Whole Genome DNA Methylation Variations in Mammary Gland Tissues from Holstein Cattle Producing Milk with Various Fat and Protein Contents
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Sample Collection
2.2. Milk Component Analysis
2.3. DNA Isolation, WGBS Library Construction and Sequencing
2.4. Identification of Methyl-Cytosine
2.5. Global Comparison of Methylated Sites between Different Groups
2.6. DMCs and Quantitative Trait Loci (QTL) Co-Location Analysis
2.7. Statistical Overrepresentation Analysis for DMC Genes
3. Results
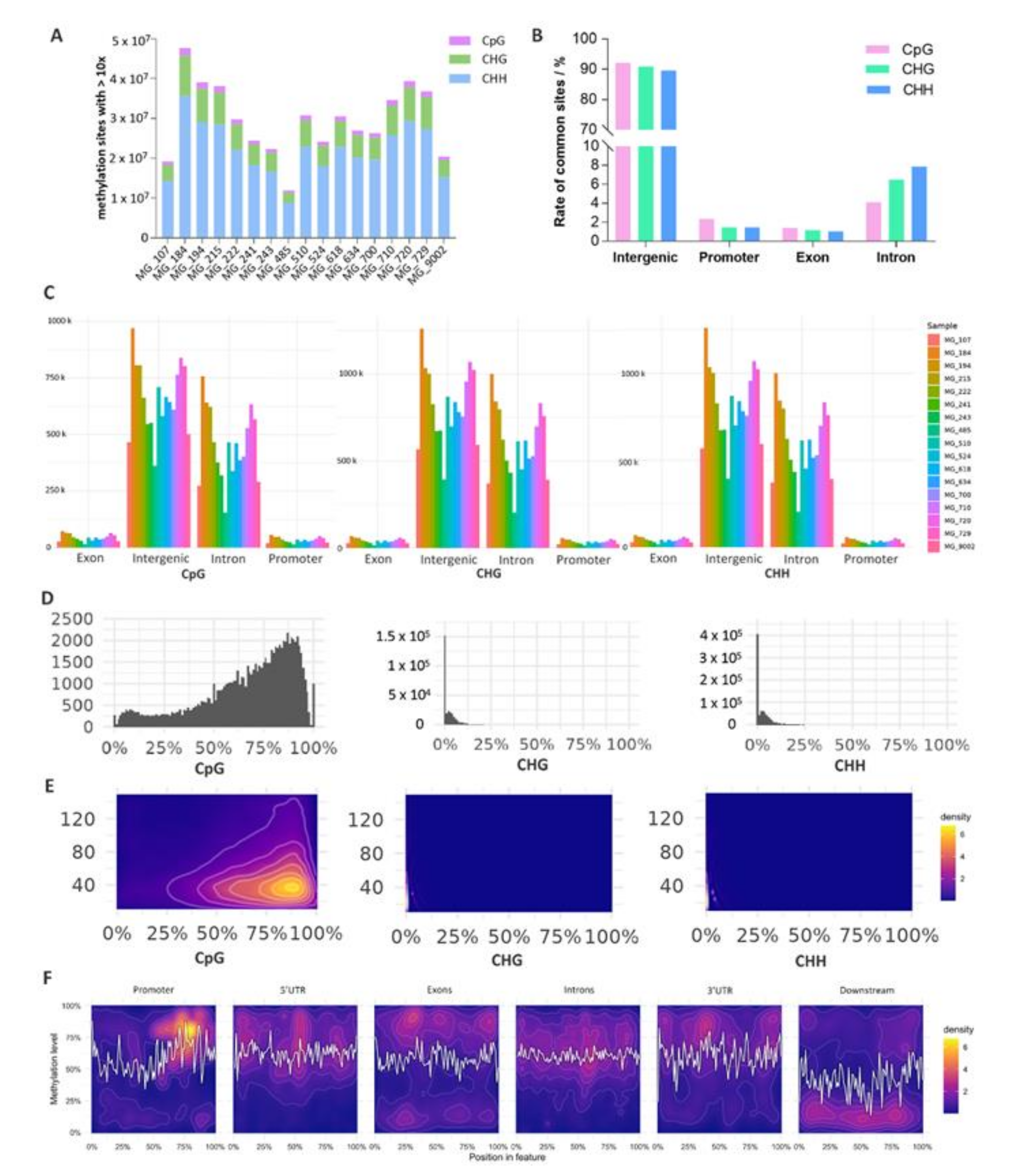
3.1. Genome-Wide DNA Methylation Landscape of Mammary Gland Tissue
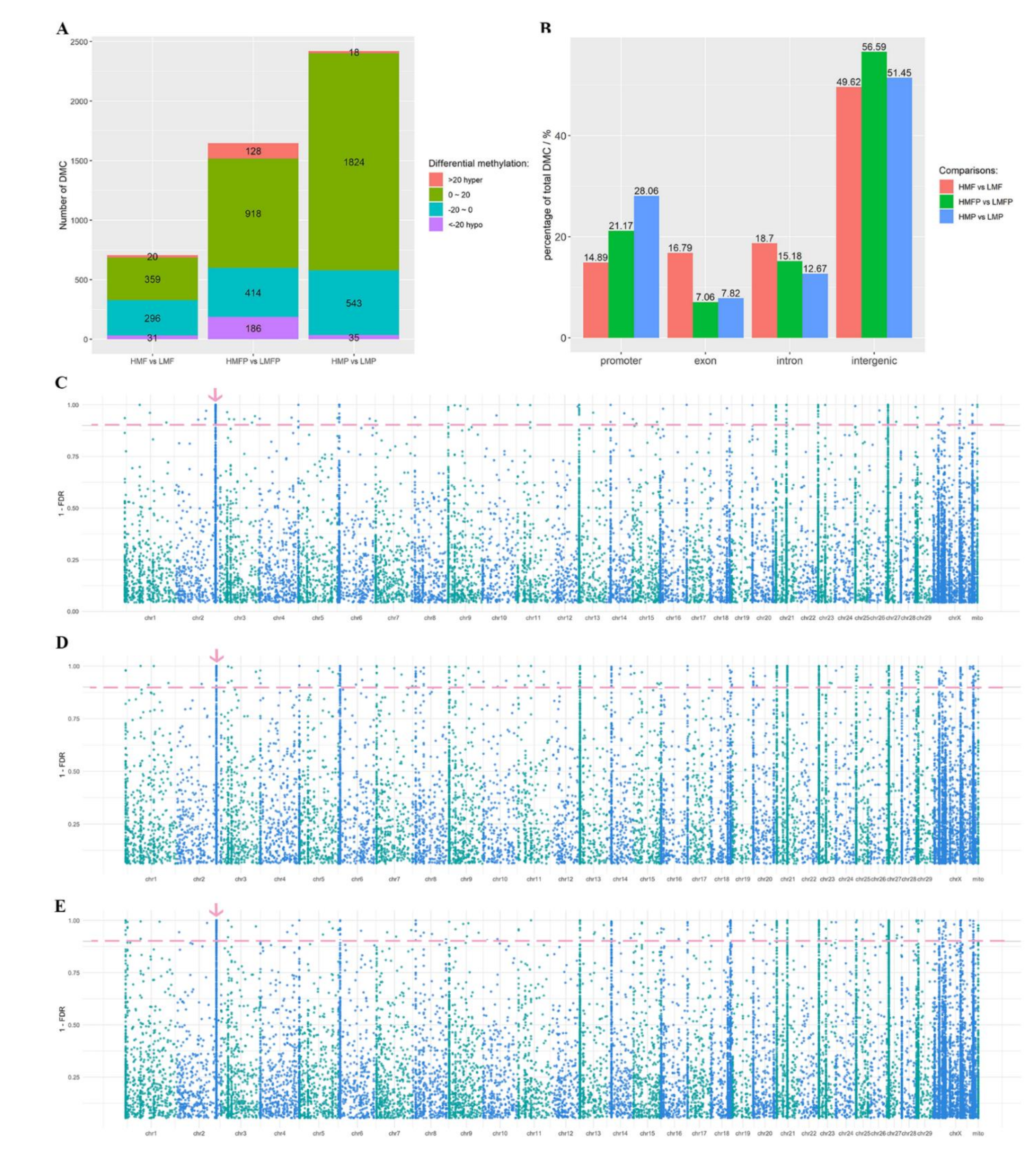
3.2. Differential Methylation between High and Low Milk Fat Producing Cows
3.3. Differential Methylation between High and Low Milk Protein Producing Cows
3.4. Differential Methylation between Cows Producing Milk with High Fat/Protein and Low Fat/Protein Contents
3.5. Overrepresented Gene Ontology Terms and Pathways by DMC Genes
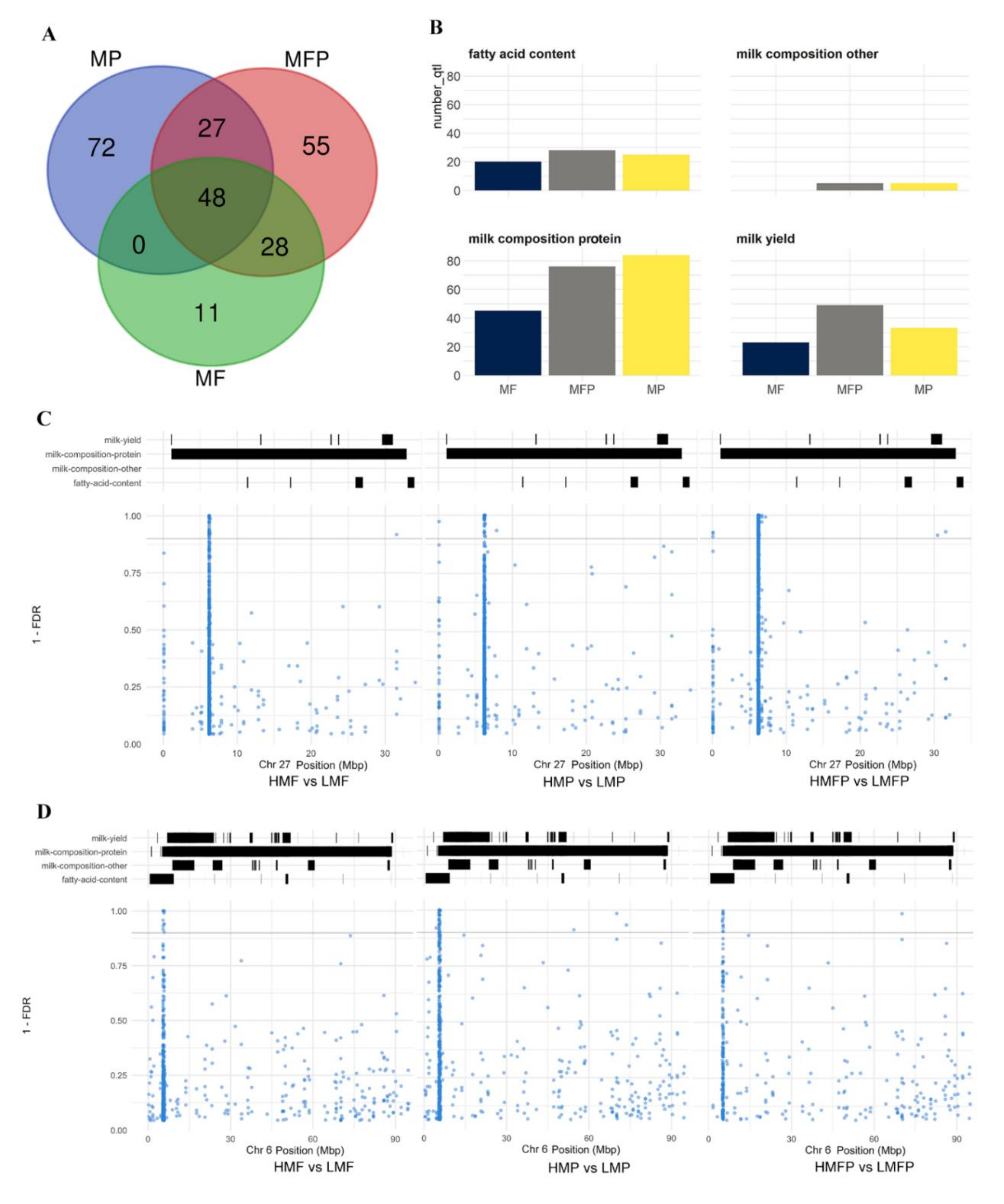
3.6. Overlapping or Co-Located DMCs with QTLs Related to Milk Production Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miglior:, F.; Fleming, A.; Malchiodi, F.; Brito, L.F.; Martin, P.; Baes, C. A 100-Year Review: Identification and genetic selection of economically important traits in dairy cattle. J. Dairy Sci. 2017, 100, 10251–10271. [Google Scholar] [CrossRef] [PubMed]
- Soyeurt, H.; Dardenne, P.; Gillon, A.; Croquet, C.; Vanderick, S.; Mayeres, P.; Bertozzi, C.; Gengler, N. Variation in Fatty Acid Contents of Milk and Milk Fat Within and Across Breeds. J. Dairy Sci. 2006, 89, 4858–4865. [Google Scholar] [CrossRef]
- Bobe, G.; Beitz, D.; Freeman, A.; Lindberg, G. Effect of Milk Protein Genotypes on Milk Protein Composition and Its Genetic Parameter Estimates. J. Dairy Sci. 1999, 82, 2797–2804. [Google Scholar] [CrossRef]
- Analla, M.; Jiménez-Gamero, I.; Muñoz-Serrano, A.; Serradilla, J.; Falagán, A. Estimation of Genetic Parameters for Milk Yield and Fat and Protein Contents of Milk from Murciano-Granadina Goats. J. Dairy Sci. 1996, 79, 1895–1898. [Google Scholar] [CrossRef]
- Dillon, P.; Berry, D.; Evans, R.; Buckley, F.; Horan, B. Consequences of genetic selection for increased milk production in European seasonal pasture based systems of milk production. Livest. Sci. 2006, 99, 141–158. [Google Scholar] [CrossRef]
