Quantitative Phosphoproteomics of cipk3/9/23/26 Mutant and Wild Type in Arabidopsis thaliana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Protein Extraction and Trypsin Digestion
2.3. TMT Labeling and Reversed-Phase High-Performance Liquid Chromatography Fractionation
2.4. Modified Enrichment and LC-MS/MS Analyses
2.5. Database Search and LC-MS/MS Data Analyses
3. Results
3.1. Quantitative Phosphoproteomic Data Analysis
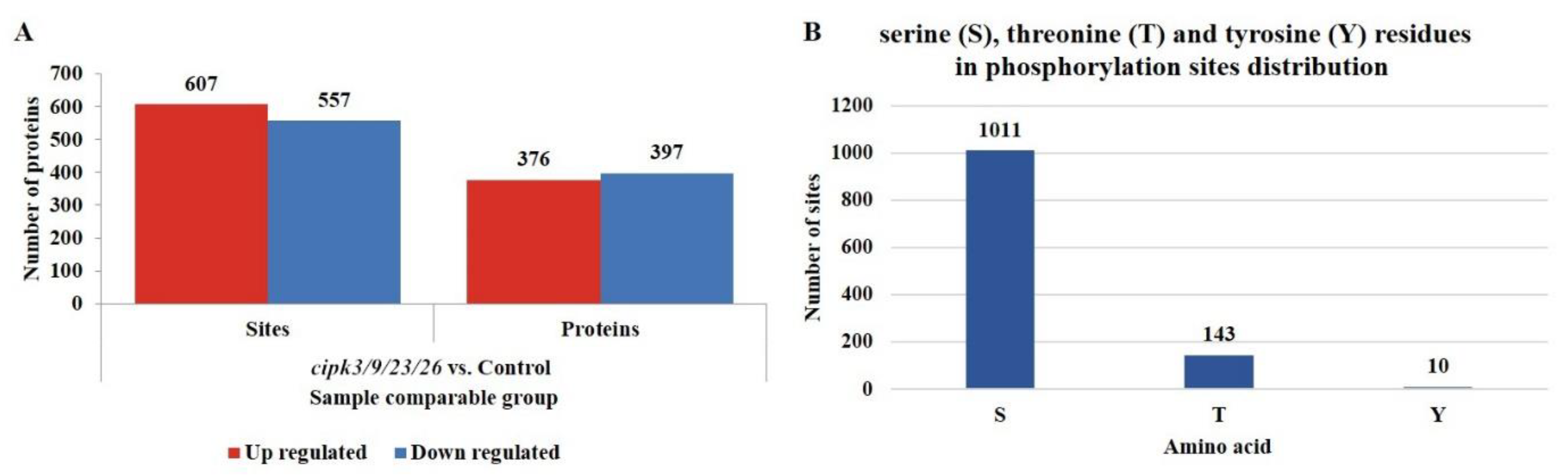
3.2. Identification of Differential Phosphorylation Modification Sites and Proteins in the cipk3/9/23/26 Mutant and WT
3.3. Motif Analysis of cipk3/9/23/26 Mutant Phosphoprotein Modification
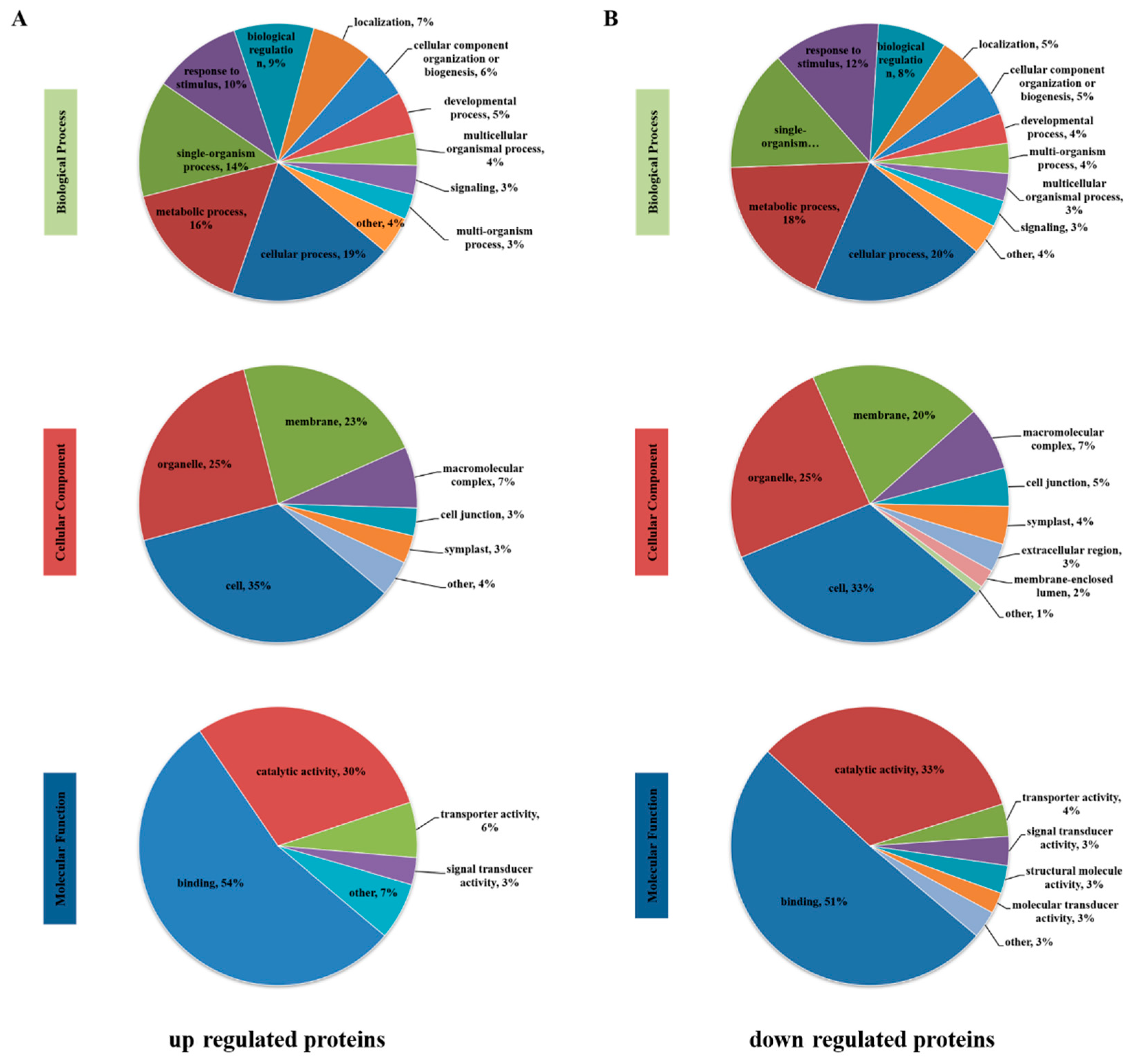
3.4. Functional Classification of Differentially Phosphorylated Proteins
3.5. Differential Expression of Related Phosphoproteins between the cipk3/9/23/26 Mutant and WT
3.6. Enrichment Analyses of DPPs
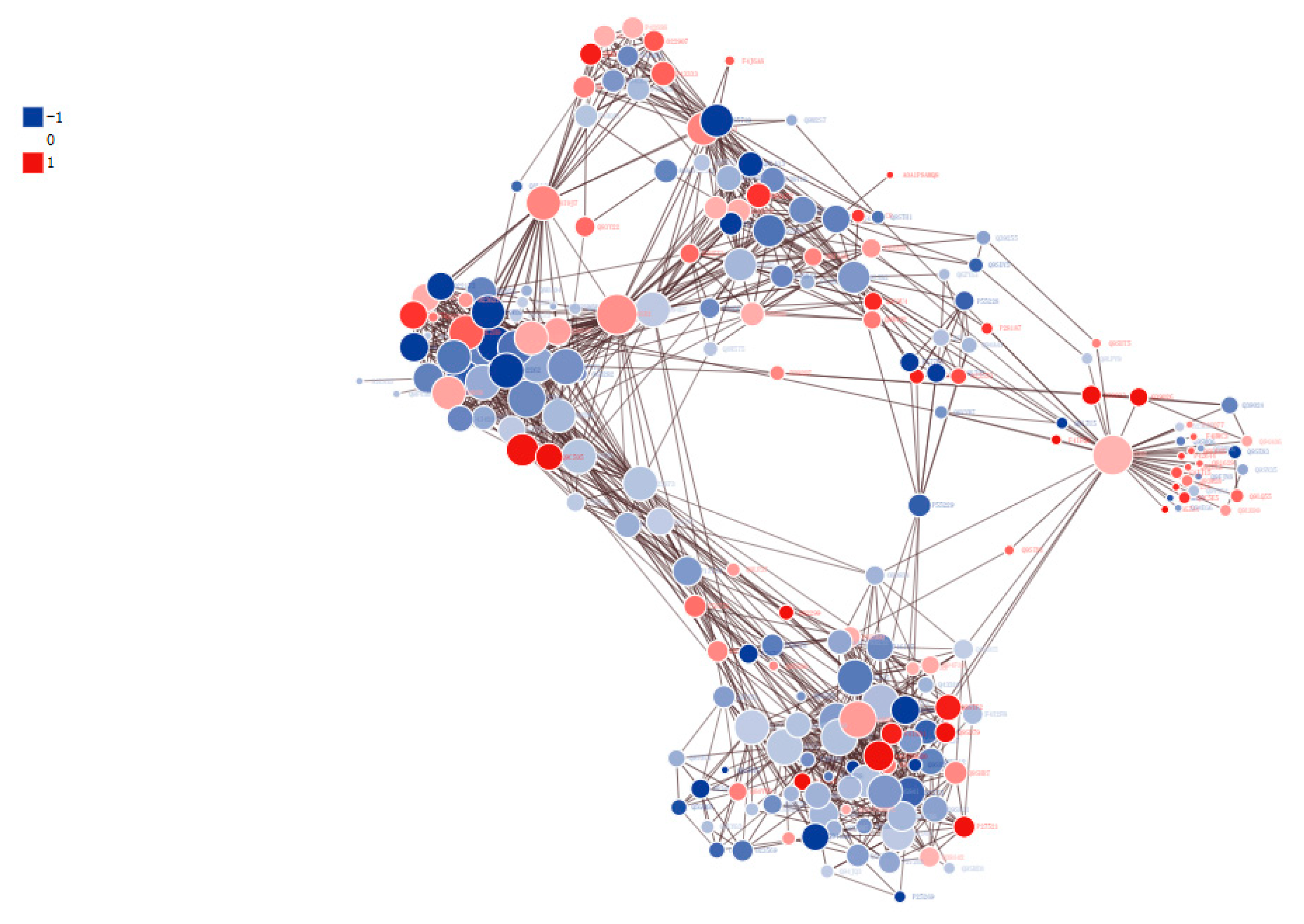
3.7. Protein–Protein Interaction (PPI) Networks of DPPs
4. Discussion
4.1. Response of the Cipk3/9/23/26 Mutant to Magnesium Toxicity
4.2. Transport-Related Proteins
4.3. Signal Transduction and Protein Kinases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grusak, M.A.; Broadley, M.R.; White, P.J. Plant macro-and micronutrient minerals. In Encyclopedia Life Sciences; Wiley & Sons: Hoboken, NJ, USA, 2016; pp. 1–6. [Google Scholar]
- Luan, S. The CBL–CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Kanwar, P.; Singh, A.; Steinhorst, L.; Pandey, A.; Yadav, A.K.; Tokas, I.; Sanyal, S.K.; Kim, B.-G.; Lee, S.-C.; et al. Calcineurin B-like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol. 2015, 169, 780–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanyal, S.K.; Kanwar, P.; Yadav, A.K.; Sharma, C.; Kumar, A.; Pandey, G.K. Arabidopsis CBL interacting protein kinase 3 interacts with ABR1, an APETALA2 domain transcription factor, to regulate ABA responses. Plant Sci. 2017, 254, 48–59. [Google Scholar] [CrossRef]
- Gerendás, J.; Führs, H. The significance of magnesium for crop quality. Plant Soil 2013, 368, 101–128. [Google Scholar] [CrossRef] [Green Version]
- Tang, R.; Zhao, F.-G.; Garcia, V.J.; Kleist, T.J.; Yang, L.; Zhang, H.-X.; Luan, S. Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 3134–3139. [Google Scholar] [CrossRef] [Green Version]
- Mogami, J.; Fujita, Y.; Yoshida, T.; Tsukiori, Y.; Nakagami, H.; Nomura, Y.; Fujiwara, T.; Nishida, S.; Yanagisawa, S.; Ishida, T.; et al. Two distinct families of protein kinases are required for plant growth under high external Mg2+ concentrations in Arabidopsis. Plant Physiol. 