MicroRNAs May Play an Important Role in Sexual Reversal Process of Chinese Soft-Shelled Turtle, Pelodiscus sinensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Maintenance of Chinese Soft-Shelled Turtles
2.2. Estradiol Treatment of Chinese Soft-Shelled Turtles
2.3. Sample Collection and RNA Extraction
2.4. Library Preparation for Small RNA
2.5. Bioinformatic Analysis
2.6. Quantitative Real-Time PCR (qPCR)
2.7. Analysis of Differentially Expressed microRNAs and Target Genes
2.8. Statistical Analysis
3. Results
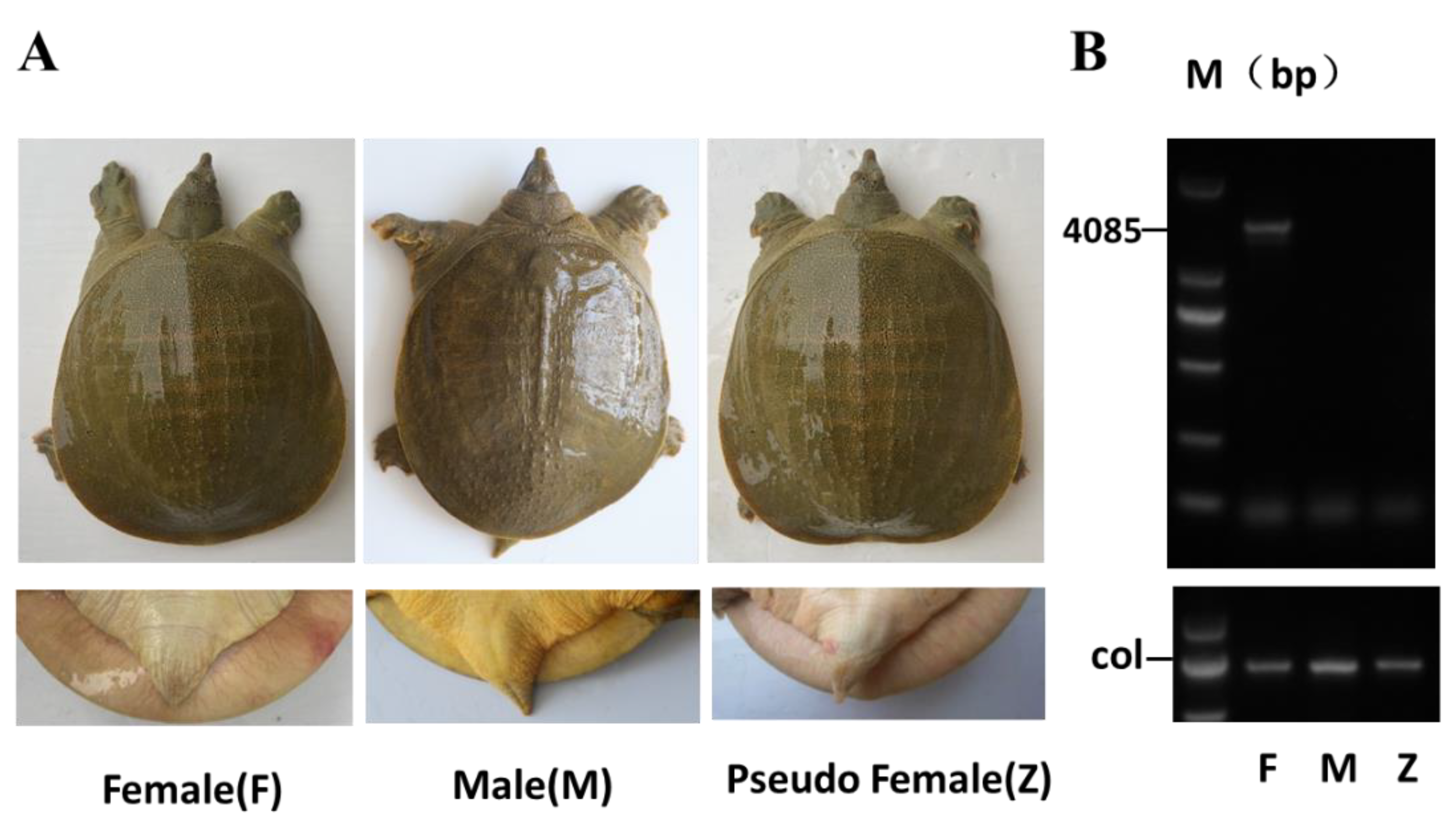
3.1. Pseudo-Female Chinese Soft-Shelled Turtles Were Obtained by E2 Treatment
3.2. The Distribution and Number of Small RNA among Males, Females and Pseudo-Females
3.3. The Character of miRNAs Identified by Males, Females and Pseudo-Females
3.4. Differential miRNAs Analysis of Males, Females and Pseudo-Females (F, M and Z)
3.5. Cluster Analysis of Differential miRNAs and Validation of the Target Genes
3.6. Construction of the Gonadal Related miRNAs and Some Target Genes Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, T.H.; Chang, H.C.; Lue, K.Y. Unregulated Trade in Turtle Shells for Chinese Traditional Medicine in East and Southeast Asia: The Case of Taiwan. Chelonian Conserv. Biol. 2009, 8, 11–18. [Google Scholar] [CrossRef]
- Shao, M.L.; Newman, C.; Buesching, C.D.; Macdonald, D.W.; Zhou, Z.M. Understanding wildlife crime in China: Socio-demographic profiling and motivation of offenders. PLoS ONE 2021, 16, e0246081. [Google Scholar] [CrossRef]
- Gong, S.; Vamberger, M.; Auer, M.; Praschag, P.; Fritz, U. Millennium-old farm breeding of Chinese softshell turtles (Pelodiscus spp.) results in massive erosion of biodiversity. Sci. Nat. 2018, 105, 34. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Wang, L.; Sha, H.; Zou, G. Development and Validation of Sex-Specific Markers in Pelodiscus Sinensis Using Restriction Site-Associated DNA Sequencing. Genes 2019, 10, 302. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhang, G.; Shao, C.; Huang, Q.; Liu, G.; Zhang, P.; Song, W.; An, N.; Chalopin, D.; Volff, J.N.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Sex determination: Why so many ways of doing it? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef] [Green Version]
- Nagahama, Y.; Chakraborty, T.; Prasanth, B.P.; Ohta, K.; Nakamura, M. Sex Determination, Gonadal Sex Differentiation and Plasticity. Physiol. Rev. 2021, 101, 1237–1308. [Google Scholar] [CrossRef]
- Tanaka, K.; Takehana, Y.; Naruse, K.; Hamaguchi, S.; Sakaizumi, M. Evidence for different origins of sex chromosomes in closely related Oryzias fishes: Substitution of the master sex-determining gene. Genetics 2007, 177, 2075–2081. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, M.; Shinomiya, A.; Kinoshita, M.; Suzuki, A.; Kobayashi, T.; Paul-Prasanth, B.; Lau, E.; Hamaguchi, S.; Sakaizumi, M.; Nagahama, Y. The DMY gene induces male development in genetically female (XX) medaka fish. Proc. Natl. Acad. Sci. USA 2007, 104, 3865–3870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, R.; Mank, J.E. The role of sex chromosomes in sexual dimorphism: Discordance between molecular and phenotypic data. J. Evol. Biol. 2014, 27, 1443–1453. [Google Scholar] [CrossRef]