- Rahayu, A.P.; Johari, S.; Kurnianto, E. Genetic gains of milk yield and milk composition as realized response to dairy cow selection in bbptu-hpt baturraden, indonesia. J. Indones. Trop. Anim. Agric. 2015, 40. [Google Scholar] [CrossRef]
- Wiggans, G.R.; Cole, J.B.; Hubbard, S.M.; Sonstegard, T.S. Genomic Selection in Dairy Cattle: The USDA Experience. Annu. Rev. Anim. Biosci. 2017, 5, 309–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibeagha-Awemu, E.M.; Khatib, H. Epigenetics of livestock breeding. In Handbook of Epigenetics, The New Molecular and Medical Genetics, 2nd ed.; Tollefsbol, T.O., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 441–463. [Google Scholar] [CrossRef]
- Ibeagha-Awemu, E.M.; Zhao, X. Epigenetic marks: Regulators of livestock phenotypes and conceivable sources of missing variation in livestock improvement programs. Front. Genet. 2015, 6, 302. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.; Zhu, X.; Li, R.; Chen, D.; Xin, S.; Zhu, Y.; Liao, X.; Wang, X.; Zhang, H.; Yang, Z.; et al. Methylation analysis of CXCR1 in mammary gland tissue of cows with mastitis induced by Staphylococcus aureus. Genet. Mol. Res. 2015, 14, 12606–12615. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Y.; Sun, Y.; Dong, X.; Wang, Z.; Zhang, Z.; Xiao, Y.; Dong, G. Bacterial Lipopolysaccharide Induced Alterations of Genome-Wide DNA Methylation and Promoter Methylation of Lactation-Related Genes in Bovine Mammary Epithelial Cells. Toxins 2019, 11, 298. [Google Scholar] [CrossRef] [Green Version]
- Rijnkels, M.; Freeman-Zadrowski, C.; Hernandez, J.; Potluri, V.; Wang, L.; Li, W.; Lemay, D. Epigenetic Modifications Unlock the Milk Protein Gene Loci during Mouse Mammary Gland Development and Differentiation. PLoS ONE 2013, 8, e53270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, K.; Molenaar, A.J.; Swanson, K.M.; Gudex, B.; Arias, J.A.; Erdman, R.A.; Stelwagen, K. Epigenetics: A possible role in acute and transgenerational regulation of dairy cow milk production. Animal 2012, 6, 375–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osorio, J.; Jacometo, C.; Zhou, Z.; Luchini, D.; Cardoso, F.; Loor, J.J. Hepatic global DNA and peroxisome proliferator-activated receptor alpha promoter methylation are altered in peripartal dairy cows fed rumen-protected methionine. J. Dairy Sci. 2016, 99, 234–244. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Ibeagha-Awemu, E.M. Impacts of Epigenetic Processes on the Health and Productivity of Livestock. Front. Genet. 2021, 11. [Google Scholar] [CrossRef]