2015, 167, 1039–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, J.; Hou, Y.; Tong, X.; Wang, Y.; Lin, H.; Liu, Q.; Zhang, W.; Li, Z.; Nallamilli, B.R.; Zhang, J. Quantitative phosphoproteomic analysis of early seed development in rice (Oryza sativa L.). Plant Mol. Biol. 2016, 90, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Li, J.; Koh, J.; Dufresne, C.; Yang, N.; Qi, S.; Zhang, Y.; Ma, C.; Duong, B.V.; Chen, S.; et al. Quantitative proteomics and phosphoproteomics of sugar beet monosomic addition line M14 in response to salt stress. J. Proteom. 2016, 143, 286–297. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tong, X.; Qiu, J.; Li, Z.; Zhao, J.; Hou, Y.; Tang, L.; Zhang, J. A phosphoproteomic landscape of rice (Oryza sativa) tissues. Physiol. Plant 2017, 160, 458–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graus, D.; Konrad, K.R.; Bemm, F.; Nebioglu, M.G.P.; Lorey, C.; Duscha, K.; Güthoff, T.; Herrmann, J.; Ferjani, A.; Cuin, T.A.; et al. High V-PPase activity is beneficial under high salt loads, but detrimental without salinity. New Phytol. 2018, 219, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Park, J.Y.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC transporters. Arab. Book 2011, 9, e0153. [Google Scholar] [CrossRef] [Green Version]
- Sidler, M.; Hassa, P.O.; Hasan, S.; Ringli, C.; Dudler, R. Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell. 1998, 10, 1623–1636. [Google Scholar] [CrossRef] [Green Version]
- Lewis, D.R.; Miller, N.D.; Splitt, B.L.; Wu, G.; Spalding, E.P. Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis multidrug resistance-like ABC transporter genes. Plant Cell. 2007, 19, 1838–1850. [Google Scholar] [CrossRef] [Green Version]
- Kaneda, M.; Schuetz, M.; Lin, B.S.; Chanis, C.; Hamberger, B.; Western, T.L.; Ehlting, J.; Samuels, A.L. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J. Exp. Bot. 2011, 62, 2063–2077. [Google Scholar] [CrossRef] [Green Version]
- Kubes, M.; Yang, H.; Richter, G.L.; Cheng, Y.; Mlodzinska, E.; Wang, X.; Blakeslee, J.J.; Carraro, N.; Petrasek, J.; Zazimalova, E.; et al. The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J. 2012, 69, 640–654. [Google Scholar] [CrossRef]
- Jenness, M.K.; Carraro, N.; Pritchard, C.A.; Murphy, A.S. The Arabidopsis ATP-BINDING CASSETTE transporter ABCB21 Regulates auxin levels in cotyledons, the root pericycle, and leaves. Front. Plant Sci. 2019, 10, 806. [Google Scholar] [CrossRef] [Green Version]
- Wanke, D.; Üner Kolukisaoglu, H. An update on the ABCC transporter family in plants: Many genes, many proteins, but how many functions? Plant Biol. 2010, 12, 15–25. [Google Scholar] [CrossRef]
