UMOD Polymorphisms Associated with Kidney Function, Serum Uromodulin and Risk of Mortality among Patients with Chronic Kidney Disease, Results from the C-STRIDE Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Samples
2.2. Genotyping
2.3. Uromodulin Measurement
2.4. Measurement of Covariates
2.5. Outcomes
2.5.1. ESKD, CVD Events and All-Cause and CVD-Specific Mortality
2.5.2. eGFR Slope
2.5.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rampoldi, L.; Scolari, F.; Amoroso, A.; Ghiggeri, G.; Devuyst, O. The rediscovery of uromodulin (Tamm-Horsfall protein): From tubulointerstitial nephropathy to chronic kidney disease. Kidney Int. 2011, 80, 338–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serafini-Cessi, F.; Malagolini, N.; Cavallone, D. Tamm-Horsfall glycoprotein: Biology and clinical relevance. Am. J. Kidney Dis. 2003, 42, 658–676. [Google Scholar] [CrossRef]
- Youhanna, S.; Weber, J.; Beaujean, V.; Glaudemans, B.; Sobek, J.; Devuyst, O. Determination of uromodulin in human urine: Influence of storage and processing. Nephrol. Dial. Transplant. 2014, 29, 136–145. [Google Scholar] [CrossRef] [Green Version]
- Delgado, G.E.; Kleber, M.E.; Scharnagl, H.; Kramer, B.K.; Marz, W.; Scherberich, J.E. Serum Uromodulin and Mortality Risk in Patients Undergoing Coronary Angiography. J. Am. Soc. Nephrol. 2017, 28, 2201–2210. [Google Scholar] [CrossRef]
- Lv, L.; Wang, J.; Gao, B.; Wu, L.; Wang, F.; Cui, Z.; He, K.; Zhang, L.; Chen, M.; Zhao, M.H. Serum uromodulin and progression of kidney disease in patients with chronic kidney disease. J. Transl. Med. 2018, 16, 316. [Google Scholar] [CrossRef]
- Hart, T.C.; Gorry, M.C.; Hart, P.S.; Woodard, A.S.; Shihabi, Z.; Sandhu, J.; Shirts, B.; Xu, L.; Zhu, H.; Barmada, M.M.; et al. Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J. Med. Genet. 2002, 39, 882–892. [Google Scholar] [CrossRef] [Green Version]
- Köttgen, A.; Glazer, N.L.; Dehghan, A.; Hwang, S.J.; Katz, R.; Li, M.; Yang, Q.; Gudnason, V.; Launer, L.J.; Harris, T.B.; et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet. 2009, 41, 712–717. [Google Scholar] [CrossRef] [Green Version]
- Chambers, J.C.; Zhang, W.; Lord, G.M.; van der Harst, P.; Lawlor, D.A.; Sehmi, J.S.; Gale, D.P.; Wass, M.N.; Ahmadi, K.R.; Bakker, S.J.; et al. Genetic loci influencing kidney function and chronic kidney disease. Nat. Genet. 2010, 42, 373–375. [Google Scholar] [CrossRef] [Green Version]
