Abstract
Y chromosomes play important roles in sex determination and male fertility. In several groups (e.g., mammals) there is strong evidence that they evolved through gene loss from a common X-Y ancestor, but in Drosophila the acquisition of new genes plays a major role. This conclusion came mostly from studies in two species. Here we report the identification of the 22 Y-linked genes in D. willistoni. They all fit the previously observed pattern of autosomal or X-linked testis-specific genes that duplicated to the Y. The ratio of gene gains to gene losses is ~25 in D. willistoni, confirming the prominent role of gene gains in the evolution of Drosophila Y chromosomes. We also found four large segmental duplications (ranging from 62 kb to 303 kb) from autosomal regions to the Y, containing ~58 genes. All but four of these duplicated genes became pseudogenes in the Y or disappeared. In the GK20609 gene the Y-linked copy remained functional, whereas its original autosomal copy degenerated, demonstrating how autosomal genes are transferred to the Y chromosome. Since the segmental duplication that carried GK20609 contained six other testis-specific genes, it seems that chance plays a significant role in the acquisition of new genes by the Drosophila Y chromosome.
1. Introduction
Y chromosomes nearly always contain male-fertility genes, and in many species also carry the master sex-determining genes [1,2,3,4]. They usually contain a small number of genes but can be physically large due to the accumulation of repetitive DNA. For example, the D. melanogaster Y has ~40 Mbp and ~20 genes, whereas the X has ~42 Mbp and ~2300 genes [5]. The fact that X and Y pair and segregate during meiosis, as autosomal homologs do, suggests that the sex-chromosomes are also homologs and presumably derived from a regular pair of autosomes. The paucity of genes on the Y would be explained by massive gene losses (“degeneration”), as first suggested by Sturtevant and Muller (cited in [6]. Further theoretical and empirical studies (reviewed in [7,8]) supported these initial suggestions.
Briefly, the current view asserts that X and Y originated when one homolog from a regular autosomal pair acquires one or more sex-determining genes, becoming a proto Y, and its homolog become a proto X. A combination of evolutionary forces then favors the suppression of recombination between the proto-Y and the proto-X, allowing for further differentiation. The absence of recombination in the proto-Y reduces the efficacy of natural selection and ultimately leads to gene degeneration and loss, and to the accumulation of repetitive DNA [9], leading to the evolution of a mature Y chromosome. X chromosomes still recombine in females and suffer comparatively small changes in their gene content [10,11]. Mammals illustrate this canonical pathway well: their X and Y are derived from an autosomal pair, and the Y contains the master sex-determining gene SRY [12]. The human Y (the best-known case) encodes around 27 different proteins, 16 of which have close counterparts on the X, being relics of the ancient autosomal pair [3]. Hence, the hallmarks of the canonical model for the origins of Y chromosomes are ancestral X-Y homology and evolution by gene loss. Interestingly, three of the 27 genes of the human Y originated by duplication of autosomal or X-linked genes.
Drosophila Y chromosomes are also gene-poor but do not fit this pattern of relic X-Y homology and evolution by gene losses: all their known genes were acquired through gene duplications from the autosomes, and gene gains are ~10-fold more frequent than gene losses, at least in the period where data is available (in the last ~30–60 Mya [13,14]. These conclusions came mostly from the study of two species (D. melanogaster and D. virilis) in which Y-linked protein-coding genes were thoroughly identified [13,15,16,17,18]. The small number of well-studied Y chromosomes is not restricted to Drosophila: in general, Y chromosomes are poorly known because their characteristics thwart many genetic and genomic methods [19]. For example, Y chromosome sequences are usually fragmented in genome assemblies due to their richness of repetitive DNA and end up in a collection of unmapped fragments, which require specific methods for proper identification and assembly [13,20]. Exons of the same gene are frequently scattered in different scaffolds, making gene identification and annotation challenging. Hence it is not surprising that few Y chromosomes have been well studied.
It has been customary to refer to all gene movements to the Drosophila Y chromosome as “new genes” (e.g., [15,21]). In contrast, the field of new gene evolution adopts a more stringent definition that focuses on the creation of novelties (e.g., “we can define new genes as those that are present in all members of a monophyletic group but absent from all outgroup species” [22]). Simple gene movements have been called “positionally relocated genes (PRGs)” [23] and seem to be excluded from the above definition of “new genes”. Hence, it is relevant to ask if we should call the genes that move to the Drosophila Y “new genes”. We argue that the answer is yes because, for several reasons, these movements always entail significant biological novelty: (i) genes that move to the Y become completely invisible to selection in females; (ii) they lose recombination, because Drosophila males are achiasmatic and hence genes that move to the Y chromosome automatically fail to recombine; (iii) they move to a heterochromatic environment. These changes have important consequences: two distinctive features of Drosophila Y-linked genes—reduced codon bias and gigantic introns—most likely reflect the lack of recombination and the heterochromatic environment [24,25,26,27]. Furthermore, the study of Drosophila Y-linked genes and sensu stricto “new genes” share many features and concepts such as gene traffic and sex-antagonistic selection. Thus, it seems appropriate to call the genes that move to the Drosophila Y chromosome new genes.
The prominence of gene gains in Drosophila Y chromosomes makes them an attractive model to study the acquisition of new genes in Y chromosomes. We aim to address here two broad sets of questions: the “how” (i.e., the mechanics of acquisition of new genes by the Drosophila Y) and the “why” (e.g., the role of chance and natural selection). Regarding the first set of questions (“How”), acquisition of new genes in general occurs by de novo origin (e.g., from non-coding DNA) or by gene duplications. The available evidence strongly supports gene duplications as the primary mechanism in most species, and in Drosophila ~25% of them are mediated by an mRNA intermediate (“retro-transposition”), the remaining 75% being DNA-based duplications [28]. Anecdotal evidence [15,21,29] indicates that DNA-mediated duplication is also widespread among Drosophila Y-linked genes, but this issue has not been systematically studied until now. A related question is how exactly an ancestrally autosomal gene is “transferred” to the Y.
Much less is known about the second set of questions (the “Why”). Fisher [30] stated that “close linkage with sex may have enabled certain variants, beneficial in the male, to have established genetic stability, for, had they been autosomal, their deleterious effect in the female might have definitely outweighed their genetic advantages”. Putting this into the context of gene movements, one can say that Y chromosomes are expected to accumulate male-related genes because male–female antagonistic effects of genes may hamper their evolution unless they are located in a male-specific region of the genome. While this selective hypothesis could explain the strongly male-biased content of Y chromosomes, we should note that a neutral process can also do this, as follows (see also the SI in [14]). Gene duplications create genetic redundancy, which can be “resolved” by several mechanisms, including the degeneration of either the original or the new copy [31]. Suppose this happened with an autosomal or X-linked male-specific gene that duplicated to the Y: the result may be a gene transfer to the Y, even if there is no selection favoring Y-linkage. Now suppose that the gene that duplicated to the Y was a housekeeping or female-specific gene: selection will keep the original copy, since females need those genes, and the Y-linked copy will most likely be lost. Given that Y chromosomes have few genes and the autosomes and X have many, the expected outcome of the above mechanisms over evolutionary time is a “gene traffic” of male-specific genes from the other chromosomes to the Y, even if there is no immediate fitness advantage for these male-specific genes being Y-linked. Which of these two hypotheses for the accumulation of male genes in the Y chromosomes (natural selection or chance) is correct, or more prevalent, is a hard question. Our aim here is necessarily humbler: to call attention to this question and provide some elements of the answer (namely, the role of chance events).