- Tang, Z.; Chen, Y.; Miller, A.J.; Zhao, F.-J. The C-type ATP-binding cassette transporter OsABCC7 is involved in the root-to-shoot translocation of arsenic in rice. Plant Cell Physiol. 2019, 60, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.A.; Hanselaer, S.; Soares, E.V. ABCC subfamily vacuolar transporters are involved in Pb (lead) detoxification in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 2015, 175, 65–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jasinski, M.; Banasiak, J.; Radom, M.; Kalitkiewicz, A.; Figlerowicz, M. Full-size ABC transporters from the ABCG subfamily in Medicago truncatula. Mol. Plant-Microbe Interact. 2009, 22, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Ruocco, M.; Ambrosino, P.; Lanzuise, S.; Woo, S.L.; Lorito, M. Four potato (Solanum tuberosum) ABCG transporters and their expression in response to abiotic factors and Phytophthora infestans infection. J. Plant Physiol. 2011, 168, 2225–2233. [Google Scholar] [CrossRef] [PubMed]
- Dhara, A.; Raichaudhuri, A. ABCG transporter proteins with beneficial activity on plants. Phytochemistry. 2021, 184, 112663. [Google Scholar] [CrossRef] [PubMed]
- Bickerton, P.D.; Pittman, J.K. Role of cation/proton exchangers in abiotic stress signaling and stress tolerance in plants. In Elucidation of Abiotic Stress Signaling in Plants; Springer: New York, NY, USA, 2015; pp. 95–117. [Google Scholar]
- Yang, L.; Ji, W.; Gao, P.; Li, Y.; Cai, H.; Bai, X.; Chen, Q.; Zhu, Y. GsAPK, an ABA-activated and calcium-independent SnRK2-type kinase from G. soja, mediates the regulation of plant tolerance to salinity and ABA stress. PLoS ONE 2012, 7, e33838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, M.; Liu, J.; Bi, D.; Zhang, Z.; Cheng, F.; Chen, S.; Zhang, Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008, 18, 1190–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, Q.; Qu, N.; Gao, M.H.; Zhang, Z.B.; Ding, X.J.; Yang, F.; Li, Y.Z.; Dong, O.X.; Chen, S.; Li, X.; et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 2012, 24, 2225–2236. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wu, Y.; Gao, M.; Zhang, J.; Kong, Q.; Liu, Y.; Ba, H.; Zhou, J.; Zhang, Y. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 2012, 11, 253–263. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.Z.; Wang, P.C.; Si, T.; Hsu, C.C.; Wang, L.; Zayed, O.; Yu, Z.P.; Zhu, Y.F.; Dong, J.; Tao, W.A.; et al. MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 2017, 43, 618–629.e5. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.Y.; Zhang, C.; Li, Z.C.; Wang, Z.-R.; Jiang, X.-X.; Shi, Y.-F.; Tian, S.-N.; Braun, E.; Mei, Y.; Qiu, W.-L.; et al. The MAPK kinase kinase GmMEKK1 regulates cell death and defense responses. Plant Physiol. 2018, 178, 907–922. [Google Scholar] [CrossRef] [Green Version]
- Xue, X.; Bian, C.; Guo, X.; Di, R.; Dong, J. The MAPK substrate MASS proteins regulate stomatal development in Arabidopsis. PLoS Genet. 2020, 16, e1008706. [Google Scholar] [CrossRef] [Green Version]