- Wang, L.; Sun, H.; Guan, L.; Liu, J. Short communication: Relationship of blood DNA methylation rate and milk performance in dairy cows. J. Dairy Sci. 2019, 102, 5208–5211. [Google Scholar] [CrossRef]
- Dechow, C.D.; Liu, W.-S. DNA methylation patterns in peripheral blood mononuclear cells from Holstein cattle with variable milk yield. BMC Genom. 2018, 19, 744. [Google Scholar] [CrossRef]
- Platenburg, G.J.; Vollebregt, E.J.; Karatzas, C.N.; Kootwijk, E.P.; De Boer, H.A.; Strijker, R. Mammary gland-specific hypomethylation ofhpa ii sites flanking the bovine αs1-casein gene. Transgenic Res. 1996, 5, 421–431. [Google Scholar] [CrossRef]
- Vanselow, J.; Yang, W.; Herrmann, J.; Zerbe, H.; Schuberth, H.-J.; Petzl, W.; Tomek, W.; Seyfert, H.-M. DNA-remethylation around a STAT5-binding enhancer in the αS1-casein promoter is associated with abrupt shutdown of αS1-casein synthesis during acute mastitis. J. Mol. Endocrinol. 2006, 37, 463–477. [Google Scholar] [CrossRef]
- Nguyen, M.; Boutinaud, M.; Petridou, B.; Gabory, A.; Pannetier, M.; Chat, S.; Bouet, S.; Jouneau, L.; Jaffrezic, F.; Laloë, D.; et al. DNA Methylation and Transcription in a Distal Region Upstream from the Bovine AlphaS1 Casein Gene after Once or Twice Daily Milking. PLoS ONE 2014, 9, e111556. [Google Scholar] [CrossRef]
- Wang, H.; Shi, H.; Luo, J.; Yi, Y.; Yao, D.; Zhang, X.; Ma, G.; Loor, J.J. MiR-145 Regulates Lipogenesis in Goat Mammary Cells Via TargetingINSIG1and Epigenetic Regulation of Lipid-Related Genes. J. Cell. Physiol. 2016, 232, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Luo, Y.; Li, X.; Tian, J.; Tao, S.; Hua, C.; Geng, Y.; Ni, Y.; Zhao, R. Negative effects of long-term feeding of high-grain diets to lactating goats on milk fat production and composition by regulating gene expression and DNA methylation in the mammary gland. J. Anim. Sci. Biotechnol. 2017, 8, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. Babraham bioinformatics. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 22 November 2020).
