+ B-cell acute lymphoblastic leukemia. Blood 2015, 126, 2676–2685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Smith, A.J.; Lavoie, G.; Walsh, K.M.; Aujla, S.; Evans, E.; Hansen, H.M.; Smirnov, I.; Kang, A.Y.; Zenker, M.; Ceremsak, J.J.; et al. Predisposing germline mutations in high hyperdiploid acute lymphoblastic leukemia in children. Genes Chromosom. Cancer 2019, 58, 723–730. [Google Scholar] [CrossRef]
- Capraro, V.; Zane, L.; Poncet, D.; Perol, D.; Galia, P.; Preudhomme, C.; Bonnefoy-Berard, N.; Gilson, E.; Thomas, X.; El-Hamri, M.; et al. Telomere deregulations possess cytogenetic, phenotype, and prognostic specificities in acute leukemias. Exp. Hematol. 2011, 39, 195–202.e2. [Google Scholar] [CrossRef]
- Jones, S.; Hruban, R.H.; Kamiyama, M.; Borges, M.; Zhang, X.; Parsons, D.W.; Cheng, J.; Lin, H.; Palmisano, E.; Brune, K.; et al. Exomic Sequencing Identifies PALB2 as a Pancreatic Cancer Susceptibility Gene. Science 2009, 324, 217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabbour, E.; O’Brien, S.; Konopleva, M.; Kantarjian, H. New insights into the pathophysiology and therapy of adult acute lymphoblastic leukemia. Cancer 2015, 121, 2517–2528. [Google Scholar] [CrossRef] [PubMed]
- Fiorini, E.; Santoni, A.; Colla, S. Dysfunctional telomeres and hematological disorders. Differentiation 2018, 100, 1–11. [Google Scholar] [CrossRef] [PubMed]
- M’Kacher, R.; Colicchio, B.; Borie, C.; Junker, S.; Marquet, V.; Heidingsfelder, L.; Soehnlen, K.; Najar, W.; Hempel, W.M.; Oudrhiri, N.; et al. Telomere and Centromere Staining Followed by M-FISH Improves Diagnosis of Chromosomal Instability and Its Clinical Utility. Genes 2020, 11, 475. [Google Scholar] [CrossRef] [PubMed]
- McNally, E.J.; Luncsford, P.J.; Armanios, M. Long telomeres and cancer risk: The price of cellular immortality. J. Clin. Investig. 2019, 129, 3474–3481. [Google Scholar] [CrossRef] [Green Version]
- Vasko, T.; Kaifie, A.; Stope, M.B.; Kraus, T.; Ziegler, P. Telomeres and Telomerase in Hematopoietic Dysfunction: Prognostic Implications and Pharmacological Interventions. Int. J. Mol. Sci. 2017, 18, 2267. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Dai, M.; Xu, D. Telomere-related Markers for Cancer. Curr. Top. Med. Chem. 2020, 20, 410–432. [Google Scholar] [CrossRef]
- Ivancich, M.; Schrank, Z.; Wojdyla, L.; Leviskas, B.; Kuckovic, A.; Sanjali, A.; Puri, N. Treating Cancer by Targeting Telomeres and Telomerase. Antioxidants 2017, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Carmena, M.; Liskovykh, M.; Peat, E.; Kim, J.-H.; Oshimura, M.; Masumoto, H.; Teulade-Fichou, M.-P.; Pommier, Y.; Earnshaw, W.C.; et al. Systematic Analysis of Compounds Specifically Targeting Telomeres and Telomerase for Clinical Implications in Cancer Therapy. Cancer Res. 2018, 78, 6282–6296. [Google Scholar] [CrossRef] [Green Version]
- Tabori, U.; Dome, J.S. Telomere Biology of Pediatric Cancer. Cancer Investig. 2007, 25, 197–208. [Google Scholar] [CrossRef]
- Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.; Gralnick, H.R.; Sultan, C. Proposals for the classification of chronic (mature) B and T lymphoid leukaemias. French-American-British (FAB) Cooperative Group. J. Clin. Pathol. 1989, 42, 567–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conter, V.; Bartram, C.R.; Valsecchi, M.G.; Schrauder, A.; Panzer-Grümayer, R.; Möricke, A.; Aricò, M.; Zimmermann, M.; Mann, G.; De Rossi, G.; et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: Results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood 2010, 115, 3206–3214. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2–27. [Google Scholar] [CrossRef] [Green Version]
- R Core Team R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 11 September 2021).