- McAinsh, M.R.; Brownlee, C.; Hetherington, A.M. Calcium ions as second messengers in guard cell signal transduction. Physiol. Plant. 1997, 100, 16–29. [Google Scholar] [CrossRef]
- Heo, D.W.; Lee, S.H.; Kim, M.C.; Chung, W.S.; Chun, H.J.; Lee, K.J.; Park, H.C.; Choi, J.Y.; Cho, M.J. Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc. Natl. Acad. Sci. USA 1999, 96, 766–771. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Yang, T.; Li, W.M.J. Calmodulin gene expression in response to mechanical wounding and Botrytis cinerea infection in tomato fruit. Plants 2014, 3, 427–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, B.; Gilroy, S.; Chehab, E.W.; Braam, J. CML24 is involved in root mechanoresponses and cortical microtubule orientation in Arabidopsis. J. Plant Growth Regul. 2011, 30, 467–479. [Google Scholar] [CrossRef]
- Yang, X.; Wang, S.S.; Wang, M.; Qiao, Z.; Bao, C.-C.; Zhang, W. Arabidopsis thaliana calmodulin-like protein CML24 regulates pollen tube growth by modulating the actin cytoskeleton and controlling the cytosolic Ca2+ concentration. Plant Mol. Biol. 2014, 86, 225–236. [Google Scholar] [CrossRef]
- Xu, B.; Cheval, C.; Laohavisit, A.; Hocking, B.; Chiasson, D.; Olsson, T.S.G.; Shirasu, K.; Faulkner, C.; Gilliham, M. A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol. 2017, 215, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Mori, I.C.; Murata, Y.; Yang, Y.; Munemasa, S.; Wang, Y.-F.; Andreoli, S.; Tiriac, H.; Alonso, J.M.; Harper, J.F.; Ecker, J.R.; et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion-and Ca2+-permeable channels and stomatal closure. PLoS Biol. 2006, 4, e327. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.J.; Wei, F.J.; Wang, C.; Wu, J.-J.; Ratnasekera, D.; Liu, W.-X.; Wu, W.-H. Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid-and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 2010, 154, 1232–1243. [Google Scholar] [CrossRef] [Green Version]
- Franz, S.; Ehlert, B.; Liese, A.; Kurth, J.; Cazalé, A.-C.; Romeis, T. Calcium-dependent protein kinase CPK21 functions in abiotic stress response in Arabidopsis thaliana. Mol. Plant 2011, 4, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Hu, W.; Deng, X.; Zhang, Y.; Liu, X.; Zhao, X.; Luo, Q.; Jin, Z.; Zhou, S.; Sun, T.; et al. A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol. 2014, 14, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asano, T.; Hakata, M.; Nakamura, H.; Aoki, N.; Komatsu, S.; Ichikawa, H.; Hirochika, H.; Ohsugi, R. Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 2011, 75, 179–191. [Google Scholar] [CrossRef]
- Swatek, K.N.; Wilson, R.S.; Ahsan, N.; Tritz, R.L.; Thelen, J.J. Multisite phosphorylation of 14-3-3 proteins by calcium-dependent protein kinases. Biochem. J. 2014, 459, 15–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Motif | Motif Score | Foreground | Background | Fold Increase | ||