- Krueger, F. Trim Galore: A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. Babraham Bioinformatics. 2015. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 22 November 2020).
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with bwa-mem. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Sedgwick, P. Multiple significance tests: The Bonferroni correction. BMJ 2011, 344, e509. [Google Scholar] [CrossRef]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De novo assembly of the cattle reference genome with single-molecule sequencing. GigaScience 2020, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. Panther version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018, 47, D419–D426. [Google Scholar] [CrossRef]
- García-Ruiz, A.; Cole, J.B.; VanRaden, P.M.; Wiggans, G.R.; Ruiz-López, F.J.; Van Tassell, C.P. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc. Natl. Acad. Sci. USA 2016, 113, E3995–E4004. [Google Scholar] [CrossRef] [Green Version]
- Ibeagha-Awemu, E.M.; Yu, Y. Consequence of epigenetic processes on animal health and productivity: Is additional level of regulation of relevance? Anim. Front. 2021. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, S.; Hu, Y.; Fang, L.; Gao, Y.; Xia, H.; Schroeder, S.G.; Rosen, B.D.; Connor, E.E.; Li, C.-J.; et al. Comparative whole genome DNA methylation profiling across cattle tissues reveals global and tissue-specific methylation patterns. BMC Biol. 2020, 18, 85. [Google Scholar] [CrossRef]
- Hao, Y.; Cui, Y.; Gu, X. Genome-wide DNA methylation profiles changes associated with constant heat stress in pigs as measured by bisulfite sequencing. Sci. Rep. 2016, 6, 27507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Jin, H.-G.; Ma, H.-H.; Zhao, Z.-H. Comparative analysis on genome-wide DNA methylation in longissimus dorsi muscle between Small Tailed Han and Dorper×Small Tailed Han crossbred sheep. Asian-Australas. J. Anim. Sci. 2017, 30, 1529–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz, R.; Lewis, Z.A.; Goll, M.G. DNA Methylation: Shared and Divergent Features across Eukaryotes. Trends Genet. 2019, 35, 818–827. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, S.; Ma, L.; Jiang, E.; Xu, H.; Chen, R.; Yang, Q.; Chen, H.; Li, Z.; Lan, X. Reduced representation bisulfite sequencing (RRBS) of dairy goat mammary glands reveals DNA methylation profiles of integrated genome-wide and critical milk-related genes. Oncotarget 2017, 8, 115326–115344. [Google Scholar] [CrossRef] [Green Version]
- Laurent, L.; Wong, E.; Li, G.; Huynh, T.; Tsirigos, A.; Ong, C.T.; Low, H.M.; Sung, W.-K.; Rigoutsos, I.; Loring, J.; et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010, 20, 320–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sati, S.; Tanwar, V.S.; Kumar, K.A.; Patowary, A.; Jain, V.; Ghosh, S.; Ahmad, S.; Singh, M.; Reddy, S.U.; Chandak, G.R.; et al. High Resolution Methylome Map of Rat Indicates Role of Intragenic DNA Methylation in Identification of Coding Region. PLoS ONE 2012, 7, e31621. [Google Scholar] [CrossRef]
- Zhang, Y.; Yanli, Z.; FengZhe, L.; Yang, H.; Zhu, A.; Pang, J.; Han, L.; Zhang, T.; Yao, X.; Wang, F. Genome-wide analysis of DNA Methylation profiles on sheep ovaries associated with prolificacy using whole-genome Bisulfite sequencing. BMC Genom. 2017, 18, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.; Bourc’His, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Razin, A.; Kantor, B. DNA Methylation in Epigenetic Control of Gene Expression. In Epigenetics and Chromatin; Jeanteur, P., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 38, pp. 151–167. [Google Scholar] [CrossRef]
- Deaton, A.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Liang, Y.; Ibeagha-Awemu, E.M.; Li, M.; Zhang, H.; Chen, Z.; Sun, Y.; Karrow, N.A.; Yang, Z.; Mao, Y. Genome-wide DNA methylation analysis of mammary gland tissues from chinese holstein cows with staphylococcus aureus induced mastitis. Front. Genet. 2020, 11, 1295. [Google Scholar] [CrossRef]
