Integrity and Stability of PTC Bearing CFTR mRNA and Relevance to Future Modulator Therapies in Cystic Fibrosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Treatments
2.2. RNA Isolation, Reverse Transcription of cDNA
2.3. Primers
2.4. Real-Time Quantitative RT-PCR
2.5. mRNA Integrity Assay
2.6. mRNA Stability Assay
2.7. Western Blot (WB) Analysis
2.8. Statistical Analysis
3. Results
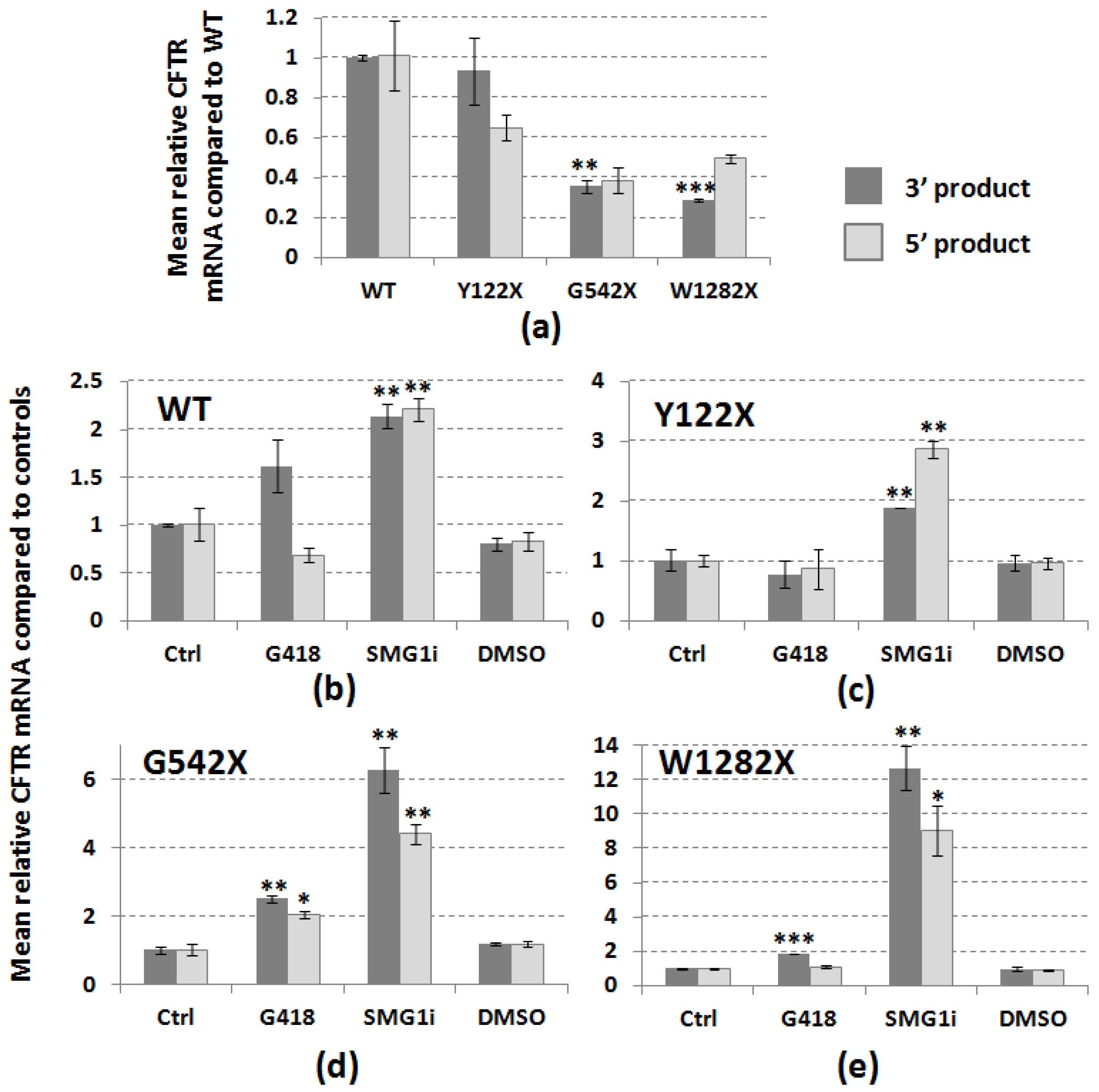
3.1. PTC Mutations Cause a Reduction in CFTR mRNA Abundance
3.2. PTC Readthrough and NMD Inhibition Can Restore PTC-Associated CFTR mRNA Levels
3.3. PTC Readthrough and NMD Inhibition Do Not Significantly Restore CFTR Protein Expression
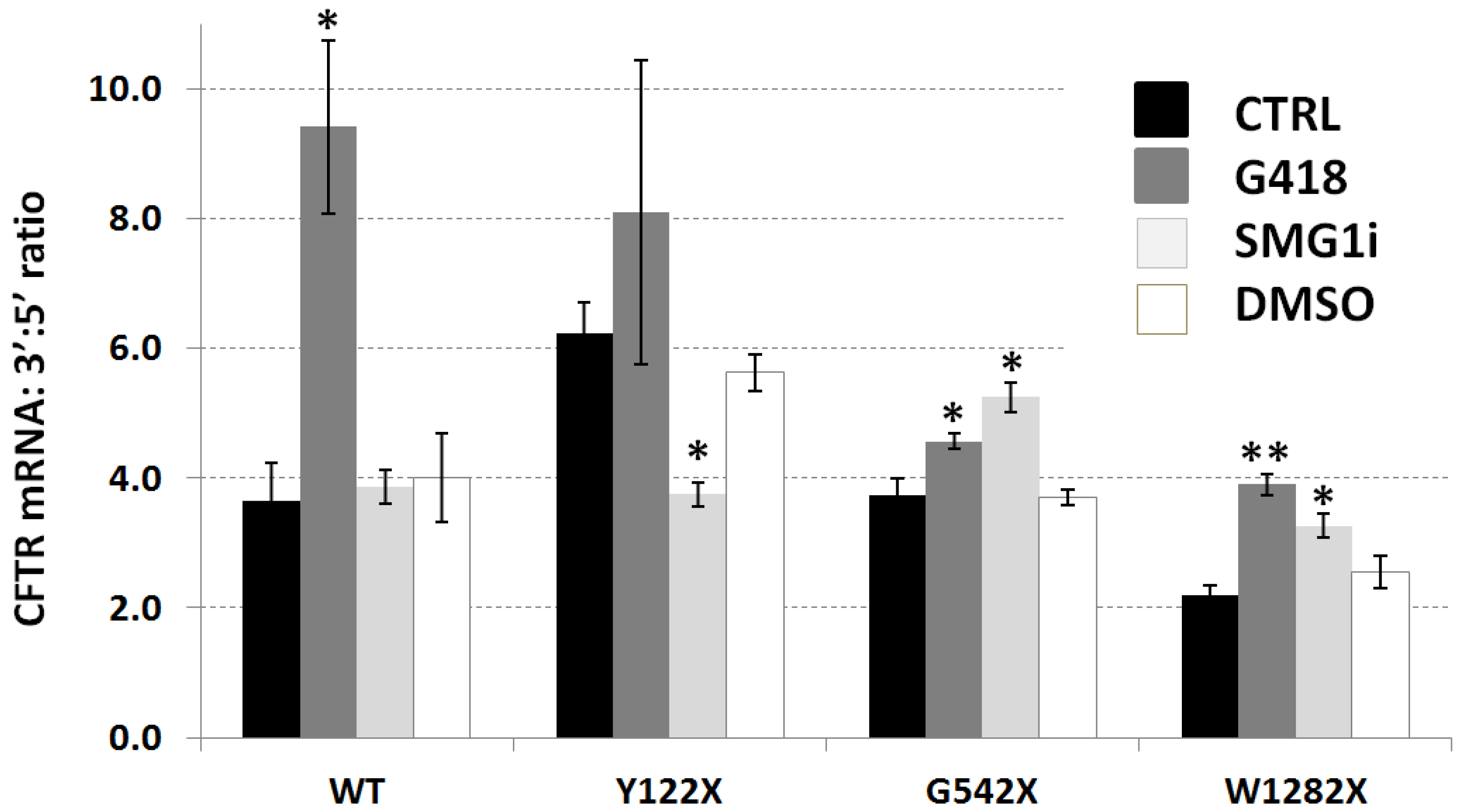