|---|---|---|---|---|---|---|
| Matches | Size | Matches | Size | |||
| xxxxPx_S_PxRxxx | 39.87 | 61 | 8351 | 255 | 952,659 | 27.3 |
| xxxxPx_S_PRxxxx | 40.29 | 53 | 8290 | 219 | 952,404 | 27.8 |
| xxxxPx_S_PKxxxx | 39.63 | 41 | 8237 | 169 | 952,185 | 28.0 |
| xxxxxx_S_PRxxxx | 32.00 | 300 | 8196 | 2586 | 952,016 | 13.5 |
| xxxxPx_S_Pxxxxx | 32.00 | 281 | 7896 | 3314 | 949,430 | 10.2 |
| xxxxxx_S_PxRxxx | 32.00 | 246 | 7615 | 2235 | 946,116 | 13.7 |
| xxxRxx_S_PxPxxx | 38.69 | 47 | 7369 | 229 | 943,881 | 26.3 |
| xxxxxx_S_PxxxRx | 32.00 | 199 | 7322 | 2288 | 943,652 | 11.2 |
| xxxxxx_S_PKxxxx | 29.51 | 163 | 7123 | 2114 | 941,364 | 10.2 |
| xxxRSx_S_xPxxxx | 38.04 | 45 | 6960 | 388 | 939,250 | 15.7 |
| xxxxxx_S_PxxxxR | 30.02 | 149 | 6915 | 1948 | 938,862 | 10.4 |
| xLxRxx_S_xxxxxx | 32.00 | 292 | 6766 | 5590 | 936,914 | 7.2 |
| xxxxxR_S_Pxxxxx | 27.82 | 112 | 6474 | 1486 | 931,324 | 10.8 |
| xxxxxG_S_Pxxxxx | 27.98 | 124 | 6362 | 1797 | 929,838 | 10.1 |
| xxxRSx_S_xxxxxx | 32.00 | 234 | 6238 | 5119 | 928,041 | 6.8 |
| xxxxxx_S_PxxRxx | 25.38 | 93 | 6004 | 1423 | 922,922 | 10.0 |
| xxxxxx_S_Pxxxxx | 16.00 | 930 | 5911 | 29,886 | 921,499 | 4.9 |
| xxxRxx_S_Fxxxxx | 32.00 | 116 | 4981 | 1886 | 891,613 | 11.0 |
| xxxRxx_S_xDxxxx | 28.35 | 85 | 4865 | 1907 | 889,727 | 8.2 |
| xxxxxx_S_DDExxx | 38.28 | 46 | 4780 | 478 | 887,820 | 17.9 |
| xxxxxx_S_DGExxx | 39.07 | 28 | 4734 | 235 | 887,342 | 22.3 |
| xMxRxx_S_xxxxxx | 25.45 | 48 | 4706 | 986 | 887,107 | 9.2 |
| xxxxxx_S_DxExxx | 32.00 | 112 | 4658 | 3395 | 886,121 | 6.3 |
| xxxRxx_S_xPxxxx | 24.23 | 63 | 4546 | 1741 | 882,726 | 7.0 |
| xxxxxG_S_Gxxxxx | 25.21 | 99 | 4483 | 5462 | 880,985 | 3.6 |
| xLxKSx_S_xxxxxx | 38.37 | 29 | 4384 | 497 | 875,523 | 11.7 |
| xxxxxD_S_DxDxxx | 39.45 | 37 | 4355 | 422 | 875,026 | 17.6 |
| xxxRxx_S_xExxxx | 23.85 | 65 | 4318 | 2050 | 874,604 | 6.4 |
| xxxKxx_S_Fxxxxx | 32.00 | 85 | 4253 | 2802 | 872,554 | 6.2 |
| xxxRxx_S_xGxxxx | 32.00 | 66 | 4168 | 2312 | 869,752 | 6.0 |
| xxxxxx_S_ExExxx | 32.00 | 105 | 4102 | 5008 | 867,440 | 4.4 |
| xxxxxx_S_DxGxxx | 24.52 | 55 | 3997 | 2966 | 862,432 | 4.0 |
| xxxxxx_S_DxDxxx | 32.00 | 86 | 3942 | 3168 | 859,466 | 5.9 |
| xxxxxG_S_Fxxxxx | 25.25 | 52 | 3856 | 2487 | 856,298 | 4.6 |
| xxxxRx_S_xDxxxx | 26.26 | 54 | 3804 | 2257 | 853,811 | 5.4 |
| xxxxxx_S_ExGxxx | 24.29 | 53 | 3750 | 3143 | 851,554 | 3.8 |
| xLxKxx_S_xxxxxx | 24.76 | 71 | 3697 | 4574 | 848,411 | 3.6 |
| xxxxSx_S_Fxxxxx | 29.03 | 68 | 3626 | 4094 | 843,837 | 3.9 |
| xxxxxG_S_xxxxxx | 16.00 | 338 | 3558 | 47,187 | 839,743 | 1.7 |
| xxxRxx_S_xxxxxx | 16.00 | 314 | 3220 | 25,377 | 792,556 | 3.0 |
| xxxxSR_S_xxxxxx | 24.33 | 54 | 2906 | 4112 | 767,179 | 3.5 |
| xxxxxx_S_xExxxx | 16.00 | 303 | 2852 | 43,739 | 763,067 | 1.9 |