- Zhao, Z.; Fu, Y.-X.; Hewett-Emmett, D.; Boerwinkle, E. Investigating single nucleotide polymorphism (SNP) density in the human genome and its implications for molecular evolution. Gene 2003, 312, 207–213. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, Y.; Mukiibi, R.; Chen, L.; Vinsky, M.; Plastow, G.; Basarab, J.; Stothard, P.; Li, C. Genetic architecture of quantitative traits in beef cattle revealed by genome wide association studies of imputed whole genome sequence variants: I: Feed efficiency and component traits. BMC Genom. 2020, 21, 36. [Google Scholar] [CrossRef]
- Ibeagha-Awemu, E.M.; Peters, S.O.; Akwanji, K.A.; Imumorin, I.G.; Zhao, X. High density genome wide genotyping-by-sequencing and association identifies common and low frequency snps, and novel candidate genes influencing cow milk traits. Sci. Rep. 2016, 6, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Osorio, J.S.; Lohakare, J.; Bionaz, M. Biosynthesis of milk fat, protein, and lactose: Roles of transcriptional and posttran-scriptional regulation. Physiol. Genom. 2016, 48, 231–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emery, R. Feeding For Increased Milk Protein. J. Dairy Sci. 1978, 61, 825–828. [Google Scholar] [CrossRef]
- Bionaz, M.; Hurley, W.; Loor, J. Milk Protein Synthesis in the Lactating Mammary Gland: Insights from Transcriptomics Analyses. Milk Protein 2012. [Google Scholar] [CrossRef] [Green Version]
- Lock, A.; Teles, B.; Perfield, J., II; Bauman, D.; Sinclair, L. A conjugated linoleic acid supplement containing trans-10, cis-12 reduces milk fat synthesis in lactating sheep. J. Dairy Sci. 2006, 89, 1525–1532. [Google Scholar] [CrossRef] [Green Version]
- Fardi, Masoumeh, Saeed Solali, and Majid Farshdousti Hagh. "Epigenetic mechanisms as a new approach in cancer treatment: An updated review. Genes Dis. 2018, 5, 304–311. [CrossRef] [PubMed]
- Ghoshal, K.; Majumder, S.; Datta, J.; Motiwala, T.; Bai, S.; Sharma, S.M.; Frankel, W.; Jacob, S.T. Role of human ribosomal rna (rrna) promoter methylation and of methyl-cpg-binding protein mbd2 in the suppression of rrna gene expression. J. Biol. Chem. 2004, 279, 6783–6793. [Google Scholar] [CrossRef] [Green Version]
- Gagnon-Kugler, T.; Langlois, F.; Stefanovsky, V.; Lessard, F.; Moss, T. Loss of human ribosomal gene cpg methylation en-hances cryptic rna polymerase ii transcription and disrupts ribosomal rna processing. Mol. Cell 2009, 35, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, R.J.; Pikaard, C. Chromatin Turn Ons and Turn Offs of Ribosomal RNA Genes. Cell Cycle 2004, 3, 878–881. [Google Scholar] [CrossRef]
- Espada, J.; Ballestar, E.; Santoro, R.; Fraga, M.; Garea, A.V.; Németh, A.; Lopez-Serra, L.; Ropero, S.; Aranda, A.; Orozco, H.; et al. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007, 35, 2191–2198. [Google Scholar] [CrossRef] [PubMed]
- Gensous, N.; Ravaioli, F.; Pirazzini, C.; Gramignoli, R.; Ellis, E.; Storci, G.; Capri, M.; Strom, S.; Laconi, E.; Franceschi, C.; et al. Aging and Caloric Restriction Modulate the DNA Methylation Profile of the Ribosomal RNA Locus in Human and Rat Liver. Nutrients 2020, 12, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Aquila, P.; Montesanto, A.; Mandalà, M.; Garasto, S.; Mari, V.; Corsonello, A.; Bellizzi, D.; Passarino, G. Methylation of the ribosomal RNA gene promoter is associated with aging and age-related decline. Aging Cell 2017, 16, 966–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietrzak, M.; Rempala, G.; Nelson, P.T.; Zheng, J.-J.; Hetman, M. Epigenetic silencing of nucleolar rrna genes in alzheimer’s disease. PloS ONE 2011, 6, e22585. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Tsuneoka, M. Control of ribosomal rna transcription by nutrients. In Gene Expression and Regulation in Mammalian cells—Transcription toward the Establishment of Novel Therapeutics; Fumiaki, U., Ed.; IntechOpen: London, UK, 2018; pp. 25–41. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.-Y.; Yan, X.-L.; Han, P.-P.; Tang, Y.-J.; Li, H.-X. Microarray and Bioinformatics Analysis of Differentially-Expressed MicroRNAs in Wnt/Β-Catenin Signaling-Activated Bovine Skeletal Muscle Satellite Cells. Pak. J. Zool. 2020, 52. [Google Scholar] [CrossRef]