- Gudbjartsson, D.F.; Holm, H.; Indridason, O.S.; Thorleifsson, G.; Edvardsson, V.; Sulem, P.; de Vegt, F.; d’Ancona, F.C.; den Heijer, M.; Wetzels, J.F.; et al. Association of Variants at UMOD with Chronic Kidney Disease and Kidney Stones—Role of Age and Comorbid Diseases. PLoS Genet. 2010, 6, e1001039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, Y.; Sim, X.; Go, M.J.; Wu, J.Y.; Gu, D.; Takeuchi, F.; Takahashi, A.; Maeda, S.; Tsunoda, T.; Chen, P.; et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat. Genet. 2012, 44, 904–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, B.; Zhang, L.; Wang, H.; Zhao, M. Chinese cohort study of chronic kidney disease: Design and methods. Chin. Med. J. 2014, 127, 2180–2185. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.; Ziaugra, L.; Tabbaa, D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet. 2009. [Google Scholar] [CrossRef] [PubMed]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.L.; Castro, A.F., 3rd; Feldman, H.I.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Greene, T.; et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Stevens, P.E.; Levin, A. Evaluation and management of chronic kidney disease: Synopsis of the kidney disease: Improving global outcomes 2012 clinical practice guideline. Ann. Intern. Med. 2013, 158, 825–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padmanabhan, S.; Melander, O.; Johnson, T.; Di Blasio, A.M.; Lee, W.K.; Gentilini, D.; Hastie, C.E.; Menni, C.; Monti, M.C.; Delles, C.; et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010, 6, e1001177. [Google Scholar] [CrossRef] [PubMed]
- Trudu, M.; Janas, S.; Lanzani, C.; Debaix, H.; Schaeffer, C.; Ikehata, M.; Citterio, L.; Demaretz, S.; Trevisani, F.; Ristagno, G.; et al. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat. Med. 2013, 19, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England) 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Liu, Y.; Rao, F.; Nievergelt, C.M.; O’Connor, D.T.; Wang, X.; Liu, L.; Bu, D.; Liang, Y.; Wang, F.; et al. Common genetic variants of the human uromodulin gene regulate transcription and predict plasma uric acid levels. Kidney Int. 2013, 83, 733–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedak, D.; Kuźniewski, M.; Fugiel, A.; Wieczorek-Surdacka, E.; Przepiórkowska-Hoyer, B.; Jasik, P.; Miarka, P.; Dumnicka, P.; Kapusta, M.; Solnica, B.; et al. Serum uromodulin concentrations correlate with glomerular filtration rate in patients with chronic kidney disease. Pol. Arch. Med. Wewn. 2016, 126, 995–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steubl, D.; Block, M.; Herbst, V.; Nockher, W.A.; Schlumberger, W.; Satanovskij, R.; Angermann, S.; Hasenau, A.L.; Stecher, L.; Heemann, U.; et al. Plasma Uromodulin Correlates With Kidney Function and Identifies Early Stages in Chronic Kidney Disease Patients. Medicine 2016, 95, e3011. [Google Scholar] [CrossRef] [PubMed]