Given the small number of well-studied Drosophila Y chromosomes, adding new species is a worthwhile effort. In this paper we describe the identification of the Y-linked protein-coding genes of D. willistoni. As D. melanogaster, D. willistoni belongs to the Sophophora subgenus; they diverged shortly after the split between the Sophophora and Drosophila subgenera (30–60 Mya; [32,33]). Hence their divergence time is quite large, similar to their divergence from D. virilis, which belongs to the Drosophila subgenus.
2. Materials and Methods
2.1. Fly Strains and Genome Assemblies
Unless otherwise noted, we used the reference D. willistoni strain 14030-0811.24, which was sequenced as part of the 12 Drosophila Genomes Project [34]. This strain was originally collected in the Guadeloupe Island (Caribbean) and belongs to the D. willistoni willistoni subspecies [35]. We used the reference assembly (GCA_000005925.1). This Sanger assembly came from DNA collected from unsexed embryos [34] and hence contains the sequences from the Y chromosome. We also used two recent long read assemblies. Assembly GCA_018902025.1 employed PacBio technology; the current NCBI info states that it came from females, but the long reads actually came from males (José Ranz, personal communication), and indeed we found all Y-linked genes there. Assembly GCA_018903445.1 is based on Nanopore technology [36] and used male DNA from strain 14030-0811.17, which was collected in Uruguay, and hence belongs to the subspecies D. willistoni winge [35]. The genomes of D. paulistorum (two different strains), D. equinoxialis and D. tropicalis came from [36]. These species are closely related to D. willistoni, and we used their genomes as outgroups in several analyses.
2.2. Illumina Reads and Testis Transcriptome Assembly
As detailed below, Y-linked scaffolds can be identified by comparing an assembled genome with Illumina reads from genomic DNA of males and females. We used the accessions SRR9426110 and SRR9426117 for this purpose. To estimate gene expression we used RNA-seq reads from different organs and body parts (accessions listed in Table S1). A de novo testis transcriptome is very helpful in annotating Y-linked genes; we used the assembly GJOF01000000. The testis transcriptome and all Illumina reads came from the reference strain 14030-0811.24.
2.3. Identification of Y-Linked Scaffolds and Annotation of Y-Linked Genes
We used the YGS method to computationally identify the Y-linked scaffolds by comparing the assembled genome with male and female Illumina reads [13]. This procedure identifies Y-linked sequences but not the genes they contain. For example, in D. virilis, most Y-linked scaffolds contain only repetitive DNA and other non-coding sequences [13]. In order to identify and annotate the protein-encoding genes we did blast searches in the Y-linked sequences identified by the YGS method against databases of Dipteran proteins, bacterial and yeast proteins (to identify contaminant scaffolds), and transposable elements. These standard procedures were applied considering that, in many Y-linked genes, exons of the same gene are scattered in different scaffolds due to assembly failure in long, repeat-rich introns [29]. Similar challenges are met during the annotation of heterochromatic genes in general [37,38]. Furthermore, Y-linked genes frequently have gaps due to low coverage in Sanger reads: the reference genome of D. willistoni was sequenced at 8.4× depth from unsexed embryos, so the expected coverage of the Y chromosome is 2.1×. We fixed the fragmentation and gaps in the candidate genes by comparing the assembled genes with the testis transcriptome assembly. We detected both functional genes and pseudogenes during annotation. We adopted two criteria to classify them: functional genes have intact CDS (without premature stop codons and frame-shift indels) and significant expression at the RNA level (defined here as contributing more than 10% of the functional product). Y-linkage of all candidate genes was confirmed by PCR using male and female DNA.
2.4. Timeline of Gene Acquisitions by the Y Chromosome
The Y-linked genes found in D. willistoni might have been acquired at any point in the phylogeny. We identified this point by examining species progressively more distant from D. willistoni (within the willistoni group) and going up to D. saltans and D. melanogaster. For the genes transferred to the Y chromosome (i.e., no autosomal copy present), we used standard PCR in the other species with primers designed with the D. willistoni genes. In cases of amplification failure, we repeated the PCR using degenerate primers ([39]; Table S2). Recent duplications to the Y which left an intact autosomal copy cannot be reliably mapped outside D. willistoni with PCR because the few mismatches we used to design Y-specific primers are not guaranteed to have persisted across species. In these cases we used the following preliminary procedure. We performed blastN searches using as queries the D. willistoni Y-linked genes and as databases the recently sequenced male genomes of D. paulistorum, D. equinoxialis, D. tropicalis, D. nebulosa, and D. saltans [36]. If we find only one hit in a large scaffold (>~5 Mbp) which contains many other genes (and hence is euchromatic), we safely conclude that the gene was not duplicated in the target species. In contrast, two hits (one in a large scaffold, the second in a smaller one) suggest that duplication to the Y had already occurred in the target species.
2.5. Measurement of Gene Expression
We measured gene expression of the Y-linked and some other genes using the RSEM program [40] and RNA-seq databases from different body parts and organs of D. willistoni (Table S1). Y-linked genes are frequently misannotated due to the assembly problems mentioned above, so we needed to build a custom reference transcriptome. We started with the FlyBase annotation release 1.3 (downloaded in 12 October 2011from http://ftp.flybase.net/genomes/dwil/dwil_r1.3_FB2010_02/fasta/dwil-all-CDS-r1.3.fasta.gz), and removed from it all sequences matching the correctly annotated 22 D. willistoni Y-linked genes (the eight previously known, plus those identified in the present paper), using the cd-hit-est-2d program [41]. We then added back the correctly annotated 22 Y-linked genes and ran the RSEM program.
2.6. Detection of Segmental Duplications in the Y Chromosome
We separately aligned male and female Illumina genomic reads (accessions SRR9426110 and SRR9426117) against the reference Sanger assembly using the bwa software [42], and obtained coverage estimates using the bedtools package [43]. We then inspected coverage in suspicious scaffolds (which have many pseudogenes), looking for increases in coverage. We obtained the exact coordinates of the duplications by examining the confirmed duplications with the IGV browser [44], looking for increases in coverage and presence of male-specific SNPs.