| xxxxxx_S_xGxxxx | 16.00 | 285 | 2549 | 48,192 | 719,328 | 1.7 |
| xxxxxx_S_xPxSPx | 39.01 | 29 | 2264 | 388 | 671,136 | 22.2 |
| xxxxxx_S_xDxxxx | 16.00 | 261 | 2235 | 37,147 | 670,748 | 2.1 |
| xxxxxx_S_GPLxxx | 39.06 | 24 | 1974 | 245 | 633,601 | 31.4 |
| xxRxxx_S_xPxxxx | 23.73 | 34 | 1950 | 2060 | 633,356 | 5.4 |
| xxxxRx_S_xSxxxx | 29.92 | 74 | 1916 | 4763 | 631,296 | 5.1 |
| xxxxxx_S_FRxxxx | 27.42 | 33 | 1842 | 1453 | 626,533 | 7.7 |
| RxxSxx_S_xxxxxx | 20.27 | 49 | 1809 | 4205 | 625,080 | 4.0 |
| xxxxxD_S_xxxxxx | 12.49 | 167 | 1760 | 32,542 | 620,875 | 1.8 |
| xxxxxx_S_xPxxxx | 13.05 | 172 | 1593 | 34,988 | 588,333 | 1.8 |
| xxxKxx_S_xxxxxx | 10.30 | 139 | 1421 | 30,182 | 553,345 | 1.8 |
| xxxxxx_S_xRxxxx | 12.48 | 149 | 1282 | 32,477 | 523,163 | 1.9 |
| xxxxxx_S_Fxxxxx | 9.38 | 99 | 1133 | 21,946 | 490,686 | 2.0 |
| xxxxSx_S_xNxxxx | 15.13 | 31 | 1034 | 3443 | 468,740 | 4.1 |
| xxxxxx_S_Dxxxxx | 7.27 | 82 | 1003 | 20,145 | 465,297 | 1.9 |
| xxxxxx_S_xxGxxx | 7.96 | 98 | 921 | 25,916 | 445,152 | 1.8 |
| xxxxxx_S_xKxxxx | 8.29 | 115 | 823 | 33,536 | 419,236 | 1.7 |
| xxxxPx_T_Pxxxxx | 32.00 | 143 | 1057 | 2335 | 529,615 | 30.7 |
| xxxxxx_T_PxRxxx | 27.88 | 63 | 914 | 1278 | 527,280 | 28.4 |
| xxxxxx_T_PRxxxx | 24.65 | 48 | 851 | 1115 | 526,002 | 26.6 |
| xxxxxx_T_PTxxxx | 24.66 | 54 | 803 | 1470 | 524,887 | 24.0 |
| xxxxxx_T_PKxxxx | 23.80 | 47 | 749 | 1406 | 523,417 | 23.4 |
| xxxxxx_T_Pxxxxx | 16.00 | 239 | 702 | 19,445 | 522,011 | 9.1 |
| xxxRxx_T_xxxxxx | 16.00 | 83 | 463 | 25,391 | 502,566 | 3.5 |
| xxxxxx_T_xExxxx | 10.57 | 63 | 380 | 31,767 | 477,175 | 2.5 |
| xxxxxx_T_Dxxxxx | 6.44 | 38 | 317 | 21,640 | 445,408 | 2.5 |
| Protein Accession | Position | Ratio | Protein Description | Modified Sequence |
|---|---|---|---|---|
| Q9ZR72 | 1014 | 1.49 | ABC transporter B family member 1 | KTEIEPDDPDT(0.203)T(0.797)PVPDR |
| Q9ZR72 | 642 | 1.386 | ABC transporter B family member 1 | NS(0.016)S(0.979)Y(0.005)GRS(0.965)PY(0.005)S(0.03)R |
| Q9LJX0 | 624 | 1.262 | ABC transporter B family member 19 | T(0.157)RS(0.831)T(0.094)RLS(0.917)HS(0.32)LS(0.672)T(0.008)K |
| Q9LJX0 | 620 | 1.405 | ABC transporter B family member 19 | T(0.157)RS(0.831)T(0.094)RLS(0.917)HS(0.32)LS(0.672)T(0.008)K |
| Q9LJX0 | 611 | 1.335 | ABC transporter B family member 19 | DFS(0.999)NPS(0.001)TR |
| Q9M1Q9 | 660 | 1.554 | ABC transporter B family member 21 | LSMES(1)MKR |
| Q0WML0 | 639 | 1.252 | ABC transporter B family member 27 | QLQS(0.009)S(0.086)S(0.889)S(0.016)VTTL |
| Q9C8G9 | 1485 | 1.778 | ABC transporter C family member 1 | S(0.007)IT(0.993)LENKR |
| Q9LZJ5 | 897 | 1.403 | ABC transporter C family member 14 | SIS(1)IES(1)PRQPKS(1)PK |
| Q9LZJ5 | 894 | 1.615 | ABC transporter C family member 14 | SIS(1)IES(1)PRQPKS(1)PK |