3.4. Ratio between Abundance of CFTR Transcript at 3′ and 5′ Ends
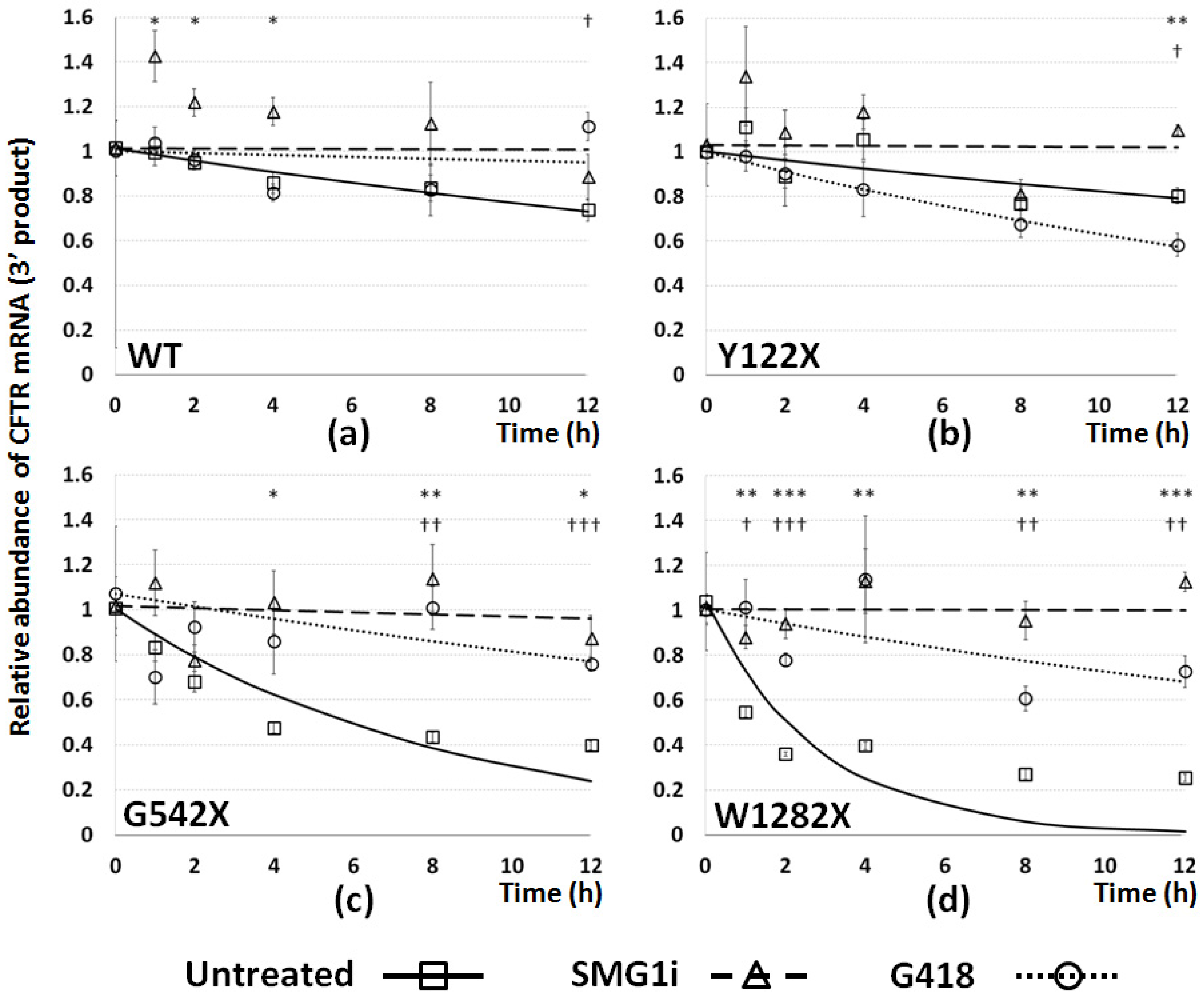
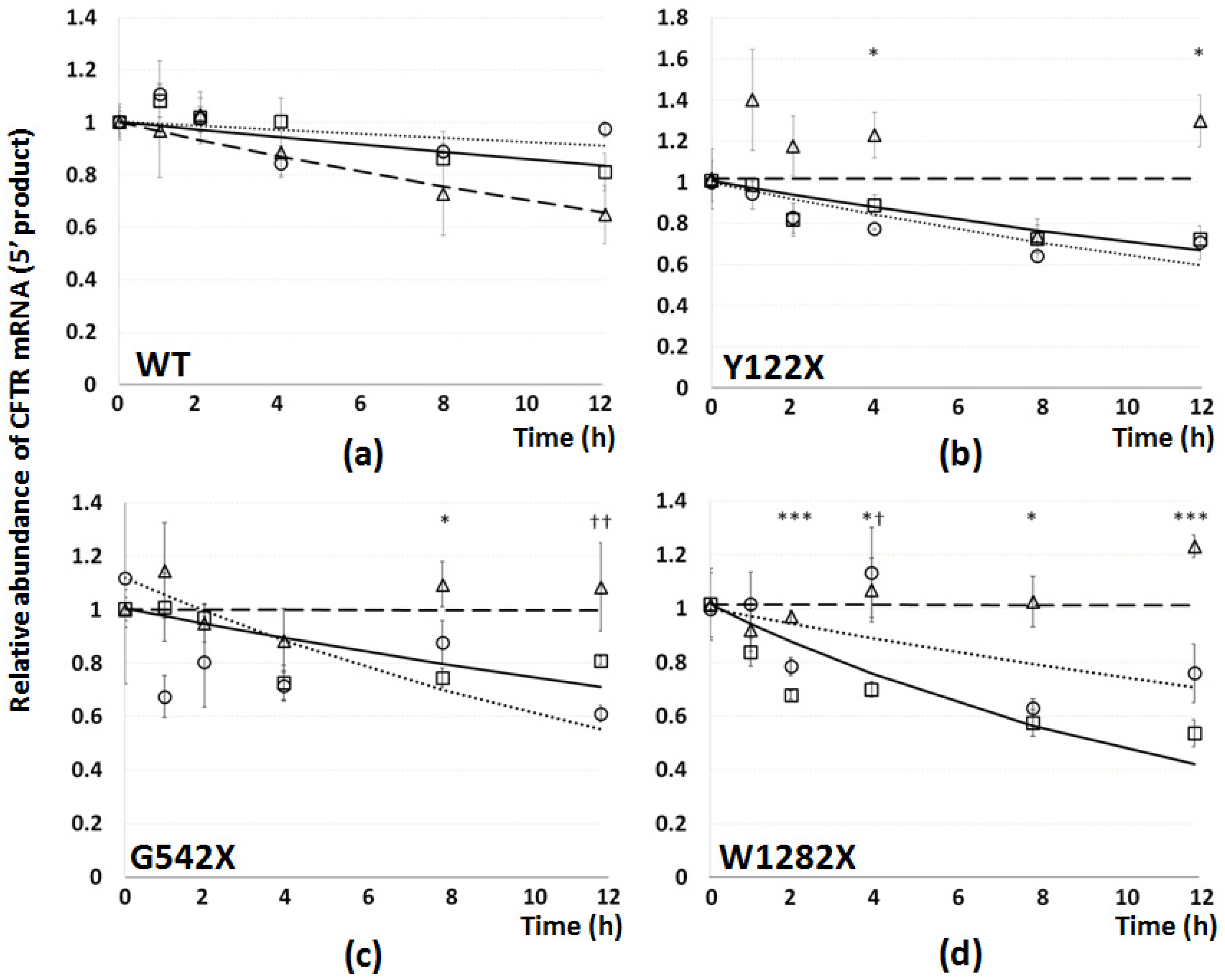
3.5. The Effect of PTC Readthrough and NMD Inhibition on Stability of PTC-Bearing CFTR mRNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
References
- Riordan, J.R.; Rommens, J.M.; Kerem, B.; Alon, N.; Rozmahel, R.; Grzelczak, Z.; Zielenski, J.; Lok, S.; Plavsic, N.; Chou, J.L. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 1989, 245, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, B.P.; Freedman, S.D. Cystic fibrosis. Lancet 2009, 373, 1891–1904. [Google Scholar] [CrossRef]
- Cystic Fibrosis Mutation Database. Available online: http://genet.sickkids.on.ca/ (accessed on 30 August 2021).