3. Results
3.1. Identification of Y-Linked Sequences of D. willistoni
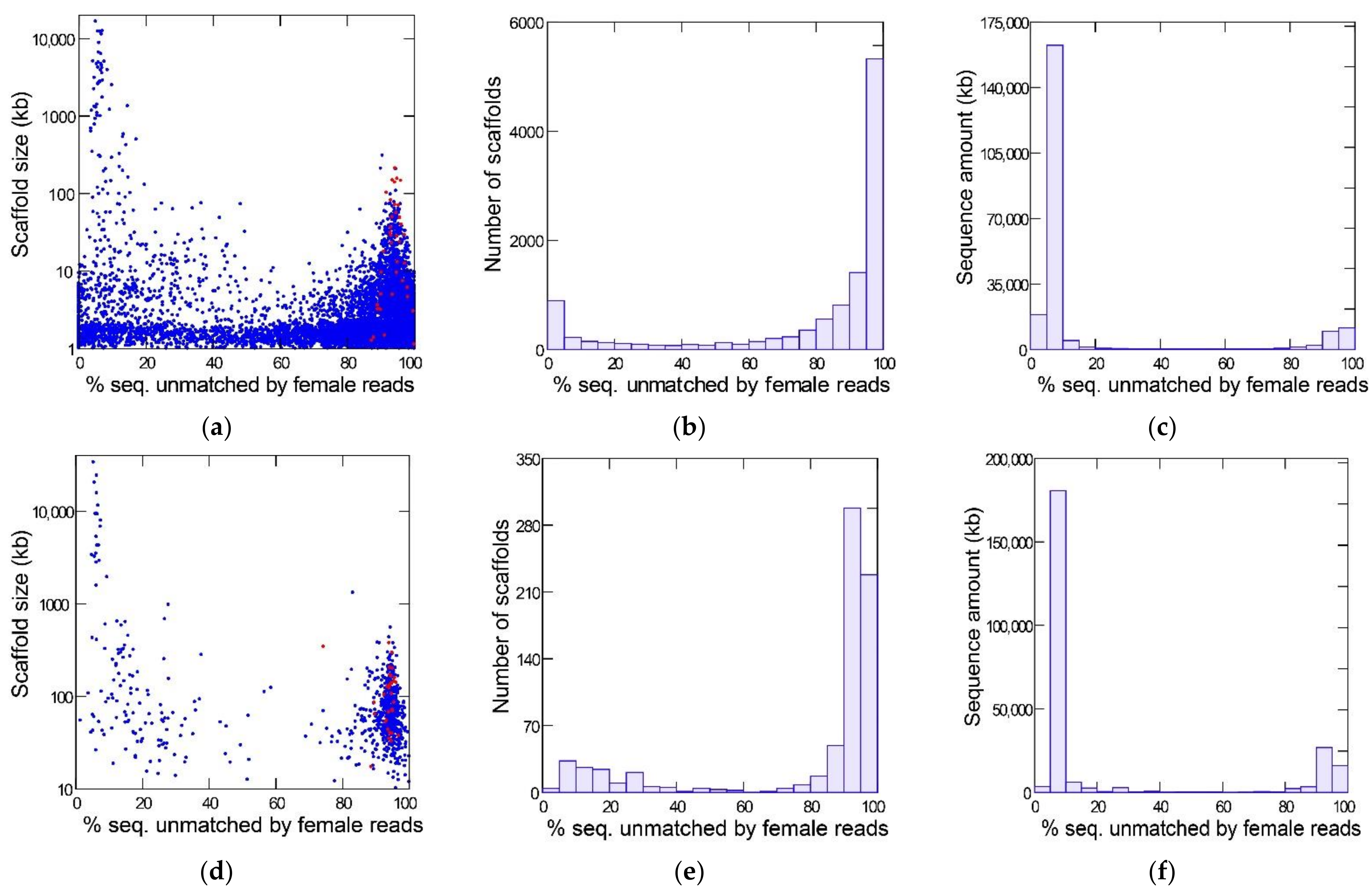
Figure 1 shows the results of the YGS method as applied to the reference Sanger assembly (top row) and a PacBio assembly (bottom row) of the same strain of D. willistoni. The bimodal distribution separates the Y-linked sequences (peak on the right-hand, with a large amount of sequence unmatched by female reads) from the X and autosomes (left-hand peak). The red dots mark the scaffolds matching the eight previously known Y-linked genes of D. willistoni [13,14]. All these sequences are located in the right-hand peak (average for Sanger assembly: 94%; range: 87–100%); they served as a positive control and show that the YGS method reliably identifies Y-linked sequences in D. willistoni. In the intermediate region of the graph, between the two peaks, there are numerous small sequences in the Sanger assembly; they correspond to misassembled repetitive regions [13]. YGS results are usually presented as in the first two columns of Figure 1, which may mislead the reader to conclude that Y-linked sequences represent a large proportion of the genome. The last column, which shows the amount of sequence in the scaffolds, clarifies that the Y chromosome is relatively small; because it is a repeat-rich region, Y-linked sequences are prone to assembly fragmentation and tend to get scattered in numerous small scaffolds. If we adopt the cut-off of 60% unmatched sequences, we find that the Y-chromosome in the Sanger assembly contains 28.5 Mbp (in 9031 scaffolds), whereas the X and autosomes amount to 189.8 Mbp (in 2107 scaffolds); the corresponding values for the PacBio assembly are 49.4 Mbp (in 605 scaffolds) and 197.6 Mbp (in 139 scaffolds). As suggested by the sharp bimodality shown in Figure 1 (panels c and f), the above estimates are quite robust in relation to the 60% cutoff; adopting an 80% cut-off would reduce the estimated size of the Y chromosome to 26.7 Mbp (Sanger assembly) and 48.6 Mbp (PacBio assembly). PacBio assemblies tend to be larger because they have a better representation of repetitive sequences; note that assembled size of the Y chromosome (being repeat-rich) increased much more than that of the X and autosomes. Another advantage of PacBio and Nanopore assemblies compared to Sanger or Illumina is that they are much less fragmented: the Sanger assembly contains 14,838 scaffolds, the PacBio contains 748, and the Nanopore contains 336. The smaller fragmentation helps the identification of Y-linked scaffolds: in the Sanger assembly, 3700 scaffolds, amounting to 5.3 Mbp, could not be classified by the YGS method because they are not informative (contain less than 20 valid single-copy k-mers; [13]); the corresponding figures for the PacBio assembly are four scaffolds and 71 kb. Despite these advantages, we will use the Sanger assembly in most analyses because it is the reference assembly.
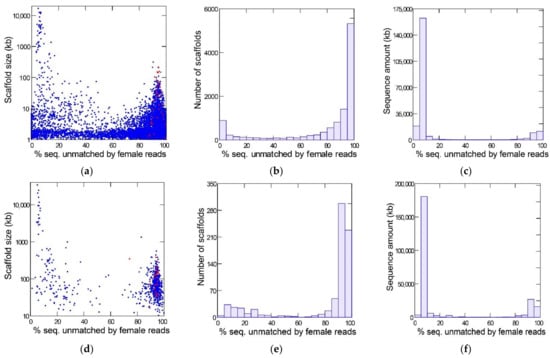
Figure 1.
Detection of Y-linked sequences in the reference D. willistoni genome with the YGS method. (a–c): reference Sanger assembly; (d–f), PacBio assembly. The X-axis in all graphs is the evidence of Y-linkage; scaffolds with a high proportion of sequence unmatched by female reads are Y-linked. In the left-hand panels (a,d), each dot represents a scaffold, and the red dots mark the previously known Y-linked sequences. The center panels (b,e) show the scaffold distribution as the number of scaffolds, and the right-hand panels (c,f) show the scaffold distribution as the sequence amount. Note that the bulk of the sequence can be reliably classified as Y-linked (peak on the right-hand) or X/autosomal (left-hand peak). Non-informative scaffolds (less than 20 single-copy k-mers) were excluded from the analysis (see text). The raw YGS results are available in the Supplementary Data.