- Mu, Y.; Zhao, B.; Tang, W.Q.; Sun, B.J.; Zeng, Z.G.; Valenzuela, N.; Du, W.G. Temperature-dependent sex determination ruled out in the Chinese soft-shelled turtle (Pelodiscus sinensis) via molecular cytogenetics and incubation experiments across populations. Sex. Dev. 2015, 9, 111–117. [Google Scholar] [CrossRef]
- Zhu, D.Y. Morphplogical and histological observation of gonadal development in soft-shelled turtle. ACTA Hydrobiol. Sin. 2009, 33, 925–930. [Google Scholar] [CrossRef]
- Wang, L.; Sun, W.; Chu, J.; Liu, Y.; Shi, S.; Ge, C.; Qian, G. The Expression Pattern of Sox9 Gene During Embryonic Development and Its Expression Changes in Sex Reversal in Pelodiscus Sinensis. J. Fish. China 2014, 38, 1286–1293. [Google Scholar]
- Liang, H.W.; Meng, Y.; Cao, L.H.; Li, X.; Zou, G.W. Expression and characterization of the cyp19a gene and its responses to estradiol/letrozole exposure in Chinese soft-shelled turtle (Pelodiscus sinensis). Mol. Reprod. Dev. 2019, 86, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Meng, Y.; Cao, L.; Li, X.; Zou, G. Effect of exogenous hormones on R-spondin 1 (RSPO1) gene expression and embryo development in Pelodiscus sinensis. Reprod. Fertil. Dev. 2019, 31, 1425–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.M.; Cheng, S.Y.; Ye, J.B.; Chen, Z.X.; Liao, Y.L.; Zhang, W.W.; Kim, S.U.; Xu, F. Screening and identification of miRNAs related to sexual differentiation of strobili in Ginkgo biloba by integration analysis of small RNA, RNA, and degradome sequencing. BMC Plant. Biol. 2020, 20, 387. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Sun, L.; Shi, H.; Cheng, Y.; Jiang, D.; Fu, B.; Conte, M.A.; Gammerdinger, W.J.; Kocher, T.D.; Wang, D. Integrated analysis of miRNA and mRNA expression profiles in tilapia gonads at an early stage of sex differentiation. BMC Genom. 2016, 17, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikert, H.; Lynch, M.D.J.; Doxey, A.C.; Giesy, J.P.; Servos, M.R.; Katzenback, B.A.; Craig, P.M. High Throughput Sequencing of MicroRNA in Rainbow Trout Plasma, Mucus, and Surrounding Water Following Acute Stress. Front. Physiol. 2020, 11, 588313. [Google Scholar] [CrossRef] [PubMed]
- Saadeldin, I.M.; Oh, H.J.; Lee, B.C. Embryonic-maternal cross-talk via exosomes: Potential implications. Stem Cells Cloning 2015, 8, 103–107. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. microRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhu, W.; Dong, Z.; Song, F.; Dong, J.; Fu, J. Comparative microRNA-seq Analysis Depicts Candidate miRNAs Involved in Skin Color Differentiation in Red Tilapia. Int. J. Mol. Sci. 2018, 19, 1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Noorolyai, S.; Mokhtarzadeh, A.; Baghbani, E.; Asadi, M.; Baghbanzadeh Kojabad, A.; Mogaddam, M.M.; Baradaran, B. The role of microRNAs involved in PI3-kinase signaling pathway in colorectal cancer. J. Cell Physiol. 2019, 234, 5664–5673. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [Green Version]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Plasterk, R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell 2006, 11, 441–450. [Google Scholar] [CrossRef] [Green Version]