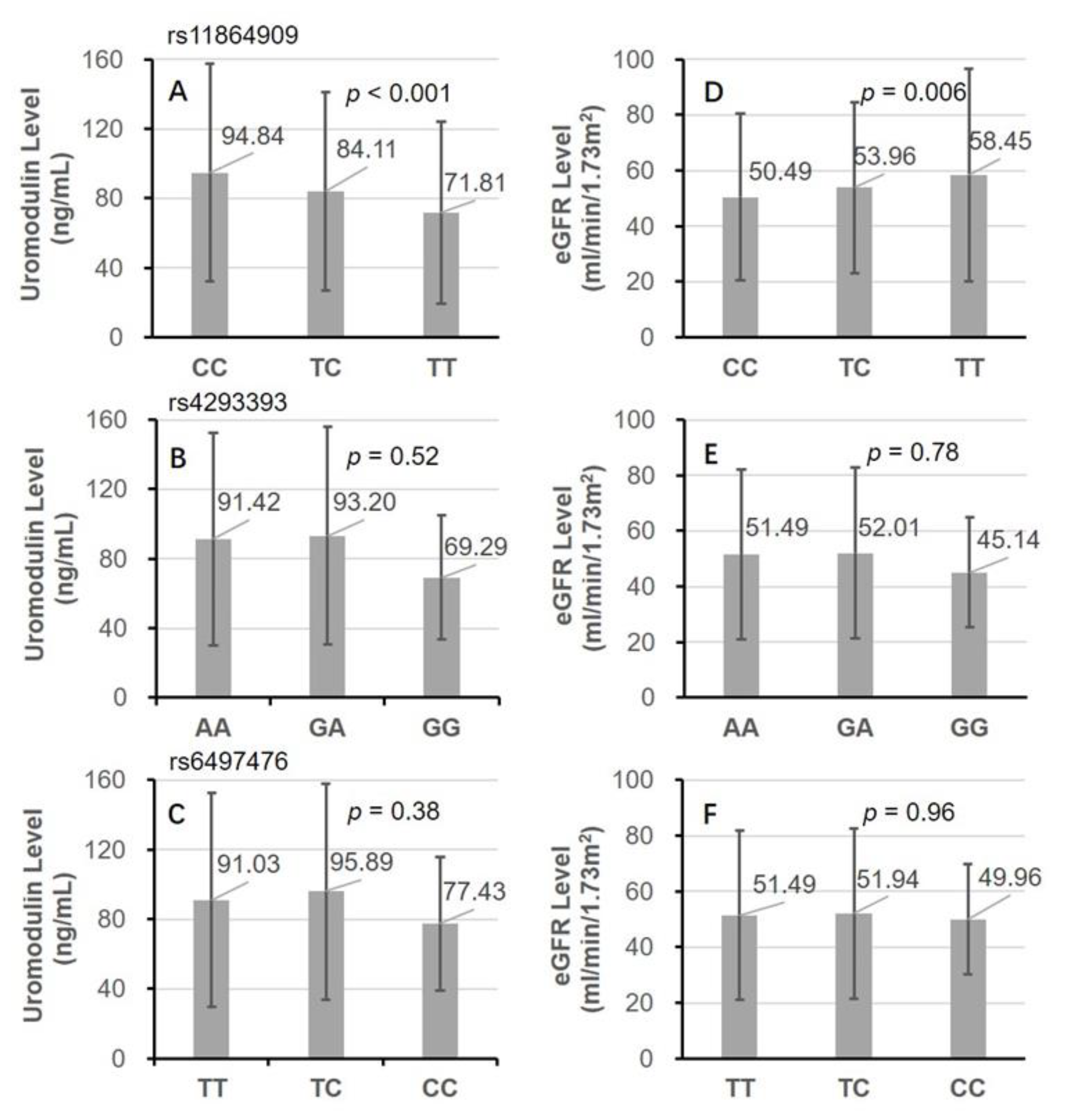
| Characteristics | Total | C/C | T/C & T/T | p-Value |
|---|---|---|---|---|
| n = 2731 | n = 1988 | n = 743 | ||
| Age, years | 48.94 ± 13.81 | 49.13 ± 13.78 | 48.42 ± 13.88 | 0.23 |
| Male, n (%) | 1634 (59.83%) | 1160 (58.35%) | 474 (63.80%) | 0.01 |
| High school and above, n (%) | 1509 (55.70%) | 1090 (55.16%) | 419 (57.16%) | 0.35 |
| Current and ever smoking, n (%) | 1043 (39.40%) | 737 (38.35%) | 306 (42.21%) | 0.07 |
| Body mass index, kg/m2 | 24.50 ± 3.62 | 24.42 ± 3.57 | 24.71 ± 3.74 | 0.07 |
| Systolic blood pressure, mmHg | 129.58 ± 17.98 | 129.71 ± 17.74 | 129.25 ± 18.59 | 0.57 |
| Diastolic blood pressure, mmHg | 80.97 ± 10.95 | 81.08 ± 10.95 | 80.67 ± 10.94 | 0.42 |
| Using anti-hypertensive medication, n (%) | 1620 (73.01%) | 1169 (73.06%) | 451 (72.86%) | 0.92 |
| Diabetes mellitus, n (%) | 625 (25.52%) | 448 (25.13%) | 177 (26.58%) | 0.46 |
| History of CVD, n (%) | 277 (10.14%) | 193 (9.71%) | 84 (11.31%) | 0.22 |
| Creatinine, μmol/L | 143 (100, 207) | 145 (102, 210) | 137 (93, 199) | 0.02 |
| eGFR, mL/min/1.73 m2 | 51.54 ± 30.44 | 50.49 ± 29.98 | 54.37 ± 31.49 | 0.003 |
| eGFR <60mL/min/1.73 m2, n (%) | 1858 (68.03%) | 1381 (69.47%) | 477 (64.20%) | 0.009 |
| ACR, mg/g | 435.59 (114.00, 991.20) | 434.85 (116.82, 985.43) | 437.38 (108.20, 1018.61) | 0.92 |
| Albuminuria groups, n(%) | 0.88 | |||
| <30 mg/g | 315 (11.81%) | 233(11.99%) | 82 (11.33%) | |
| 30–299 mg/g | 770 (28.86%) | 562 (28.91%) | 208 (28.73%) | |
| ≥300 mg/g | 1583 (59.33%) | 1149 (59.10%) | 434 (59.94%) | |
| Uromodulin, ng/mL | 91.60 ± 61.37 | 94.84 ± 62.79 | 82.99 ± 56.57 | <0.001 |
| Etiology of CKD | 0.63 | |||
| Diabetic nephropathy | 392 (14.77%) | 286 (14.83%) | 106 (14.60%) | |
| Glomerulonephritis | 1605 (60.47%) | 1156 (59.96%) | 449 (61.85%) | |
| Others | 657 (24.76%) | 486 (25.21%) | 171 (23.55%) | |
| Genotype of rs13333226 | <0.001 1 | |||
| AA | 2333 (85.43%) | 1668 (83.90%) | 665 (89.50%) | |
| GA | 389 (14.24%) | 313 (15.74%) | 76 (10.23%) | |
| GG | 9 (0.33%) | 7 (0.35%) | 2 (0.27%) | |
| Genotype of rs4293393 | <0.001 1 | |||
| AA | 2333 (85.43%) | 1668 (83.90%) | 665 (89.50%) | |
| GA | 389 (14.24%) | 313 (15.74%) | 76 (10.23%) | |
| GG | 9 (0.33%) | 7 (0.35%) | 2 (0.27%) | |
| Genotype of rs6497476 | <0.001 1 | |||
| TT | 2382 (87.22%) | 1693 (85.16%) | 689 (92.73%) | |
| TC | 342 (12.52%) | 288 (14.49%) | 54 (7.27%) | |
| CC | 7 (0.26%) | 7 (0.35%) | 0 (0.00%) |
| Association Model | β (95% CI) for eGFR | β (95% CI) for Uromodulin 1 |