3.2. Identification of Y-Linked Protein Coding Genes
We found all eight previously known Y-linked genes of D. willistoni while annotating the 28.5 Mbp (9031 scaffolds) predicted to be Y-linked (their orthologs in D. melanogaster are: kl-2, kl-3, PRY, PPrY, ORY, CCY, ARY, and JYalpha; [13,14]. Besides these, we found 14 protein-coding genes, reported here as Y-linked genes for the first time (Table 1).

Table 1.
D. willistoni Y-linked genes found in this work.
We confirmed Y-linkage of these genes with PCR in male and female DNA (e.g., [39]); no false-positives were found. Eleven out of these 14 genes have been automatically annotated during the sequencing of the D. willistoni genome [34], and the remaining three by Yang et al. [45]. None of these have been manually curated before (Table S7). We adopted a conservative nomenclature by keeping their original names, even if only a small portion of their coding sequence had been identified before. Due to the gaps and fragmentation in the genome assembly in most cases the original annotation was incomplete. In the Piezo-like ortholog parts of the gene were annotated as three different genes (GK27406, GK18657, and GK27211; we kept the name GK27406, which is the largest gene). A similar situation happened with YOgnWI000172. The main problems we found during gene annotation were the fragmentation and the gaps, but the genomic sequence of the multicopy genes GK20618/GK20619 and GK18510 may have mis-assemblies. Due to these uncertainties the classification of GK18510 as a functional gene should be seen as a preliminary result (Supplementary Material).
All 14 genes were acquired by gene duplications from ancestrally autosomal or X- linked genes which already had testis-specific expression (Table 1 and Table S3). The evidence of gene duplication is unequivocal for recent movements because, in these cases, we still found the original autosomal copy, either as a functional gene (as occurred with GK18510, GK20591, YOgnWI018045, and GK20619) or as a pseudogene (as occurred with GK20609 and GK21220; see Discussion). For older acquisitions, the evidence is based on parsimony. For example, as detailed in Section 3.5, the Y-linked gene GK27406 has autosomal orthologs in five progressively more distant outgroups (D. insularis, D. nebulosa, D. sucinea, D. saltans, and D. melanogaster), so its ancestral location must be autosomal. Its Y-linkage in D. willistoni must be the result of one gene duplication to the Y, followed by the loss of the original autosomal copy. The alternative hypothesis (ancestral Y-linkage) would imply five duplications, coupled with the loss of the original Y-linked copy in each case (see ref. [14] for details).
The pattern of gene duplications from autosomal (or X-linked) testis-specific genes we observed in D. willistoni fits what is found in other Drosophila species, except that D. willistoni is the first species in which X-linked genes were found as the source of duplications (we confirmed the X-linkage by looking at the D. paulistorum genome; see below). Ancestrally X-linked genes that move to the Y have potential implications for hybrid sterility, on which we will comment further in the Discussion. The 14 genes span a range of functions, such as enzymes and structural proteins of the spermatozoon. The first 10 genes showed in Table 1 were fully transferred to the Y, in the sense that the original autosomal copy was lost or reduced to a pseudogene. Therefore, they will most likely stay in the Y chromosome. The fate of the last four genes (GK20591, YOgnWI018045, and the multicopy GK20618/GK20619 and GK18510) is less certain because a functional autosomal copy remained at the original location (the genes GK19651, GK14595, YOgnWI012342, and GK18077, respectively), and both copies have a similar level of expression (Table S3; Table S4). The high similarity between their Y and autosomal copies (equal or greater than 96% identity at the nucleotide level) implies a recent origin (see section “When did the D. willistoni Y chromosome acquire its genes?”). It seems that, for these four genes, all outcomes are possible: loss of Y-copy, loss of autosomal copy, or retention of both, perhaps via subfunctionalization [31]. Some of them, in particular the multicopy with pseudogenes GK18510 and GK20618/GK20619, may be examples of what Betrán [46] called ‘‘live fast, die young’’: genes that participate in arms races such as male–male competition or male–female antagonism, and that do not last long in the genome.
Besides functional genes, Drosophila Y chromosomes contain a number of pseudogenes, as happens in other heterochromatic regions [13,15,37,47,48]. As detailed in the Material and Methods, we used the intactness of the coding regions and the expression at the RNA level as criteria to sort pseudogenes from functional genes. Such classifications may be too arbitrary, particularly if there is a large gray zone. This was not the case in D. willistoni. Besides the GK18510 gene mentioned above (and detailed in Appendix A), the two most doubtful cases we found are: (i) the annotated “gene” GK22179 (which has an expression level of ~3% of its autosomal counterpart, GK11098), and (ii) an unannotated sequence in scaffold CH961348 (similar to the autosomal gene GK11572), which most likely was an artifact in the Sanger assembly: PCR repeatedly fail to amplify it in males and females, and it is absent from the PacBio assembly. We found 45 pseudogenes (Table S5), by far the largest number ever found in a Drosophila Y chromosome [13,15,37,47,48]. This finding has an interesting explanation: the D. willistoni Y chromosome harbors four large segmental duplications from autosomes, which originally contained ~58 protein-coding genes. All but four became Y-linked pseudogenes or disappeared.