- Yakovlev, A.F. Epigenetic Effects in Livestock Breeding. Russ. J. Genet. 2018, 54, 897–909. [Google Scholar] [CrossRef]
- Chhabra, R. miRNA and Methylation: A Multifaceted Liaison. ChemBioChem 2014, 16, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bian, Y.; Wang, Z.; Li, D.; Wang, C.; Li, Q.; Gao, X. Microrna-152 regulates DNA methyltransferase 1 and is involved in the development and lactation of mammary glands in dairy cows. PLoS ONE 2014, 9, e101358. [Google Scholar] [CrossRef] [PubMed]
- Imgenberg-Kreuz, J.; Almlöf, J.C.; Leonard, D.; Alexsson, A.; Nordmark, G.; Eloranta, M.-L.; Rantapää-Dahlqvist, S.; A Bengtsson, A.; Jönsen, A.; Padyukov, L.; et al. DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2018, 77, 736–743. [Google Scholar] [CrossRef] [Green Version]
- Banovich, N.; Lan, X.; McVicker, G.; Van De Geijn, B.; Degner, J.F.; Blischak, J.; Roux, J.; Pritchard, J.K.; Gilad, Y. Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels. PLoS Genet. 2014, 10, e1004663. [Google Scholar] [CrossRef] [Green Version]
- Maldonado, M.B.C.; Neto, N.B.D.R.; Nagamatsu, S.T.; Carazzolle, M.F.; Hoff, J.L.; Whitacre, L.K.; Schnabel, R.D.; Behura, S.K.; McKay, S.D.; Taylor, J.F.; et al. Identification of bovine CpG SNPs as potential targets for epigenetic regulation via DNA methylation. PLoS ONE 2019, 14, e0222329. [Google Scholar] [CrossRef]
- Zhi, D.; Aslibekyan, S.; Irvin, M.R.; Claas, S.A.; Borecki, I.B.; Ordovas, J.M.; Absher, D.M.; Arnett, D.K. SNPs located at CpG sites modulate genome-epigenome interaction. Epigenetics 2013, 8, 802–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.; Dusza, M.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Distinguishing pleiotropy from linked QTL between milk production traits and mastitis resistance in Nordic Holstein cattle. Genet. Sel. Evol. 2020, 52, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Atashi, H.; Salavati, M.; De Koster, J.; Ehrlich, J.; Crowe, M.; Opsomer, G.; Hostens, M. Genome-wide association for milk production and lactation curve parameters in Holstein dairy cows. J. Anim. Breed. Genet. 2020, 137, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-W.; Lee, K.-T.; Liao, X.; Stothard, P.; An, H.-S.; Ahn, S.; Lee, S.; Lee, S.-Y.; Moore, S.S.; Kim, T.-H. Genome-wide copy number variation in Hanwoo, Black Angus, and Holstein cattle. Mamm. Genome 2013, 24, 151–163. [Google Scholar] [CrossRef]
| Animal ID | Phenotype/Analysis Group 1 | Parity | DIM 2 | Fat (%) 2 | Protein (%) 2 |
|---|---|---|---|---|---|
| High milk fat (HMF) Group | |||||
| 241 | HMF-HMP | 3 | 262 | 5.55 ± 0.38 | 4.1 ± 0.22 |
| 194 | HMF-HMP | 4 | 216 | 5.20 ± 0.18 | 3.83 ± 0.22 |
| 184 | HMF | 5 | 264 | 5.09 ± 0.29 | 3.40 ± 0.14 |
| 215 | HMF | 4 | 89 | 5.05 ± 0.07 | 2.98 ± 0.21 |
| 524 | HMF | 3 | 145 | 4.93 ± 0.66 | 3.28 ± 0.35 |
| 222 | HMF | 4 | 230 | 4.90 ± 0.655 | 3.39 ± 0.096 |
| 700 | HMF-HMP | 1 | 318 | 4.83 ± 1.18 | 3.83 ± 0.29 |
| Low milk fat (LMF) group | |||||
| 618 | LMF-LMP | 2 | 155 | 3.40 ± 0.22 | 3.15 ± 0.13 |
| 729 | LMF-LMP | 1 | 99 | 3.55 ± 0.37 | 2.93 ± 0.22 |
| 243 | LMF-LMP | 3 | 205 | 3.64 ± 0.15 | 3.11± 0.13 |
| 634 | LMF | 2 | 58 | 3.70 ± 0.31 | 3.3 ± 0.377 |
| 107 | LMF | 7 | 203 | 3.70 ± 2.05 | 3.28 ± 0.28 |
| High milk protein (HMP)group | |||||
| 241 | HMF-HMP | 3 | 262 | 5.55 ± 0.38 | 4.1 ± 0.22 |
| 194 | HMF-HMP | 4 | 216 | 5.20 ± 0.18 | 3.83 ± 0.22 |
| 700 | HMF-HMP | 1 | 318 | 4.83 ± 1.18 | 3.83 ± 0.29 |
| 9002 | HMP | 3 | 48 | 4.58 ± 0.49 | 3.70 ± 0.848 |
| 710 | HMP | 1 | 352 | 4.38 ± 0.096 | 3.63 ± 0.08 |