- De Boeck, K.; Amaral, M.D. Progress in therapies for cystic fibrosis. Lancet Respir. Med. 2016, 4, 662–674. [Google Scholar] [CrossRef]
- Laselva, O.; Eckford, P.D.W.; Bartlett, C.; Ouyang, H.; Gunawardena, T.N.A.; Gonska, T.; Moraes, T.J.; Bear, C.E. Functional rescue of c.3846G>A (W1282X) in patient-derived nasal cultures achieved by inhibition of nonsense mediated decay and protein modulators with complementary mechanisms of action. J. Cyst. Fibros. 2020, 19, 717–727. [Google Scholar] [CrossRef] [Green Version]
- Keenan, M.M.; Huang, L.; Jordan, N.J.; Wong, E.; Cheng, Y.; Valley, H.C.; Mahiou, J.; Liang, F.; Bihler, H.; Mense, M.; et al. Nonsense-mediated RNA decay pathway inhibition restores expression and function of W1282X CFTR. Am. J. Respir. Cell Mol. Biol. 2019, 61, 290–300. [Google Scholar] [CrossRef]
- Frischmeyer, P.A.; Dietz, H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999, 8, 1893–1900. [Google Scholar] [CrossRef] [Green Version]
- Will, K.; Dörk, T.; Stuhrmann, M.; Hardt, H.V.D.; Ellemunter, H.; Tümmler, B.; Schmidtke, J. Transcript analysis of CFTR nonsense mutations in lymphocytes and nasal epithelial cells from cystic fibrosis patients. Hum. Mutat. 1995, 5, 210–220. [Google Scholar] [CrossRef]
- Sharma, J.; Keeling, K.M.; Rowe, S.M. Pharmacological approaches for targeting cystic fibrosis nonsense mutations. Eur. J. Med. Chem. 2020, 200, 112436. [Google Scholar] [CrossRef]
- Linde, L.; Boelz, S.; Neu-Yilik, G.; Kulozik, A.E.; Kerem, B. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur. J. Hum. Genet. 2007, 15, 1156–1162. [Google Scholar] [CrossRef] [Green Version]
- Clarke, L.A.; Awatade, N.T.; Felício, V.M.; Silva, I.A.; Calucho, M.; Pereira, L.; Azevedo, P.; Cavaco, J.; Barreto, C.; Bertuzzo, C.; et al. The effect of premature termination codon mutations on CFTR mRNA abundance in human nasal epithelium and intestinal organoids: A basis for read-through therapies in cystic fibrosis. Hum. Mutat. 2019, 40, 326–334. [Google Scholar] [CrossRef]
- Linde, L.; Boelz, S.; Nissim-Rafinia, M.; Oren, Y.S.; Wilschanski, M.; Yaacov, Y.; Virgilis, D.; Neu-Yilik, G.; Kulozik, A.E.; Kerem, E.; et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J. Clin. Investig. 2007, 117, 683–692. [Google Scholar] [CrossRef] [Green Version]
- Ramalho, A.S.; Beck, S.; Meyer, M.; Penque, D.; Cutting, G.R.; Amaral, M.D. Five percent of normal cystic fibrosis transmembrane conductance regulator mRNA ameliorates the severity of pulmonary disease in cystic fibrosis. Am. J. Respir. Cell Mol. Biol. 2002, 27, 619–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoek, T.A.; Khuperkar, D.; Lindeboom, R.G.; Sonneveld, S.; Verhagen, B.M.; Boersma, S.; Vermeulen, M.; Tanenbaum, M.E. Single-molecule imaging uncovers rules governing nonsense-mediated mRNA decay. Mol. Cell 2019, 75, 324–339. [Google Scholar] [CrossRef] [PubMed]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntzinger, E.; Kashima, I.; Fauser, M.; Saulière, J.; Izaurralde, E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 2008, 14, 2609–2617. [Google Scholar] [CrossRef] [Green Version]
- Eberle, A.B.; Lykke-Andersen, S.; Muhlemann, O.; Jensen, T.H. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009, 16, 49–55. [Google Scholar] [CrossRef]
- Unterholzner, L.; Izaurralde, E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 2004, 16, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Pacheco, M. CFTR modulators: The changing face of cystic fibrosis in the era of precision medicine. Front. Pharmacol. 2020, 10, 1662. [Google Scholar] [CrossRef] [Green Version]
- McHugh, D.R.; Cotton, C.U.; Hodges, C.A. Synergy between readthrough and nonsense mediated decay inhibition in a murine model of cystic fibrosis nonsense mutations. Int. J. Mol. Sci. 2021, 22, 344. [Google Scholar] [CrossRef]