3.3. Y-Linked Segmental Duplications from Autosomes
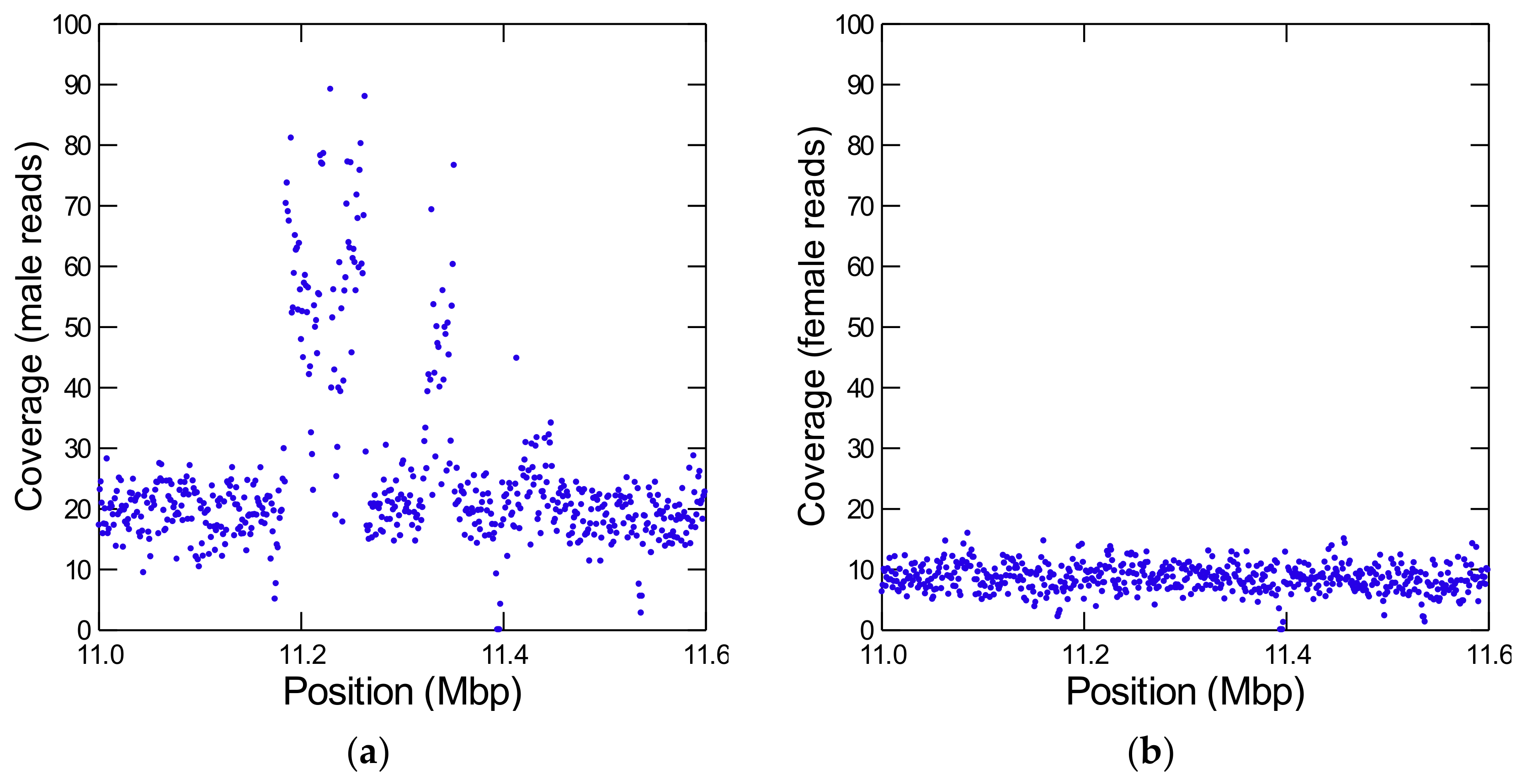
During the annotation of Y-linked scaffolds it came to our attention that most pseudogenes have ancestral locations close to each other (Table S5), suggesting that they originated from segmental duplications of autosomal or X-linked chromosomal regions. We confirmed this suspicion by looking at the coverage across these regions (Figure 2; [49]). We identified four large segmental duplications (Table 2), and there are probably additional smaller ones.
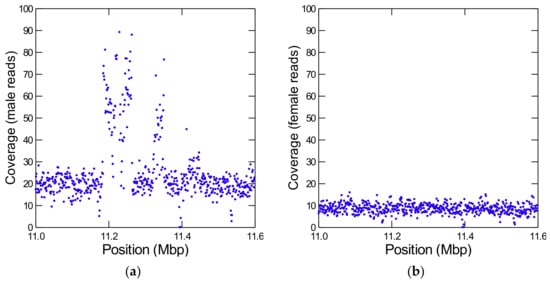
Figure 2.
Detection of a Y-linked segmental duplication from an autosome. The figure shows the coverage of Illumina reads from males (a) and females (b) in a part of the autosomal scaffold CH964272. Note the huge increase in male coverage between positions ~11.2–11.4 Mbp, which is absent in the female reads. It corresponds to a segmental duplication of at least 265 kb from CH964272 to the Y chromosome. The coverage is very high and irregular, indicating many partial duplications and deletions inside the Y-chromosome. Each dot is the average depth of 1 kb interval (to reduce scattering). In the raw data, some regions have at least ten copies.

Table 2.
Y-linked segmental duplications in D. willistoni.
The very high coverage in these regions shows that they have multiple copies inside the Y chromosome (in some cases ~10 copies), and the unevenness of the coverage implies that they suffered partial deletions and duplications. Given the high copy number, it is not surprising that this region is heavily fragmented in the Sanger assembly (and to a lesser extent in PacBio). Indeed, among the 9031 Y-linked scaffolds that we found in the Sanger assembly, 3033 match these four regions (the matching regions span ~1.7 Mbp). This duplication to the Y, followed by additional duplication and deletions, is reminiscent of the FDY region found before in the D. melanogaster Y [15], except that its 11 kb are dwarfed by the size of the D. willistoni segmental duplications.
These segmental duplications created four functional genes (GK20609, YOgnWI018045, and the multicopy genes GK20618/GK20619 and GK18510), while ~54 became pseudogenes or disappeared. This finding raises the question of what factor(s) influenced the fate of these duplicated genes. The autosomal or X-linked ancestors of all four surviving genes are testis-specific genes, as judged by their orthologs in D. melanogaster (Table 1) and their gene expression in D. willistoni (Tables S3 and S4). The same pattern has been previously found in the well-studied D. melanogaster and D. virilis [13,16]. Hence, testis-specific expression is necessary for the retention of a gene in the Drosophila Y chromosome. However, it is not sufficient: among the 54 lost genes, 10 have testis-specific expression in D. willistoni, and in most cases the same is true for their D. melanogaster orthologs. We will return to this point in the Discussion
3.4. Mechanism of Duplications to the Y: DNA or RNA-Based?
In order to identify the duplication mechanism, we used two criteria. First, genes that are part of a segmental duplication certainly originated through a DNA duplication. Second, we looked for conserved intron positions between the Y-linked gene, and its autosomal (or X-linked) ortholog in another species, since RNA-mediated duplications lead to intron loss. We initially used only D. melanogaster as the outgroup. However, we found one case (the GK20609 gene) suggesting complete intron loss in a gene duplicated as part of a segmental duplication, which does not make sense. It is known that the D. willistoni lineage underwent many intron losses [50], which are unrelated to the movement of genes. We circumvented this problem by looking at the intron–exon structure in the species of the willistoni group that have been recently sequenced by Kim et al. [36]. In these species, the GK20609 ortholog is autosomal and already intron-less. We cannot ascertain the duplication mechanism for seven out of the 14 genes (they are intron-less in the outgroup species and are not part of a segmental duplication). The remaining seven genes were DNA-based duplications: three have conserved intron positions (GK21041, YOgnWI000172, and GK27406), and four are part of segmental duplications (GK20609, GK18510, YOgnWI018045, and GK20618/GK20619).