| Low milk protein (LMP) group | |||||
| 720 | LMP | 1 | 111 | 4.7 ± 0.65 | 2.9 ± 0.32 |
| 729 | LMF-LMP | 1 | 99 | 3.55 ± 0.37 | 2.93 ± 0.22 |
| 215 | HMF-LMP | 4 | 89 | 5.05 ± 0.07 | 2.98 ± 0.21 |
| 510 | LMP | 3 | 192 | 4.58 ± 0.63 | 3.00 ± 0.19 |
| 485 | LMP | 4 | 132 | 4.33 ± 0.32 | 3.00 ± 0.43 |
| 243 | LMF-LMP | 3 | 205 | 3.64 ± 0.15 | 3.11 ± 0.13 |
| 618 | LMF-LMP | 2 | 155 | 3.40 ± 0.22 | 3.15 ± 0.13 |
| High milk fat and protein (HMFP) group | |||||
| 241 | HMF-HMP | 3 | 262 | 5.55 ± 0.38 | 4.1 ± 0.22 |
| 194 | HMF-HMP | 4 | 216 | 5.20 ± 0.18 | 3.83 ± 0.22 |
| 700 | HMF-HMP | 1 | 318 | 4.83 ± 1.18 | 3.83 ± 0.29 |
| Low milk fat and protein (LMFP) group | |||||
| 618 | LMF-LMP | 2 | 155 | 3.40 ± 0.22 | 3.15 ± 0.13 |
| 729 | LMF-LMP | 1 | 99 | 3.55 ± 0.37 | 2.93 ± 0.22 |
| 243 | LMF-LMP | 3 | 205 | 3.64 ± 0.15 | 3.11 ± 0.13 |
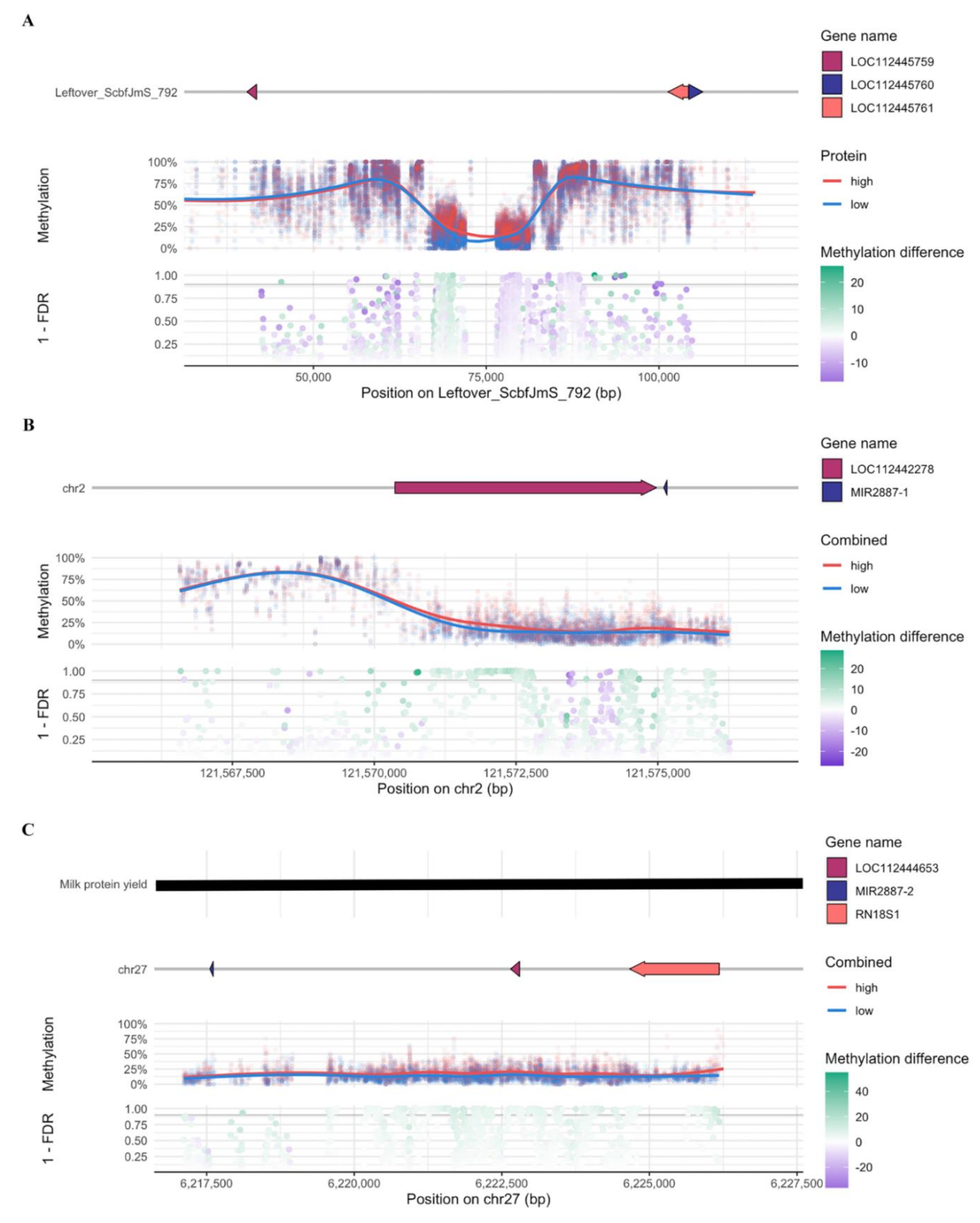
| Comparison 1 | Gene Name | Gene Description | Chr 2 | DMCs with Increased Methylation Level | DMCs with Decreased Methylation Level |
|---|---|---|---|---|---|
| HMF vs. LMF | LOC112442278 | Basic proline-rich protein-like | chr2 | 96 | 6 |
| LOC112444653 | 5.8S ribosomal RNA | chr27 | 12 | 1 | |
| PDE5A | Phosphodiesterase 5A | chr6 | 3 | 3 | |
| MIR2887-1 | MicroRNA mir-2887-1 | chr2 | 1 | 3 | |
| HMP vs. LMP | LOC112442278 | Basic proline-rich protein-like | chr2 | 328 | 0 |
| LOC112444653 | 5.8S ribosomal RNA | chr27 | 211 | 0 | |
| MIR2887-1 | MicroRNA mir-2887-1 | chr2 | 84 | 0 | |
| RN18S1 | 18S ribosomal RNA | chr27 | 60 | 0 | |
| MIR2887-2 | MicroRNA mir-2887-2 | chr27 | 14 | 0 | |
| LOC112443250 | 5S ribosomal RNA | chr21 | 9 | 1 | |
| PDE5A | Phosphodiesterase 5A | chr6 | 3 | 1 | |
| HMFP vs. LMFP | LOC112442278 | Basic proline-rich protein-like | chr2 | 168 | 6 |
| LOC112444653 | 5.8S ribosomal RNA | chr27 | 108 | 0 | |
| RN18S1 | 18S ribosomal RNA | chr27 | 35 | 0 | |
| MIR2887-1 | MicroRNA mir-2887-1 | chr2 | 30 | 0 | |
| ENSBTAG00000052622.1 | Novel gene (protein coding) | chr14 | 1 | 9 | |
| PDE5A | Phosphodiesterase 5A | chr6 | 2 | 3 | |
| MIR2887-2 | MicroRNA mir-2887-2 | chr27 | 5 | 0 | |
| FBXO16 | F-box protein 16 | chr8 | 1 | 3 | |
| DEFB7 | Befensin β 7 | chr27 | 0 | 3 | |
| COX1 | Cytochrome c oxidase subunit I | chrM | 2 | 1 | |
| ENSBTAG00000043565.1 | Novel gene (Mt tRNA) | chrM | 2 | 1 |
| Comparison 1 | GO Identification | GO Term Name | GO Category 2 | Fold Enrichment | + 3 | Adjust p Value |
|---|---|---|---|---|---|---|
| HMP vs. LMP | GO:0019646 | Aerobic electron transport chain | BP | 37.81 | + | 0.0355 |
| GO:0009060 | Aerobic respiration | BP | 20.97 | + | 0.0335 | |
| GO:0006119 | Oxidative phosphorylation | BP | 28.02 | + | 0.0085 | |
| GO:0046034 | ATP metabolic process | BP | 18.53 | + | 0.0066 | |
| GO:0042775 | Mitochondrial ATP synthesis coupled electron transport | BP | 35.18 | + | 0.0466 | |
| GO:0009055 | Electron transfer activity | MF | 25.33 | + | 0.0046 | |
| GO:0070469 | Respirasome | CC | 20.56 | + | 0.0062 | |
| HMFP vs. LMFP | GO:0042773 | ATP synthesis coupled electron transport | BP | 34.88 | + | 0.0032 |
| GO:0006119 | Oxidative phosphorylation | BP | 28.15 | + | 0.0007 | |
| GO:0022904 | Respiratory electron transport chain | BP | 26.25 | + | 0.0124 | |
| GO:0009060 | Aerobic respiration | BP | 21.07 | + | 0.0036 | |
| GO:0022900 | Electron transport chain | BP | 18.71 | + | 0.0070 | |
| GO:0046034 | ATP metabolic process | BP | 18.1 | + | 0.0010 | |
| GO:0045333 | Cellular respiration | BP | 17.58 | + | 0.0100 | |
| GO:0015980 | Energy derivation by oxidation of organic compounds | BP | 13.65 | + | 0.0411 | |
| GO:0009055 | Electron transfer activity | MF | 25.45 | + | 0.0004 | |
| GO:0098803 | Respiratory chain complex | CC | 23.56 | + | 0.0003 | |
| GO:0070469 | Respirasome | CC | 24.1 | + | 0.0000 | |
| GO:0005746 | Mitochondrial respirasome | CC | 20.55 | + | 0.0066 | |