- Xue, X.; Mutyam, V.; Thakerar, A.; Mobley, J.; Bridges, R.J.; Rowe, S.M.; Keeling, K.M.; Bedwell, D.M. Identification of the amino acids inserted during suppression of CFTR nonsense mutations and determination of their functional consequences. Hum. Mol. Genet. 2017, 26, 3116–3129. [Google Scholar] [CrossRef] [Green Version]
- Mutyam, V.; Sharma, J.; Li, Y.; Peng, N.; Chen, J.; Tang, L.P.; Falk Libby, E.; Singh, A.K.; Conrath, K.; Rowe, S.M. Novel correctors and potentiators enhance translational readthrough in CFTR nonsense mutations. Am. J. Respir. Cell Mol. Biol. 2021, 64, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Evans, T.A.; Pellicore, M.J.; Davis, E.; Aksit, M.A.; McCague, A.F.; Joynt, A.T.; Lu, Z.; Han, S.T.; Anzmann, A.F.; et al. Capitalizing on the heterogeneous effects of CFTR nonsense and frameshift variants to inform therapeutic strategy for cystic fibrosis. PLoS Genet. 2018, 14, e1007723. [Google Scholar] [CrossRef] [Green Version]
- Valley, H.C.; Bukis, K.M.; Bell, A.; Cheng, Y.; Wong, E.; Jordan, N.J.; Allaire, N.E.; Sivachenko, A.; Liang, F.; Bihler, H.; et al. Isogenic cell models of cystic fibrosis-causing variants in natively expressing pulmonary epithelial cells. J. Cyst Fibros. 2019, 18, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Bidou, L.; Bugaud, O.; Belakhov, V.; Baasov, T.; Namy, O. Characterization of new-generation aminoglycoside promoting premature termination codon readthrough in cancer cells. RNA Biol. 2017, 14, 378–388. [Google Scholar] [CrossRef] [Green Version]
- Primerbank—Harvard University. Available online: https://pga.mgh.harvard.edu/primerbank/ (accessed on 30 August 2021).
- Vermeulen, J.; De Preter, K.; Lefever, S.; Nuytens, J.; De Vloed, F.; Derveaux, S.; Hellemans, J.; Speleman, F.; Vandesompele, J. Measurable impact of RNA quality on gene expression results from quantitative PCR. Nucleic Acids Res. 2011, 39, e63. [Google Scholar] [CrossRef]
- Padhi, B.K.; Singh, M.; Rosales, M.; Pelletier, G.; Cakmak, S. A PCR-based quantitative assay for the evaluation of mRNA integrity in rat samples. Biomol. Detect. Quant. 2018, 15, 18–23. [Google Scholar] [CrossRef]
- Perry, R.P.; Kelley, D.E. Inhibition of RNA synthesis by actinomycin D: Characteristic dose-response of different RNA species. J. Cell Physiol. 1970, 76, 127–139. [Google Scholar] [CrossRef]
- Yang, E.; van Nimwegen, E.; Zavolan, M.; Rajewsky, N.; Schroeder, M.; Magnasco, M.; Darnell, J.E. Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res. 2003, 13, 1863–1872. [Google Scholar] [CrossRef]
- Kemmer, G.; Keller, S. Nonlinear least-squares data fitting in Excel spreadsheets. Nat. Protoc. 2010, 5, 267–281. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Rahman, K.S.; Infield, D.T.; Kuang, C.; Prince, C.Z.; McCarty, N.A. Three charged amino acids in extracellular loop 1 are involved in maintaining the outer pore architecture of CFTR. J. Genet. Physiol. 2014, 144, 159–179. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.; Aleksandrov, L.; Chang, X.B.; Hou, Y.X.; He, L.; Hegedus, T.; Gentzsch, M.; Aleksandrov, A.; Balch, W.E.; Riordan, J.R. Domain interdependence in the biosynthetic assembly of CFTR. J. Mol. Biol. 2007, 365, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, A.S.; Lewandowska, M.A.; Farinha, C.M.; Mendes, F.; Gonçalves, J.; Barreto, C.; Harris, A.; Amaral, M.D. Deletion of CFTR translation start site reveals functional isoforms of the protein in CF patients. Cell Physiol. Biochem. 2009, 24, 335–346. [Google Scholar] [CrossRef]