3.5. When Did the D. willistoni Y Chromosome Acquire Its Genes?
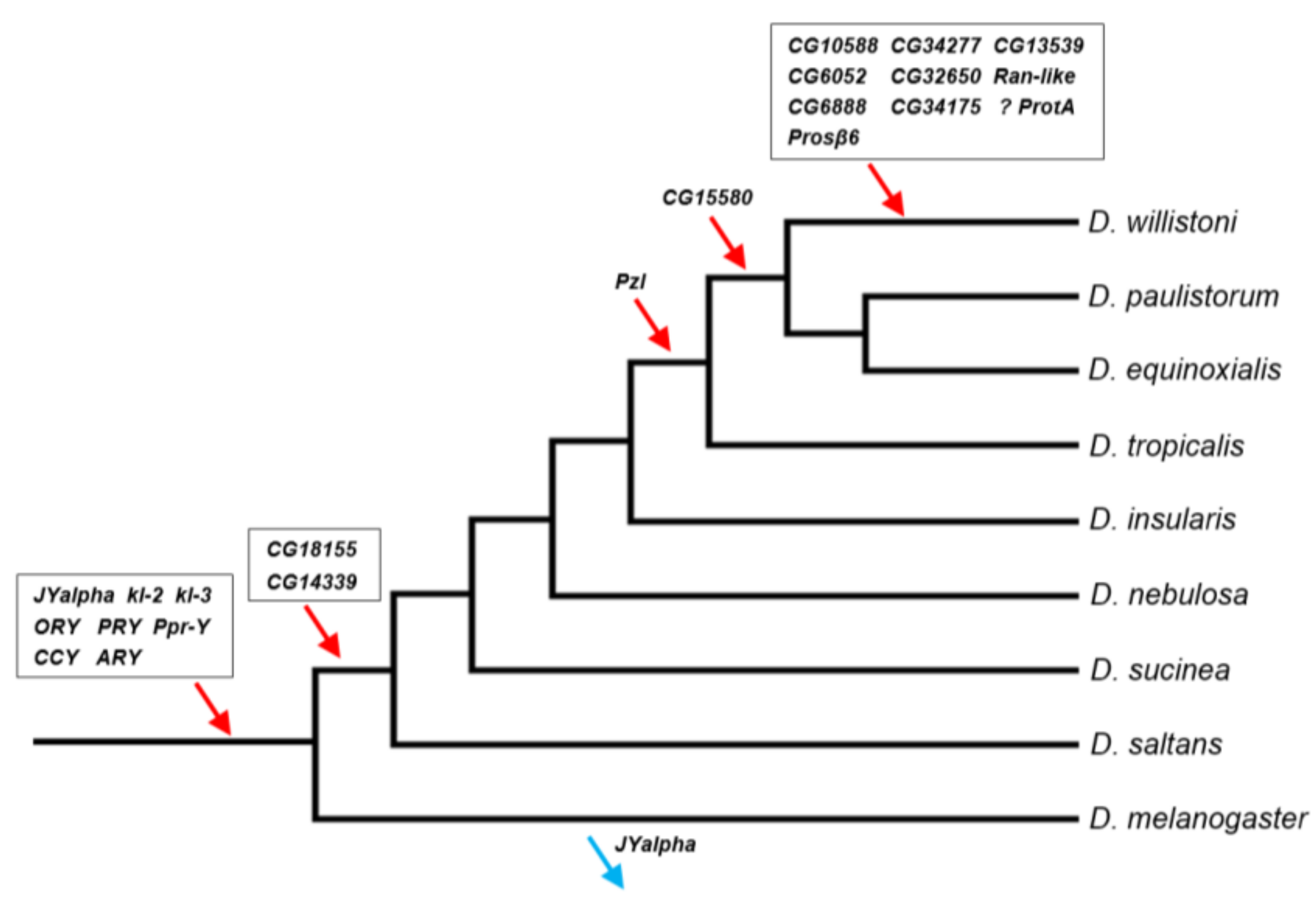
Previous investigations found eight genes in the D. willistoni Y by searching for genes found to be Y-linked in D. melanogaster and D. virilis [13,14]. Thus, almost by definition, these genes were acquired before the split between D. willistoni and these two species. In this paper, we used a de novo identification approach (YGS), so the Y chromosome might have acquired its genes at any point in the phylogeny. We investigated this issue as detailed in ref. [39] and in the Material and Methods. Similar to what has been seen in previous studies [13,14], we found that the D. willistoni Y-linked genes were acquired throughout the phylogeny (Figure 3; Table S6).
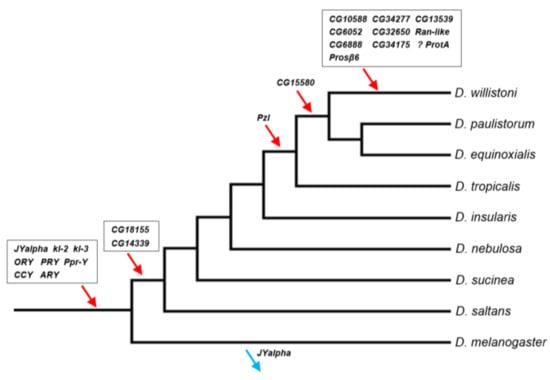
Figure 3.
Timeline of gene acquisitions by the Y chromosome of the D. willistoni lineage. Gene location (Y-linkage vs. autosomal/X-linkage) was determined by PCR, except for the four genes with very similar autosomal and Y-linked copies (CG6888, CG34175, ProtA and Prosβ6; see Material and Methods). We inferred the direction of the movements (gene gains, red arrows; gene losses, blue arrows) by synteny and parsimony [14,39]. The eight genes on the basal branch were already known to be Y-linked in D. willistoni. JYalpha is not Y-linked in D. melanogaster due to a gene movement to an autosome within the melanogaster group [13]. There is some uncertainty regarding the ProtA gene, as it may have been acquired in the previous branch (at the same point of CG15580; Table S6). Genes were labeled with the names of the D. melanogaster orthologs to facilitate cross-species comparisons. Phylogenetic relationships were taken from [51].
We also extended this investigation to the subspecies level. D. willistoni contains three subspecies with partial reproductive isolation [35,52]. Two of them have been sequenced in males: D. w. willistoni (to which the reference strain belongs) and D. w. winge [36]. We found that all 22 Y-linked genes (the eight previously known plus the 14 identified in the present work) are Y-linked in the D. w. winge strain (i.e., they are located in the right-hand peak of the YGS graph). Hence, they were acquired by the Y chromosome before the split of these two subspecies.
4. Discussion
Previous work searched in D. willistoni for protein-encoding Y-linked genes in two other Drosophila species with well-known Y chromosomes, D. melanogaster and D. virilis. These efforts identified eight genes [13,14]. Here, we used a de novo approach and found 14 additional genes, all acquired after the split between D. willistoni and these two species. Below we discuss the main implications of these findings.