| Q9LZJ5 | 903 | 1.345 | ABC transporter C family member 14 | SIS(1)IES(1)PRQPKS(1)PK |
| Q9C8K2 | 667 | 1.237 | ABC transporter G family member 12 | KVPS(0.003)LS(0.162)S(0.162)LS(0.68)S(0.994)RR |
| Q9C8K2 | 666 | 1.289 | ABC transporter G family member 12 | KVPSLS(0.003)S(0.009)LS(0.9)S(0.088)RR |
| Q9C8K2 | 661 | 1.245 | ABC transporter G family member 12 | KVPS(0.999)LS(0.002)S(0.009)LS(0.829)S(0.162)R |
| Q93YS4 | 71 | 1.29 | ABC transporter G family member 22 | LMGMS(0.996)PGRS(0.175)S(0.806)GAGT(0.022)HIR |
| Q93YS4 | 66 | 1.428 | ABC transporter G family member 22 | RLMGMS(1)PGR |
| Q9XIE2 | 40 | 1.266 | ABC transporter G family member 36 | NIEDIFSS(0.003)GS(0.997)R |
| A0A1P8AZ84 | 586 | 2.017 | p-glycoprotein 6 | QKS(0.888)NGS(0.116)DPES(0.988)PIS(0.007)PLLISDPQNER |
| A0A1P8AZ84 | 610 | 1.419 | p-glycoprotein 6 | S(0.002)HS(0.997)QT(0.001)FSRPLGHSDDTSASVK |
| A0A1P8AZ84 | 593 | 1.548 | p-glycoprotein 6 | QKS(0.888)NGS(0.116)DPES(0.988)PIS(0.007)PLLISDPQNER |
| Q9S7Z8 | 235 | 1.326 | Auxin efflux carrier component 3 | PSNLTGAEIYS(1)LS(0.003)T(0.201)T(0.797)PR |
| F4IS06 | 38 | 1.674 | Vacuolar cation/proton exchanger | TAHNMS(0.993)S(0.119)S(0.539)S(0.348)LRK |
| Q9ZV07 | 282 | 1.214 | Probable aquaporin PIP2-6 | S(1)QLHELHA |
| A0A1P8AU30 | 30 | 1.27 | Cationic amino acid transporter 9 | S(0.012)KS(0.986)LPPPS(0.001)S(0.001)QT(0.001)AVR |
| Q9ZTZ7 | 120 | 1.733 | K(+) efflux antiporter 1, chloroplastic | IGES(0.002)S(0.013)ES(0.864)S(0.121)DETEATDLK |
| Q8W4J2 | 527 | 1.374 | Mitogen-activated protein kinase 16 | T(0.237)QPCKS(0.763)NRGDEDCATAAEGPSR |
| Q94A06 | 27 | 1.235 | Mitogen-activated protein kinase kinase 1 | FLT(0.096)QS(0.9)GT(0.005)FKDGDLR |
| Q9C9U4 | 530 | 1.742 | Mitogen-activated protein kinase 15 | ASQQAEGTENGGGGGYS(1)AR |
| Q39026 | 221 | 2.126 | Mitogen-activated protein kinase 6 | VTSESDFMT(1)EY(1)VVTR |
| Q39026 | 223 | 2.106 | Mitogen-activated protein kinase 6 | VTSESDFMT(1)EY(1)VVTR |
| Q39023 | 198 | 2.156 | Mitogen-activated protein kinase 3 | PTSENDFMT(1)EY(1)VVTR |
| Q39023 | 196 | 2.309 | Mitogen-activated protein kinase 3 | PTSENDFMT(1)EY(1)VVTR |
| Q39024 | 195 | 0.637 | Mitogen-activated protein kinase 4 | T(0.036)KS(0.908)ET(0.056)DFMTEYVVTR |
| Q9SSC1 | 98 | 1.208 | MAPK kinase substrate protein At1g80180 | VS(1)PAVDPPS(1)PR |
| Q9SSC1 | 16 | 1.429 | MAPK kinase substrate protein At1g80180 | RQGS(1)S(1)GIVWDDR |
| Q9SSC1 | 17 | 1.33 | MAPK kinase substrate protein At1g80180 | RQGS(1)S(1)GIVWDDR |
| Q9SSC1 | 82 | 0.464 | MAPK kinase substrate protein At1g80180 | S(0.013)RS(0.987)NGGGAIR |
| F4IVN6 | 80 | 1.251 | Calmodulin 5 | MKDT(0.998)DS(0.002)EEELK |
| F4IVN6 | 82 | 1.21 | Calmodulin 5 | MKDT(0.017)DS(0.983)EEELK |
| P30188 | 27 | 1.812 | Probable calcium-binding protein CML35 | AS(0.008)VS(0.183)RS(0.803)EPS(0.501)S(0.501)FS(0.002)S(0.002)NASSSSSDGSYGNLK |