|---|---|---|
| rs11864909 (TT & TC vs. CC) | ||
| Model 1 | 3.88 (1.25, 6.62) | −11.78 (−16.79, −6.12) |
| Model 2 | 3.36 (0.94, 6.03) | −12.10 (−17.27, −6.68) |
| Model 3 | 2.68 (0.61, 4.96) | −12.95 (−17.59, −7.98) |
| Model 4 | 5.65(4.02, 7.67) | −18.24 (−22.27, −14.12) |
| Haplotype composed of rs11864909, rs4293393 and rs6497476 (TAT vs. other types) | ||
| Model 1 | 7.85 (3.17, 13.31) | −19.78 (−28.46, −9.80) |
| Model 2 | 6.80 (2.74, 12.17) | −20.62 (−29.38, −11.09) |
| Model 3 | 5.71 (2.01, 10.12) | −22.22 (−29.87, −13.20) |
| Model 4 | 10.90 (7.63, 14.77) | −32.73 (−39.52, −25.44) |
| Genetic Variant | ESKD | CVD Events | All-Cause Mortality | CVD Specific Mortality | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Events (%) | Rate/100 Patient-Years | p-Value for Log-Rank Test | No. of Events (%) | Rate/100 Patient-Years | p-Value for Log-Rank Test | No. of Events (%) | Rate/100 Patient-Years | p-Value for Log-Rank Test | No. of Events (%) | Rate/100 Patient-Years | p-Value for Log-Rank Test | |
| All patients | 444 (16.26%) | 3.66 | 218 (7.98%) | 1.70 | 122 (4.47%) | 0.91 | 48 (1.76%) | 0.36 | ||||
| rs11864909 | 0.97 | 0.74 | 0.50 | 0.34 | ||||||||
| TT&TC | 121 (16.29%) | 3.67 | 57 (7.67%) | 1.63 | 30 (4.04%) | 0.82 | 13 (1.75%) | 0.36 | ||||
| CC | 323 (16.25%) | 3.66 | 161 (8.1%) | 1.72 | 92 (4.63%) | 0.95 | 35 (1.76%) | 0.36 | ||||
| rs4293393 | 0.68 | 0.57 | 0.01 | 0.67 | ||||||||
| GA&GG | 68 (17.09%) | 3.82 | 29 (7.29%) | 1.53 | 8 (2.01%) | 0.41 | 4 (1.01%) | 0.20 | ||||
| AA | 376 (16.12%) | 3.63 | 189 (8.1%) | 1.73 | 114 (4.89%) | 1.00 | 44 (1.89%) | 0.39 | ||||
| rs6497476 | 0.84 | 0.31 | 0.02 | 0.90 | ||||||||
| CT&CC | 56 (16.05%) | 3.56 | 23 (6.59%) | 1.39 | 7 (2.01%) | 0.41 | 3 (0.86%) | 0.17 | ||||
| TT | 388 (16.29%) | 3.67 | 195 (8.19%) | 1.75 | 115 (4.83%) | 0.99 | 45 (1.89%) | 0.39 | ||||
| Association Model | Hazard Ratio (95% CI) |
|---|---|
| rs4293393 (GG & GA vs. AA) | |
| Model 1 | 0.388 (0.141, 0.752) |
| Model 2 | 0.399 (0.143, 0.772) |
| Model 3 | 0.370 (0.131, 0.697) |
| Model 4 | 0.341 (0.105, 0.679) |
| rs6497476 (CC & TC vs. TT) | |
| Model 1 | 0.410 (0.156, 0.748) |
| Model 2 | 0.420 (0.159, 0.755) |
| Model 3 | 0.375 (0.147, 0.699) |
| Model 4 | 0.344 (0.104, 0.671) |
| Haplotype composed of rs11864909, rs4293393 and rs6497476 (CGC vs. other types) | |
| Model 1 | 0.168 (0.025, 0.548) |
| Model 2 | 0.176 (0.026, 0.556) |
| Model 3 | 0.141 (0.022, 0.474) |
| Model 4 | 0.118 (0.011, 0.446) |