- Sun, K.; Lai, E.C. Adult-specific functions of animal microRNAs. Nat. Rev. Genet. 2013, 14, 535–548. [Google Scholar] [CrossRef]
- Bhat, R.A.; Priyam, M.; Foysal, M.J.; Gupta, S.K.; Sundaray, J.K. Role of sex-biased miRNAs in teleosts—A review. Rev. Aquac. 2020, 13, 269–281. [Google Scholar] [CrossRef]
- Presslauer, C.; Tilahun Bizuayehu, T.; Kopp, M.; Fernandes, J.M.; Babiak, I. Dynamics of miRNA transcriptome during gonadal development of zebrafish. Sci. Rep. 2017, 7, 43850. [Google Scholar] [CrossRef] [PubMed]
- Jing, J.; Wu, J.; Liu, W.; Xiong, S.; Ma, W.; Zhang, J.; Wang, W.; Gui, J.F.; Mei, J. Sex-biased miRNAs in gonad and their potential roles for testis development in yellow catfish. PLoS ONE 2014, 9, e107946. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef] [Green Version]
- Kuang, Z.; Wang, Y.; Li, L.; Yang, X. miRDeep-P2: Accurate and fast analysis of the microRNA transcriptome in plants. Bioinformatics 2019, 35, 2521–2522. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Pascual-Anaya, J.; Zadissa, A.; Li, W.; Niimura, Y.; Huang, Z.; Li, C.; White, S.; Xiong, Z.; Fang, D.; et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat. Genet. 2013, 45, 701–706. [Google Scholar] [CrossRef] [Green Version]
- Ge, G.; Long, Y.; Shi, L.; Ren, J.; Yan, J.; Li, C.; Li, Q.; Cui, Z. Transcriptomic profiling revealed key signaling pathways for cold tolerance and acclimation of two carp species. BMC Genom. 2020, 21, 539. [Google Scholar] [CrossRef]
- Yue, C.; Li, Q.; Yu, H. Integrated analysis of miRNA and mRNA expression profiles identifies potential regulatory interactions during sexual development of Pacific oyster Crassostrea gigas. Aquaculture 2022, 546, 737294. [Google Scholar] [CrossRef]
- Wang, Q.; Li, X.; Sha, H.; Luo, X.; Zou, G.; Liang, H. Identification of microRNAs in Silver Carp (Hypophthalmichthys molitrix) Response to Hypoxia Stress. Animals 2021, 11, 2917. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 2, 1101. [Google Scholar] [CrossRef]
- Witkos, T.M.; Koscianska, E.; Krzyzosiak, W.J. Practical Aspects of microRNA Target Prediction. Curr. Mol. Med. 2011, 11, 93–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Hiraki-Kajiyama, T.; Nakajo, M.; Umatani, C.; Kanda, S.; Oka, Y.; Matsumoto, K.; Ozawa, H.; Okubo, K. Sexually Dimorphic Neuropeptide B Neurons in Medaka Exhibit Activated Cellular Phenotypes Dependent on Estrogen. Endocrinology 2019, 160, 827–839. [Google Scholar] [CrossRef] [Green Version]