| P30188 | 11 | 0.765 | Probable calcium-binding protein CML35 | LAASLNRLS(1)PK |
| P30188 | 44 | 0.208 | Probable calcium-binding protein CML35 | SEPSSFSSNASSSSSDGS(1)YGNLK |
| P25070 | 46 | 1.215 | Calcium-binding protein CML24 | ALS(0.937)PT(0.059)AS(0.004)PEETVTMMK |
| Q8L3R2 | 159 | 0.593 | Probable calcium-binding protein CML41 | GSGCIT(1)PK |
| Q8L3R2 | 26 | 1.321 | Probable calcium-binding protein CML41 | LNLS(1)FQNR |
| Q8L3R2 | 47 | 0.566 | Probable calcium-binding protein CML41 | SNSS(0.001)S(0.003)T(0.017)LNS(0.98)PRS(1)NSDDNNNIK |
| Q38868 | 69 | 1.208 | Calcium-dependent protein kinase 9 | AAAAAPGLS(1)PK |
| Q38868 | 51 | 1.627 | Calcium-dependent protein kinase 9 | TTQQPEKPGS(0.997)VNS(0.003)QPPPWR |
| Q38868 | 78 | 1.889 | Calcium-dependent protein kinase 9 | S(0.001)NS(0.999)ILENAFEDVK |
| Q9ZSA2 | 244 | 0.432 | Calcium-dependent protein kinase 21 | DIVGS(1)AYYVAPEVLR |
| Q9ZSA2 | 53 | 1.322 | Calcium-dependent protein kinase 21 | PMTQPIHQQIS(0.964)T(0.036)PSSNPVSVR |
| Q9ZSA2 | 414 | 1.449 | Calcium-dependent protein kinase 21 | LGS(0.997)RLS(0.003)ETEVK |
| Q9ZSA2 | 417 | 0.744 | Calcium-dependent protein kinase 21 | LS(0.971)ET(0.029)EVK |
| Q9M9V8 | 40 | 0.394 | Calcium-dependent protein kinase 10 | LNPFAGDFT(0.002)RS(0.998)PAPIR |
| Q8W4I7 | 18 | 0.712 | Calcium-dependent protein kinase 13 | EDVKS(1)NY(0.001)S(0.999)GHDHAR |
| Q8W4I7 | 21 | 0.454 | Calcium-dependent protein kinase 13 | SNYS(1)GHDHAR |
| Q8W4I7 | 43 | 0.511 | Calcium-dependent protein kinase 13 | VLS(1)DVPKENIEDR |
| Q42479 | 18 | 1.485 | Calcium-dependent protein kinase 3 | SSDPPPSS(0.005)S(0.768)S(0.175)S(0.041)S(0.009)S(0.002)GNVVHHVKPAGER |
| Q06850 | 130 | 0.778 | Calcium-dependent protein kinase 1 | RVS(0.076)S(0.924)AGLR |
| Q06850 | 64 | 1.878 | Calcium-dependent protein kinase 1 | LSDEVQNKPPEQVT(0.997)MPKPGT(0.003)DVETK |
| Q06850 | 129 | 0.549 | Calcium-dependent protein kinase 1 | RVS(0.994)S(0.006)AGLR |
| P46077 | 248 | 1.31 | 14-3-3-like protein GF14 phi | DNLTLWTSDMQDES(1)PEEIKEAAAPKPAEEQK |
| P42644 | 238 | 1.575 | 14-3-3-like protein GF14 psi | DNLTLWTSDMT(1)DEAGDEIK |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Z.; Shi, J.; Zhen, Y. Quantitative Phosphoproteomics of cipk3/9/23/26 Mutant and Wild Type in Arabidopsis thaliana. Genes 2021, 12, 1759. https://doi.org/10.3390/genes12111759
Yin Z, Shi J, Zhen Y. Quantitative Phosphoproteomics of cipk3/9/23/26 Mutant and Wild Type in Arabidopsis thaliana. Genes. 2021; 12(11):1759. https://doi.org/10.3390/genes12111759
Chicago/Turabian StyleYin, Ziyi, Jisen Shi, and Yan Zhen. 2021. "Quantitative Phosphoproteomics of cipk3/9/23/26 Mutant and Wild Type in Arabidopsis thaliana" Genes 12, no. 11: 1759. https://doi.org/10.3390/genes12111759