| Study | rs12917707 (G > T) | rs11864909 (C > T) | rs13333226 (A > G) | rs4293393 (A > G) | rs6497476 (T >C) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | Study Type | Population | MAF | Phenotype/Outcome | MAF | Phenotype/Outcome | MAF | Phenotype/Outcome | MAF | Phenotype/Outcome | MAF | Phenotype/Outcome |
| Anna Kottgen, 2009 [7] | GWAS | Europeans | 0.18 | CKD and eGFR | NA | NA | NA | NA | NA | NA | NA | NA |
| Daniel F. Gudbjartsson, 2010 [9] | GWAS | Icelanders | NA | NA | NA | NA | NA | NA | 0.168–0.202 | CKD and serum creatinine | NA | NA |
| Sandosh Padmanabhan, 2010 [15] | GWAS | Europeans | NA | NA | NA | NA | 0.16-0.23 | Hypertension, cardiovascular events and urinary uromodulin | NA | NA | NA | NA |
| Yukinori Okada, 2012 [10] | GWAS | East Asians | NA | NA | 0.19 | Serum creatinine and eGFR | NA | NA | NA | NA | NA | NA |
| Graciela E. Delgado, 2017 [4] | Candidate gene strategy | Europeans | 0.19 | eGFR and serum uromodulin; All-cause mortality (only among participants aged > 67 years) | NA | NA | NA | NA | NA | NA | NA | NA |
| Jia Han, 2013 [18] | Candidate gene strategy | Chinese | NA | NA | NA | NA | AA: 85.4%, GA + GG: 14.6% | Urine uric acid excretion and plasma uric acid | TT: 85.3%, CT + CC: 14.7% | Urine uric acid excretion and plasma uric acid | TT: 87.3%, CT + CC: 12.7% | Urine uric acid excretion and plasma uric acid |
| Jinwei Wang, current study | Candidate gene strategy | Chinese | NA | NA | 0.148 | eGFR and serum uromodulin | 0.0745 | All-cause mortality | 0.0745 | All-cause mortality | 0.0652 | All-cause mortality |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Liu, L.; He, K.; Gao, B.; Wang, F.; Zhao, M.; Zhang, L.; on behalf of the Chinese Cohort Study of Chronic Kidney Disease (C-STRIDE). UMOD Polymorphisms Associated with Kidney Function, Serum Uromodulin and Risk of Mortality among Patients with Chronic Kidney Disease, Results from the C-STRIDE Study. Genes 2021, 12, 1687. https://doi.org/10.3390/genes12111687
Wang J, Liu L, He K, Gao B, Wang F, Zhao M, Zhang L, on behalf of the Chinese Cohort Study of Chronic Kidney Disease (C-STRIDE). UMOD Polymorphisms Associated with Kidney Function, Serum Uromodulin and Risk of Mortality among Patients with Chronic Kidney Disease, Results from the C-STRIDE Study. Genes. 2021; 12(11):1687. https://doi.org/10.3390/genes12111687
Chicago/Turabian StyleWang, Jinwei, Lili Liu, Kevin He, Bixia Gao, Fang Wang, Minghui Zhao, Luxia Zhang, and on behalf of the Chinese Cohort Study of Chronic Kidney Disease (C-STRIDE). 2021. "UMOD Polymorphisms Associated with Kidney Function, Serum Uromodulin and Risk of Mortality among Patients with Chronic Kidney Disease, Results from the C-STRIDE Study" Genes 12, no. 11: 1687. https://doi.org/10.3390/genes12111687