4.1. D. willistoni Y-Linked Genes: Comparison with Other Drosophila Species
The Y-linked gene content of D. willistoni shares several important features with the two other species with well-known Y chromosomes, D. melanogaster and D. virilis. First, in the three species nearly all genes originated by duplications from genes already strongly or exclusively expressed in testis. The only known exception is FDY, a recent Y-linked gene from D. melanogaster whose ancestral gene (vig2) is expressed in many tissues and organs (including testis) and is strongly expressed in ovaries [53]. Second, the gene duplications to the D. willistoni Y were mediated by a DNA mechanism in all seven cases that we could ascertain. This probably holds true for other Drosophila species, although they have not yet been systematically examined in this respect. If confirmed, this would be an interesting difference between genes acquired by the Y and the other chromosomes, which involve an RNA intermediate in 25% of the cases [28]. A possible explanation for the absence of RNA-based duplications to the Drosophila Y is that this chromosome (as other heterochromatic regions) is a harsh environment for arriving genes: it has been long known that euchromatic genes that move to heterochromatic regions are silenced [54], and it is likely that a gene duplication carrying flanking euchromatic sequence has a higher survival chance than a naked, promoter-less retrocopy. The third commonality among the three species is the preponderance of gene gains over gene losses, as suggested by the inspection of Figure 3 and confirmed by the statistical analysis. As detailed in the Supplementary Material, using the data of the three species we found a gain–loss ratio of 25 (p = 0.002; 95% confidence interval: 3.4–184.5), and the same qualitative result is obtained when removing the four genes whose original autosomal copies are functional (Figure S3 and Table S8). These four genes—GK20591, YOgnWI018045, and the multicopy GK18510 and GK20618/GK20619—arguably can become pseudogenes in the Y; if we remove them, we obtain a gain–loss ratio of 21; p = 0.003. It will be interesting to look at other species, particularly outgroups, to better understand what is happening. However, it is already clear that Drosophila Y chromosomes are not evolving according to the canonical theory of Y chromosome evolution (see Appendix A for an alternative view).
On the other hand, the D. willistoni Y chromosome has some features not seen in other Drosophila species. First, it seems that a recent burst in gene acquisitions happened after the split between D. willistoni and its cryptic species D. paulistorum/D. equinoxialis, which created a set of ~10 private Y-linked genes in D. willistoni. This gene gain burst is even more remarkable given the rather short time interval (4.8 Mya; Ref. [55]) and the lack of recent bursts in the well-studied D. melanogaster and D. virilis (the former has one or two private Y-linked genes [15,56]; D. virilis has none). Second, it seems that the gene gain rate is higher in D. willistoni. The previous estimates of the gain-loss ratio were 10.7 and 4.9 (using D. melanogaster and D. virilis as the focal species, respectively [13,14], and when we included D. willistoni we got 25. Furthermore, the heterogeneity in the gain–loss ratio among branches becomes statistically significant, suggesting that D. willistoni is an outlier. Third, we found four large segmental duplications that copied ~700 kb of autosomal sequence in the D. willistoni Y chromosome (Table 2); the previously known cases have a few kb [15,56]. The first and second peculiarities of the D. willistoni Y may be a consequence of the third: a higher rate of segmental duplications to the Y is expected to increase the gene gain rate and, if recent, may generate a fairly large amount of recent gene acquisitions by the Y. We got mixed results while trying to find evidence for this hypothesis in the literature. Suppose this hypothesis is correct and that segmental duplications occur in other chromosomes as well. In that case, one might expect to find increased gene movements in general, but D. willistoni does not seem to be an outlier in this respect [23]. On the other hand, Vibranovski et al. [28] analyzed the same dataset of [23] by partitioning it in A→A, A→X, and X→A movements and noted that D. willistoni is an outlier to the general pattern of excess of gene movements out of the X: it has many A→A and A→X movements, which weakly supports the idea that gene gains are more frequent in the D. willistoni lineage. Perhaps at this point, the most solid conclusion we can obtain was already outlined in the 12 Drosophila Genome Project paper: “D. willistoni is an exceptional outlier by several criteria, including its unusually skewed codon usage, increased transposable element content and potential lack of seleno-proteins” [34,57]. The list goes on: “Some clades, like the willistoni group, seem to undergo many more [intron] losses per million years than others” [50]. It will be interesting to directly investigate the occurrence of segmental duplications in the X and autosomes to verify if it is indeed increased in the D. willistoni lineage and if these duplications created new genes. In particular, one can look at the unexpected gene movements reported by Vibranovski et al. [28], and check if several genes came from adjacent chromosomal regions, which is a telltale sign of a segmental duplication. Finally, careful studies of more Drosophila Y chromosomes might help us better understand the relationship between segmental duplications and gene traffic to the Y chromosome.
4.2. Gene Movements to the Y Chromosome and Reproductive Isolation
All previously known Drosophila Y-linked genes originated from duplications of autosomal genes, whereas two D. willistoni Y-linked genes were formerly X-linked (Table 1). In itself, this may be just a coincidence: the sample size is small (there were seven gene acquisitions in the D. melanogaster lineage whose ancestral location is known, and four in D. virilis [13,14]), and male genes (which usually originate Y-linked genes) are underrepresented in the X [10,11]. Nevertheless, X-Y gene movements potentially have major biological consequences on speciation. Namely, if a gene essential for male fertility moves from the X to the Y in one species (or population), it instantly creates unidirectional hybrid male sterility (i.e., one direction of the interspecific cross will produce sterile F1 males, and the other will produce fertile F1 males). Unidirectional hybrid male sterility is indeed quite common in Drosophila [58]. The most relevant case here is the recent Y-linked gene GK28041 whose orthologs are X-linked in D. melanogaster (ortholog: CG32650) and D. paulistorum (it is flanked by Muller A genes in the D. paulistorum assemblies reported in [36]; the gene order is: CG4542-CG32649-CG32650-Pde9-CG3775). Crosses between D. willistoni and D. paulistorum yield unidirectional hybrid male sterility, but (disappointingly) in the “wrong” direction: D. willistoni male × D. paulistorum female produce sterile males, and the reciprocal cross produces fertile males [59]. So GK28041 is not essential for male fertility, and other sterility factors play the major role in those hybrids. Another interesting case is provided by three subspecies of D. willistoni, in which hybrids also display unidirectional hybrid male sterility [35,52]. As commented before, all 22 genes (the eight previously known plus the 14 identified in the present work, including GK28041) are Y-linked in the two sequenced subspecies. Hence, X–Y movements of the known Y-linked D. willistoni genes are not the cause of hybrid sterility in crosses between these two subspecies. While both cases we investigated here yield negative results, gene movements can cause hybrid sterility, as shown by Masly et al. [60], and are a particularly powerful mechanism when they occur between the X and Y.
4.3. How Do Male Genes Move to the Y Chromosome?
In most Drosophila Y-linked genes, the only sign of their autosomal (or X) origin is their location in other species. The recent acquisition of the FDY (flagrante delicto Y) by the D. melanogaster Y chromosome left more clear signs of this process: an 11 kb autosomal sequence containing five genes got duplicated to the Y, and all genes except FDY (which is a copy of vig2) became pseudogenes, their sequences becoming scrambled by deletions, duplications and point mutations [15]. However, in FDY, the autosomal copy is active, so this gene does not represent the most common case of a gene transfer to the Y chromosome. Two D. willistoni genes provide snapshots of this transfer process.