| GO:1990204 | Oxidoreductase complex | CC | 17.56 | + | 0.0138 | |
| GO:0098800 | Inner mitochondrial membrane protein complex | CC | 13.88 | + | 0.0415 |
| QTL ID | QTL Symbol 1 | QTL Trait Name | Chr 2 | Start Position | End Position | QTL Trait Class | DMC (HMF vs. LMF) 3 | DMC (HMP vs. LMP) 4 | DMC (HMFP vs. LMFP) 5 |
|---|---|---|---|---|---|---|---|---|---|
| 10104 | PY | Milk protein yield | chr27 | 1099081 | 32912109 | milk-composition-protein | 36 | 534 | 299 |
| 2611 | PY | Milk protein yield | chr27 | 5484642 | 21801052 | milk-composition-protein | 35 | 533 | 297 |
| 2592 | MY | Milk yield | chr27 | 5484642 | 21801052 | milk-yield | 35 | 533 | 297 |
| 12232 | ALD-C18:0 | Stearic aldehyde content | chr6 | 612894 | 9283340 | fatty-acid-content | 11 | 11 | 10 |
| 12233 | FA-C22:6 | Docosahexaenoic acid content | chr6 | 612894 | 9283340 | fatty-acid-content | 11 | 11 | 10 |
| 12234 | N6N3R | Omega-6 to omega-3 fatty acid ratio | chr6 | 612894 | 9283340 | fatty-acid-content | 11 | 11 | 10 |
| 10208 | PP | Milk protein percentage | chr6 | 5023293 | 18528012 | milk-composition-protein | 11 | 11 | 10 |
| 6065 | PY | Milk protein yield | chr6 | 767213 | 13254392 | milk-composition-protein | 11 | 11 | 10 |
| 10148 | PY | Milk protein yield | chr6 | 5747732 | 5928842 | milk-composition-protein | 6 | 4 | 5 |
| 2443 | PP | Milk protein percentage | chr3 | 12993207 | 44877667 | milk-composition-protein | 3 | 3 | 4 |
| Gene ID | Gene Name | Chr 1 | DMCs 2 | QTL Trait Class | QTL Trait | QTL ID | Gene Function | ||
|---|---|---|---|---|---|---|---|---|---|
| HMF vs. LMF | LMP vs. HMP | HMFP vs. LMFP | |||||||
| RN18S1 | 18S ribosomal RNA | Chr27 | 1 | 60 | 35 | Milk-composition-protein | Milk protein yield | 10104; 2611 | Structural constituent of ribosome |
| Milk yield | 2592 | ||||||||
| LOC112444653 | 5.8S ribosomal RNA | Chr27 | 1 | 17 | 16 | Milk-composition-protein | Milk protein yield | 10104; 2611 | Structural constituent of ribosome |
| Milk yield | 2592 | ||||||||
| PDE5A | Phosphodiesterase 5A | Chr6 | 6 | 4 | 5 | fatty-acid-content | Stearic aldehyde content | 12232 | Implicated in 3′,5′-cyclic-GMP phosphodiesterase activity; Involved in cGMP binding; Involved in cyclic-nucleotide phosphodiesterase activity; Involved in metal ion binding |
| Docosahexaenoic acid content | 12233 | ||||||||
| Omega-6 to omega-3 fatty acid ratio | 12234 | ||||||||
| Milk-composition-protein | Milk protein yield | 10148; 6065 | |||||||
| Milk protein percentage | 10208 | ||||||||
| FBXO16 | F-box protein 16 | Chr8 | 1 | 1 | 4 | Milk-yield | Milking speed | 3438 | Involved in protein binding |
| DEFB7 | defensin β 7 | Chr27 | 0 | 1 | 3 | Milk-composition-protein | Milk protein yield | 10104; 2611 | Involved in positive chemotaxis |
| Milk yield | 2592 | ||||||||
| ENSBTAG00000052622.1 | Novel gene (protein coding) | Chr14 | 0 | 1 | 9 | Milk-composition-protein; milk-yield | Milk protein percentage | 2604; 3413 | None |
| Milk protein yield | 10099, 10100, 10101 | ||||||||
| Milk yield | 3608 | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Bissonnette, N.; Dudemaine, P.-L.; Zhao, X.; Ibeagha-Awemu, E.M. Whole Genome DNA Methylation Variations in Mammary Gland Tissues from Holstein Cattle Producing Milk with Various Fat and Protein Contents. Genes 2021, 12, 1727. https://doi.org/10.3390/genes12111727
Wang M, Bissonnette N, Dudemaine P-L, Zhao X, Ibeagha-Awemu EM. Whole Genome DNA Methylation Variations in Mammary Gland Tissues from Holstein Cattle Producing Milk with Various Fat and Protein Contents. Genes. 2021; 12(11):1727. https://doi.org/10.3390/genes12111727
Chicago/Turabian StyleWang, Mengqi, Nathalie Bissonnette, Pier-Luc Dudemaine, Xin Zhao, and Eveline M. Ibeagha-Awemu. 2021. "Whole Genome DNA Methylation Variations in Mammary Gland Tissues from Holstein Cattle Producing Milk with Various Fat and Protein Contents" Genes 12, no. 11: 1727. https://doi.org/10.3390/genes12111727