- Smith, E.M.; Pendlebury, D.F.; Nandakumar, J. Structural biology of telomeres and telomerase. Cell. Mol. Life Sci. 2020, 77, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Rubtsova, M.; Dontsova, O. Human Telomerase RNA: Telomerase Component or More? Biomolecules 2020, 10, 873. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Sandri, S.; De Sanctis, F.; Lamolinara, A.; Boschi, F.; Poffe, O.; Trovato, R.; Fiore, A.; Sartori, S.; Sbarbati, A.; Bondanza, A.; et al. Effective control of acute myeloid leukaemia and acute lymphoblastic leukaemia progression by telomerase specific adoptive T-cell therapy. Oncotarget 2017, 8, 86987–87001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kustanovich, A.M.; Savitskaya, T.V.; Potapnev, M.P. Telomerase activity and telomere length in malignant cells of children with acute lymphoblastic leukemia. Exp. Oncol. 2003, 25, 69–73. [Google Scholar]
- Borssén, M.; Cullman, I.; Norén-Nyström, U.; Sundström, C.; Porwit, A.; Forestier, E.; Roos, G. hTERT promoter methylation and telomere length in childhood acute lymphoblastic leukemia—Associations with immunophenotype and cytogenetic subgroup. Exp. Hematol. 2011, 39, 1144–1151. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Chen, X.; Liu, S.; Wen, R.; Yuan, X.; Xu, D.; Liu, G.; Wen, F. Methylation of CDKN2B CpG islands is associated with upregulated telomerase activity in children with acute lymphoblastic leukemia. Oncol. Lett. 2017, 13, 2115–2120. [Google Scholar] [CrossRef] [PubMed]
- Cogulu, O.; Kosova, B.; Gunduz, C.; Karaca, E.; Aksoylar, S.; Erbay, A.; Karapinar, D.; Vergin, C.; Vural, F.; Tombuloglu, M.; et al. The evaluation of hTERT mRNA expression in acute leukemia children and 2 years follow-up of 40 cases. Int. J. Hematol. 2008, 87, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Ohyashiki, J.H.; Hisatomi, H.; Nagao, K.; Honda, S.; Takaku, T.; Zhang, Y.; Sashida, G.; Ohyashiki, K. Quantitative relationship between functionally active telomerase and major telomerase components (hTERT and hTR) in acute leukaemia cells. Br. J. Cancer 2005, 92, 1942–1947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karow, A.; Haubitz, M.; Leibundgut, E.O.; Helsen, I.; Preising, N.; Steiner, D.; Dantonello, T.M.; Ammann, R.A.; Roessler, J.; Kartal-Kaess, M.; et al. Targeting Telomere Biology in Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2021, 22, 6653. [Google Scholar] [CrossRef] [PubMed]
- PDQ® Pediatric Treatment Editorial Board. PDQ Childhood Acute Lymphoblastic Leukemia Treatment. Available online: https://www.cancer.gov/types/leukemia/hp/child-all-treatment-pdq (accessed on 5 October 2021).