- Sharma, J.; Du, M.; Wong, E.; Mutyam, V.; Li, Y.; Chen, J.; Wangen, J.; Thrasher, K.; Fu, L.; Peng, N.; et al. A small molecule that induces translational readthrough of CFTR nonsense mutations by eRF1 depletion. Nat. Commun. 2021, 12, 4358. [Google Scholar] [CrossRef]
- Santos, L.; Mention, K.; Cavusoglu-Doran, K.; Sanz, D.J.; Bacalhau, M.; Lopes-Pacheco, M.; Harrison, P.T.; Farinha, C.M. Comparison of Cas9 and Cas12a CRISPR editing methods to correct the W1282X-CFTR mutation. J. Cyst. Fibros. 2021, S1569-1993(21)00167-3. [Google Scholar] [CrossRef] [PubMed]
- Nomakuchi, T.; Rigo, F.; Aznarez, I.; Krainer, A.R. Antisense oligonucleotide–directed inhibition of nonsense-mediated mRNA decay. Nat. Biotechnol. 2016, 34, 164–166. [Google Scholar] [CrossRef] [Green Version]
- Venturini, A.; Borrelli, A.; Musante, I.; Scudieri, P.; Capurro, V.; Renda, M.; Pedemonte, N.; Galietta, L. Comprehensive Analysis of Combinatorial Pharmacological Treatments to Correct Nonsense Mutations in the CFTR Gene. Int. J. Mol. Sci. 2021, 22, 11972. [Google Scholar] [CrossRef] [PubMed]
- Hagedoorn, R.; Joseph, R.; Kasanmoentalib, S.; Eilers, P.; Killian, J.; Raap, A.K. Chemical RNA labeling without 3’ end bias using fluorescent cis-platin compounds. BioTechniques 2003, 34, 974–980. [Google Scholar] [CrossRef] [Green Version]
- Pratik, D.; Chao, J.A. Insights into mRNA degradation from single-molecule imaging in living cells. Curr. Opin. Struct. Biol. 2020, 65, 89–95. [Google Scholar] [CrossRef]
- Yu, J.; Russell, J.E. Structural and functional analysis of an mRNP complex that mediates the high stability of human beta-globin mRNA. Mol. Cell Biol. 2001, 21, 5879–5888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, J. mRNA stability in mammalian cells. Microbiol. Rev. 1995, 59, 423–450. [Google Scholar] [CrossRef] [PubMed]
- Sharova, L.V.; Sharov, A.A.; Nedorezov, T.; Piao, Y.; Shaik, N.; Ko, M.S. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009, 16, 45–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baudouin-Legros, M.; Hinzpeter, A.; Jaulmes, A.; Brouillard, F.; Costes, B.; Fanen, P.; Edelman, A. Cell-specific posttranscriptional regulation of CFTR gene expression via influence of MAPK cascades on 3’UTR part of transcripts. Am. J. Physiol. Cell Physiol. 2005, 289, C1240–C1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitash, N.; Mu, F.; Donovan, J.E.; Myerburg, M.M.; Ranganathan, S.; Greene, C.M.; Swiatecka-Urban, A. Transforming Growth factor-β1 selectively recruits microRNAs to the RNA-induced silencing complex and degrades CFTR mRNA under Permissive conditions in human bronchial epithelial cells. Int. J. Mol. Sci. 2019, 20, 4933. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clarke, L.A.; Luz, V.C.C.; Targowski, S.; Ramalho, S.S.; Farinha, C.M.; Amaral, M.D. Integrity and Stability of PTC Bearing CFTR mRNA and Relevance to Future Modulator Therapies in Cystic Fibrosis. Genes 2021, 12, 1810. https://doi.org/10.3390/genes12111810
Clarke LA, Luz VCC, Targowski S, Ramalho SS, Farinha CM, Amaral MD. Integrity and Stability of PTC Bearing CFTR mRNA and Relevance to Future Modulator Therapies in Cystic Fibrosis. Genes. 2021; 12(11):1810. https://doi.org/10.3390/genes12111810
Chicago/Turabian StyleClarke, Luka A., Vanessa C. C. Luz, Szymon Targowski, Sofia S. Ramalho, Carlos M. Farinha, and Margarida D. Amaral. 2021. "Integrity and Stability of PTC Bearing CFTR mRNA and Relevance to Future Modulator Therapies in Cystic Fibrosis" Genes 12, no. 11: 1810. https://doi.org/10.3390/genes12111810
APA StyleClarke, L. A., Luz, V. C. C., Targowski, S., Ramalho, S. S., Farinha, C. M., & Amaral, M. D. (2021). Integrity and Stability of PTC Bearing CFTR mRNA and Relevance to Future Modulator Therapies in Cystic Fibrosis. Genes, 12(11), 1810. https://doi.org/10.3390/genes12111810