- Campbell, J.H.; Dixon, B.; Whitehouse, L.M. The intersection of stress, sex and immunity in fishes. Immunogenetics 2021, 73, 111–129. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, L.; Sun, W.; Li, P.; Zhou, Y.; Qian, G.; Ge, C. Knockdown of R-spondin1 leads to partial sex reversal in genetic female Chinese soft-shelled turtle Pelodiscus sinensis. Gen. Comp. Endocrinol. 2021, 309, 113788. [Google Scholar] [CrossRef] [PubMed]
- Toyota, K.; Masuda, S.; Sugita, S.; Miyaoku, K.; Yamagishi, G.; Akashi, H.; Miyagawa, S. Estrogen Receptor 1 (ESR1) Agonist Induces Ovarian Differentiation and Aberrant Mullerian Duct Development in the Chinese Soft-shelled Turtle, Pelodiscus sinensi. Zool Stud. 2020, 59, e54. [Google Scholar] [CrossRef]
- Ma, X.; Cen, S.; Wang, L.; Zhang, C.; Wu, L.; Tian, X.; Wu, Q.; Li, X.; Wang, X. Genome-wide identification and comparison of differentially expressed profiles of miRNAs and lncRNAs with associated ceRNA networks in the gonads of Chinese soft-shelled turtle, Pelodiscus sinensis. BMC Genom. 2020, 21, 443. [Google Scholar] [CrossRef]
- Liu, X.; Zhu, Y.; Wang, Y.; Li, W.; Hong, X.; Zhu, X.; Xu, H. Comparative transcriptome analysis reveals the sexual dimorphic expression profiles of mRNAs and non-coding RNAs in the Asian yellow pond turtle (Meauremys mutica). Gene 2020, 750, 144756. [Google Scholar] [CrossRef]
- Choudhuri, S. Small noncoding RNAs biogenesis, function, and emerging significance in toxicology. J. Biochem. Mol. Toxicol. 2010, 24, 195–216. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef] [Green Version]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Azhar, S.; Dong, D.; Shen, W.J.; Hu, Z.; Kraemer, F.B. The role of miRNAs in regulating adrenal and gonadal steroidogenesis. J. Mol. Endocrinol. 2020, 64, R21–R43. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.; Lai, K.P.; Bao, J.Y.; Zhang, N.; Tse, A.; Tong, A.; Li, J.W.; Lok, S.; Kong, R.Y.; Lui, W.Y.; et al. Identification and expression profiling of microRNAs in the brain, liver and gonads of marine medaka (Oryzias melastigma) and in response to hypoxia. PLoS ONE 2014, 9, e110698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, W.; Zhu, Y.; Wu, Y.; Yuan, C.; Chen, K.; Li, M. Identification and expression analysis of microRNAs in medaka gonads. Gene 2018, 646, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef]
- Wang, C.; Qi, C.; Shu, L.; Li, S.; Zhao, Z.J.O. Dual inhibition of PCDH9 expression by miR-215-5p up-regulation in gliomas. Oncotarget 2017, 8, 10287. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| ID | Primer Sequence (5′-3′) | |
|---|---|---|
| miR-26a-5p | Forward | GCGCGCTTCAAGTAATCCAGGA |
| Reverse | GCAGGGTCCGAGGTATTC | |
| RT1 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCCTA | |
| miR-212-5p | Forward | GCGCACCTTGGCTCTAGACTG |
| Reverse | GCAGGGTCCGAGGTATTC | |
| RT1 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTAAG | |
| miR-202-3p | Forward | GCGCGCAGAGGTGTAGAGCATG |
| Reverse | GCAGGGTCCGAGGTATTC | |
| RT1 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTTTCC | |
| miR-301a | Forward | GCGCGCCAGTGCAATAGTATTG |
| Reverse | GCAGGGTCCGAGGTATTC | |
| RT1 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTTTGA | |
| miR-202-5p | Forward | GCGCGCTTCCTATGCATATACC |
| Reverse | GCAGGGTCCGAGGTATTC | |
| RT1 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAAAGA | |
| miR-96-5p | Forward | GCGCGCTTTGGCACTAGCACATT |
| Reverse | GCAGGGTCCGAGGTATTC | |
| RT1 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCAAA | |
| miR-181b | Forward | GCGCGCCTCACTGATCAATGAA |
| Reverse | GCAGGGTCCGAGGTATTC | |
| RT1 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTTGCA | |
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT | |
| RT1 | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACTGCTG | |
| 4085 | Forward | GTTTGAAGTGCTGCTGGGAAG |
| Reverse | TTCCCCGTATAAAGCCAGGG | |
| actin | Forward | GTGTATGCAACTCTTCCCTCTCCTATTC |
| Reverse | AGCTTCCATTCGGTCTTGTCCTG |
| Exported Data | ||||||
|---|---|---|---|---|---|---|
| Sample | Reads | Bases | Error_rate | Q20 | Q30 | GC_content |
| F | 27,912,069 | 1.396 G | 0% | 100% | 99% | 49% |
| M | 24,060,307 | 1.203 G | 0% | 100% | 99% | 51% |
| Z | 31,682,467 | 1.584 G | 0% | 100% | 99% | 50% |
| Sample | Total_Reads | Total_Bases | Uniq_Reads | Uniq_Bases |
|---|---|---|---|---|
| F | 22,867,409 | 511,199,551 | 374,086 | 9,254,425 |
| M | 14,375,684 | 311,711,063 | 428,327 | 10,622,778 |
| Z | 21,932,331 | 477,377,965 | 462,946 | 11,181,717 |
| miRNA | F | M | Z |
|---|---|---|---|
| miR-143 | 66,453.90 | 69,180.60 | 96,007.02 |
| miR-26a-5p | 70,778.96 | 55,517.76 | 77,512.06 |
| miR-22a-3p | 2.20 | 111.12 | 0.16 |
| miR-212-5p | 13.97 | 62.99 | 8.98 |
| miR-181b-3-3p | 8.28 | 44.57 | 16.66 |
| miR-212 | 10.22 | 25.55 | 4.08 |
| miR-202-3p | 3.23 | 24.36 | 1.96 |
| miR-301a | 1.04 | 18.42 | 2.61 |
| miR-202-5p | 0.13 | 8.91 | 0.65 |
| miR-25-3p | 0.00 | 8.32 | 0.00 |
| miR-96-5p | 68.58 | 7.72 | 25.81 |
| miR-181b-3p | 6.21 | 0.59 | 6.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, T.; Sha, H.; Chen, M.; Chen, G.; Zou, G.; Liang, H. MicroRNAs May Play an Important Role in Sexual Reversal Process of Chinese Soft-Shelled Turtle, Pelodiscus sinensis. Genes 2021, 12, 1696. https://doi.org/10.3390/genes12111696
Zhou T, Sha H, Chen M, Chen G, Zou G, Liang H. MicroRNAs May Play an Important Role in Sexual Reversal Process of Chinese Soft-Shelled Turtle, Pelodiscus sinensis. Genes. 2021; 12(11):1696. https://doi.org/10.3390/genes12111696
Chicago/Turabian StyleZhou, Tong, Hang Sha, Meng Chen, Guobin Chen, Guiwei Zou, and Hongwei Liang. 2021. "MicroRNAs May Play an Important Role in Sexual Reversal Process of Chinese Soft-Shelled Turtle, Pelodiscus sinensis" Genes 12, no. 11: 1696. https://doi.org/10.3390/genes12111696
APA StyleZhou, T., Sha, H., Chen, M., Chen, G., Zou, G., & Liang, H. (2021). MicroRNAs May Play an Important Role in Sexual Reversal Process of Chinese Soft-Shelled Turtle, Pelodiscus sinensis. Genes, 12(11), 1696. https://doi.org/10.3390/genes12111696