- Ohyashiki, J.H.; Ohyashiki, K.; Iwama, H.; Hayashi, S.; Toyama, K.; Shay, J.W. Clinical implications of telomerase activity levels in acute leukemia. Clin. Cancer Res. 1997, 3, 619–625. [Google Scholar]
- Leite, E.P.; Muniz, M.T.C.; Azevedo, A.D.C.A.C.D.; Souto, F.R.; Maia, Â.C.L.; Gondim, C.M.D.F.; Bandeira, F.M.G.C.; Melo, R.A.M. Fatores prognósticos em crianças e adolescentes com Leucemia Linfóide Aguda. Rev. Bras. Saúde Matern. Infant. 2007, 7, 413–421. [Google Scholar] [CrossRef]
- Kato, M.; Manabe, A. Treatment and biology of pediatric acute lymphoblastic leukemia. Pediatr. Int. 2018, 60, 4–12. [Google Scholar] [CrossRef]
- Ibagy, A.; Silva, D.B.; Seiben, J.; Winneshoffer, A.P.F.F.; Costa, T.E.J.B.; Dacoregio, J.S.; Costa, I.; Faraco, D. Acute Lymphoblastic Leukemia in Infants: 20 years of Experience. J. Pediatr. 2013, 89, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Bruedigam, C.; Lane, S.W. Telomerase in hematologic malignancies. Curr. Opin. Hematol. 2016, 23, 346–353. [Google Scholar] [CrossRef]
- Paiva, R.M.A.; Calado, R.T. Telomere Dysfunction and Hematologic Disorders, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2014; Volume 125, pp. 133–157. ISBN 9780123978981. [Google Scholar]
- Buitenkamp, T.D.; Izraeli, S.; Zimmermann, M.; Forestier, E.; Heerema, N.A.; Heuvel-Eibrink, M.M.V.D.; Pieters, R.; Korbijn, C.M.; Silverman, L.B.; Schmiegelow, K.; et al. Acute lymphoblastic leukemia in children with Down syndrome: A retrospective analysis from the Ponte di Legno study group. Blood 2014, 123, 70–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pui, C.-H.; Yang, J.J.; Hunger, S.P.; Pieters, R.; Schrappe, M.; Biondi, A.; Vora, A.; Baruchel, A.; Silverman, L.B.; Schmiegelow, K.; et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J. Clin. Oncol. 2015, 33, 2938–2948. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Mullighan, C.G. Molecular markers in ALL: Clinical implications. Best Pr. Res. Clin. Haematol. 2020, 33, 101193. [Google Scholar] [CrossRef]
- Terwilliger, T.; Abdul-Hay, M. Acute lymphoblastic leukemia: A comprehensive review and 2017 update. Blood Cancer J. 2017, 7, e577. [Google Scholar] [CrossRef] [Green Version]
- Place, A.E.; Pikman, Y.; Stevenson, K.E.; Harris, M.H.; Pauly, M.; Sulis, M.-L.; Hijiya, N.; Gore, L.; Cooper, T.M.; Loh, M.L.; et al. Phase I trial of the mTOR inhibitor everolimus in combination with multi-agent chemotherapy in relapsed childhood acute lymphoblastic leukemia. Pediatr. Blood Cancer 2018, 65, e27062. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Flores, E.; Comeaux, E.Q.; Kim, K.L.; Melnik, E.M.; Beckman, K.; Davis, K.L.; Wu, K.; Akutagawa, J.; Bridges, O.; Marino, R.; et al. Bcl-2 Is a Therapeutic Target for Hypodiploid B-Lineage Acute Lymphoblastic Leukemia. Cancer Res. 2019, 79, 2339–2351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez, D.L.M.; Armando, R.G.; Cerrudo, C.S.; Ghiringhelli, P.D.; Gomez, D.E. Telomerase as a Cancer Target. Development of New Molecules. Curr. Top. Med. Chem. 2016, 16, 2432–2440. [Google Scholar] [CrossRef] [Green Version]
- Guterres, A.N.; Villanueva, J. Targeting telomerase for cancer therapy. Oncogene 2020, 39, 5811–5824. [Google Scholar] [CrossRef]
- Valls, C.; Piñol, C.; Reñé, J.M.; Buenestado, J.; Viñas, J. Telomere length is a prognostic factor for overall survival in colorectal cancer. Colorectal Dis. 2011, 13, 1265–1272. [Google Scholar] [CrossRef]
- Fernandez-Marcelo, T.; Gómez, A.; Pascua, I.; De Juan, C.; Head, J.; Hernando, F.; Jarabo, J.-R.; Calatayud, J.; Torres-García, A.-J.; Iniesta, P. Telomere length and telomerase activity in non-small cell lung cancer prognosis: Clinical usefulness of a specific telomere status. J. Exp. Clin. Cancer Res. 2015, 34, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroupa, M.; Rachakonda, S.K.; Liska, V.; Srinivas, N.; Urbanová, M.; Jiraskova, K.; Schneiderova, M.; Vycital, O.; Vymetalkova, V.; Vodickova, L.; et al. Relationship of telomere length in colorectal cancer patients with cancer phenotype and patient prognosis. Br. J. Cancer 2019, 121, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, J.O.; Komrokji, R.S.; Cavo, M.; Martino, B.; Niederwieser, D.; Reiter, A.; Scott, B.L.; Baer, M.R.; Hoffman, R.; Odenike, O.; et al. Telomerase Activity, Telomere Length and HTERT Expression Correlate with Clinical Outcomes in Higher-Risk Myelofibrosis (MF) Relapsed/Refractory (R/R) to Janus Kinase Inhibitor Treated with Imetelstat. Hemasphere 2020, 4, 1098. [Google Scholar]
- Jebaraj, B.M.C.; Stilgenbauer, S. Telomere Dysfunction in Chronic Lymphocytic Leukemia. Front. Oncol. 2021, 10, 3062. [Google Scholar] [CrossRef]
- Bruedigam, C.; Bagger, F.O.; Heidel, F.H.; Kuhn, C.P.; Guignes, S.; Song, A.; Austin, R.; Vu, T.; Lee, E.; Riyat, S.; et al. Telomerase Inhibition Effectively Targets Mouse and Human AML Stem Cells and Delays Relapse following Chemotherapy. Cell Stem Cell 2014, 15, 775–790. [Google Scholar] [CrossRef] [Green Version]
- Bruedigam, C.; Wackrow, B.; Song, A.; Porter, A.H.; Lee, S.C.-W.; Moore, M.F.A.S.; Abdel-Wahab, O.; Lane, S.W. The Preclinical Efficacy of a Novel Telomerase Inhibitor, Imetelstat, in AML—A Randomized Trial in Patient-Derived Xenografts. Blood 2016, 128, 578. [Google Scholar] [CrossRef]
- Mascarenhas, J.; Komrokji, R.S.; Cavo, M.; Martino, B.; Niederwieser, D.; Reiter, A.; Scott, B.L.; Baer, M.R.; Hoffman, R.; Odenike, O.; et al. Imetelstat Is Effective Treatment for Patients with Intermediate-2 or High-Risk Myelofibrosis Who Have Relapsed on or Are Refractory to Janus Kinase Inhibitor Therapy: Results of a Phase 2 Randomized Study of Two Dose Levels. Blood 2018, 132, 685. [Google Scholar] [CrossRef]
- Farnebo, M. Wrap53, a novel regulator of p53. Cell Cycle 2009, 8, 2343–2346. [Google Scholar] [CrossRef]
- Mahmoudi, S.; Henriksson, S.; Corcoran, M.; Méndez-Vidal, C.; Wiman, K.G.; Farnebo, M. Wrap53, a Natural p53 Antisense Transcript Required for p53 Induction upon DNA Damage. Mol. Cell 2009, 33, 462–471. [Google Scholar] [CrossRef]
- Henriksson, S.; Farnebo, M. On the road with WRAP53β: Guardian of Cajal bodies and genome integrity. Front. Genet. 2015, 6, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venteicher, A.S.; Abreu, E.B.; Meng, Z.; McCann, K.E.; Terns, R.M.; Veenstra, T.D.; Terns, M.P.; Artandi, S.E. A Human Telomerase Holoenzyme Protein Required for Cajal Body Localization and Telomere Synthesis. Science 2009, 323, 644–648. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, S.; Henriksson, S.; Farnebo, L.; Roberg, K.; Farnebo, M. WRAP53 promotes cancer cell survival and is a potential target for cancer therapy. Cell Death Dis. 2011, 2, e114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, D.-W.; Adell, G.; Sun, X.-F. WRAP53 is an independent prognostic factor in rectal cancer- a study of Swedish clinical trial of preoperative radiotherapy in rectal cancer patients. BMC Cancer 2012, 12, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.-K.; Luo, X.-B.; Gou, Y.-P.; Hu, L.; Wang, K.; Li, C.; Xiang, Z.-T.; Zhang, P.; Kong, X.-L.; Zhang, C.-L.; et al. TCAB1: A potential target for diagnosis and therapy of head and neck carcinomas. Mol. Cancer 2014, 13, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lulkiewicz, M.; Bajsert, J.; Kopczynski, P.; Barczak, W.; Rubis, B. Telomere length: How the length makes a difference. Mol. Biol. Rep. 2020, 47, 7181–7188. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, B.M.D.; Machado, C.B.; Montenegro, R.C.; De Moraes, M.E.A.; Moreira-Nunes, C.A. Telomere Length and Hematological Disorders: A Review. In Vivo 2020, 34, 3093–3101. [Google Scholar] [CrossRef]



| N (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | WBC (×103)/mm3 | Gender | |||||||||
| >1 | 1–9 | ≥10 | p-Value | >50 | 50–100 | >100 | p-Value | M | F | p-Value | |
| Immunophenotype | 0.3853 | 0.0002 * | 0.5586 | ||||||||
| Biphenotypic | 0 (0) | 11 (11.3) | 2 (4.3) | 10 (10) | 1 (5.3) | 2 (6.9) | 7 (8.3) | 6 (9.4) | |||
| T-ALL cell | 0 (0) | 2 (2.1) | 3 (6.4) | 0 (0) | 0 (0) | 5 (17.2) | 4 (4.8) | 1 (1.6) | |||
| B-ALL cell | 4 (100) | 84 (86.6) | 42 (89.4) | 90 (90) | 18 (94.7) | 22 (75.9) | 73 (86.9) | 57 (89.1) | |||
| Genetic Fusion | <0.0001 ** | 0.0002 * | 0.9998 | ||||||||
| E2A-PBX1 | 0 (0) | 26 (26.8) | 4 (8.5) | 19 (19) | 8 (42.1) | 3 (10.3) | 17 (20.2) | 13 (20.3) | |||
| BCR-ABL | 1 (16.7) | 8 (8.2) | 10 (21.3) | 12 (12) | 1 (5.3) | 6 (20.7) | 11 (13.1) | 8 (12.5) | |||
| TEL-AML1 | 0 (0) | 9 (9.3) | 1 (2.1) | 8 (8) | 2 (10.5) | 0 (0) | 6 (7.1) | 4 (6.3) | |||
| MLL-AF4 | 2 (33.3) | 5 (5.2) | 0 (0) | 4 (4) | 1 (5.3) | 2 (6.9) | 4 (4.8) | 3 (4.7) | |||
| SIL-TAL | 0 (0) | 2 (2.1) | 3 (6.4) | 0 (0) | 0 (0) | 5 (17.2) | 3 (3.6) | 2 (3.1) | |||
| Others | 1 (16.7) | 47 (48.5) | 29 (61.7) | 57 (57) | 7 (36.8) | 13 (44.8) | 43 (51.2) | 34 (53.1) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nogueira, B.M.D.; da Costa Pantoja, L.; da Silva, E.L.; Mello Júnior, F.A.R.; Teixeira, E.B.; Wanderley, A.V.; da Silva Maués, J.H.; de Moraes Filho, M.O.; de Moraes, M.E.A.; Montenegro, R.C.; et al. Telomerase (hTERT) Overexpression Reveals a Promising Prognostic Biomarker and Therapeutical Target in Different Clinical Subtypes of Pediatric Acute Lymphoblastic Leukaemia. Genes 2021, 12, 1632. https://doi.org/10.3390/genes12101632
Nogueira BMD, da Costa Pantoja L, da Silva EL, Mello Júnior FAR, Teixeira EB, Wanderley AV, da Silva Maués JH, de Moraes Filho MO, de Moraes MEA, Montenegro RC, et al. Telomerase (hTERT) Overexpression Reveals a Promising Prognostic Biomarker and Therapeutical Target in Different Clinical Subtypes of Pediatric Acute Lymphoblastic Leukaemia. Genes. 2021; 12(10):1632. https://doi.org/10.3390/genes12101632
Chicago/Turabian StyleNogueira, Beatriz Maria Dias, Laudreísa da Costa Pantoja, Emerson Lucena da Silva, Fernando Augusto Rodrigues Mello Júnior, Eliel Barbosa Teixeira, Alayde Vieira Wanderley, Jersey Heitor da Silva Maués, Manoel Odorico de Moraes Filho, Maria Elisabete Amaral de Moraes, Raquel Carvalho Montenegro, and et al. 2021. "Telomerase (hTERT) Overexpression Reveals a Promising Prognostic Biomarker and Therapeutical Target in Different Clinical Subtypes of Pediatric Acute Lymphoblastic Leukaemia" Genes 12, no. 10: 1632. https://doi.org/10.3390/genes12101632