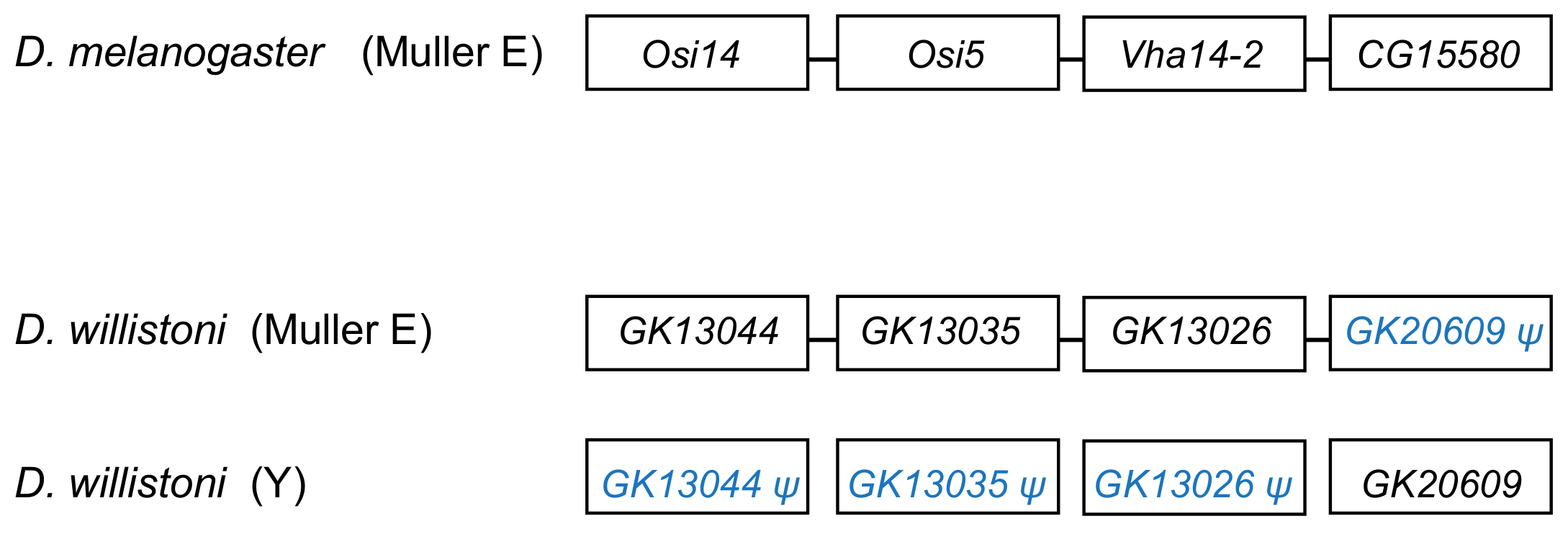
The segmental duplication involving scaffold CH964272 (Table 2) duplicated a 265 kb long autosomal sequence, which originally contained 25 genes, to the Y chromosome. Remnants of 12 genes (Table S5), and one functional gene, GK20609 (Table 1) are found in the Y chromosome. Interestingly we found a mirrored situation when we looked at the autosomal sequence: all genes are functional, except for a pseudogenized copy of GK20609, that contains many deletions and substitutions (Figure 4). This provides direct evidence of how male genes move to the Y. First, an autosomal or X-linked gene duplicates to the Y (possibly as part of a segmental duplication). Then, in some cases, the genetic redundancy is “resolved” by the degeneration of the original copy; over time, the pseudogene in the autosome (or X), and its flanking sequences in the Y chromosome tend to disappear. Eventually, a functional Y-linked gene whose ortholog is autosomal or X-linked in other Drosophila species is the only remaining sign of the original gene duplication.
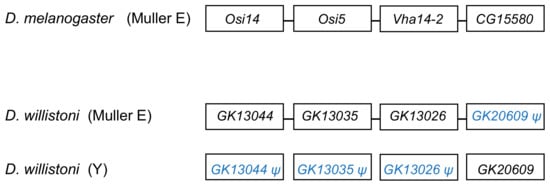
Figure 4.
Fate of genes in the CH964272 segmental duplication. The segmental duplication copied 265 kb of autosomal sequence containing 25 genes to the Y chromosome of D. willistoni (for the sake of simplicity, the figure shows only four genes and omits several intervening ones). The Y-linked copy of GK20609 (D. melanogaster ortholog: CG15580) remained functional, and the remaining 24 genes disappeared or became pseudogenes, whereas the opposite happened in the autosomal region. CG15580/GK20609 and Vha14-2/GK13026 are testis-specific genes in both species. The D. willistoni autosomal region is syntenic with D. melanogaster chromosome 3R (Muller E element). Gene order is unknown in the Y chromosome copy because it suffered duplications and deletions and the assembly is fragmented.
The second example we found caught a later stage of the above process. Blast searches with the Y-linked gene GK21220 found the Y-linked scaffolds (as expected) and a very similar 118 bp match in the autosomal scaffold CH963850 (coordinates 5158275–5158392). This small matching region is also present in the PacBio and Nanopore assemblies, ruling out assembly error. When we inspected the corresponding region in the D. paulistorum assembly [36], we found that it is autosomal (as expected) and contains the functional ortholog of the GK21220 gene (Figure S2). This shows that the 118 bp region of scaffold CH963850 in the D. willistoni genome is a remnant of the original GK21220 gene, which degenerated after its duplication to the Y. We could not find any sign of the flanking genes in the D. willistoni Y, either because they were not copied to the Y, or had degenerated beyond recognition.
4.4. Why Do Male Genes Move to the Y Chromosome?
The traffic of male genes to the Y chromosome (e.g., Figure 3) intuitively suggests that natural selection favors the Y-linkage of these genes. However, as argued in the Introduction, this is not necessarily correct. The Y-linked segmental duplications of D. willistoni are very useful in this respect because they provide replicas: many genes were transferred at the same time to the same location in the Y chromosome; a few of them became functional Y-linked genes, and most were lost. Testis-expression seems to be a necessary condition for the establishment of a functional gene in the Drosophila Y chromosome. Still, as shown by the D. willistoni data, it is not sufficient: among the 25 genes duplicated to the Y by the scaffold CH964272 segmental duplication, seven were testis-specific (GK13044, GK13038, GK14208, GK13026, GK26902, GK28338, and GK20609). Only GK20609 survived as a functional gene; the remaining six became Y-linked pseudogenes or disappeared. Why?
A possible selective explanation would be gene amplification: Y chromosomes (as other heterochromatic regions) are prone to accumulate duplications [3,15,56], and if natural selection favors multiple copies for some male genes, there may be an advantage in transferring to the Y chromosome, due to several mechanisms. For example, the gene may be more easily duplicated, or the duplications inside the Y may be more easily tolerated because there is no recombination (and hence ectopic recombination is greatly reduced), or because the Y has very low gene density (and thus there is a smaller chance of interfering with other genes). While there are examples of gene amplifications in Drosophila and human Y chromosomes Y [3,61], and two recent Y-linked genes of D. willistoni are multicopy (GK18510 and GK20618/GK20619), we can rule this out as a general explanation for Drosophila because nearly all known Drosophila Y-linked protein-encoding genes (including GK20609) are single copy [13,16,17,18]. Another popular selective explanation is that male genes may evolve more easily on the Y chromosome due to antagonistic selection in females [30]. While we cannot reject this explanation, we must note that it is not compelling either: Drosophila Y-linked genes have a myriad of functions (motor proteins, structural proteins of the sperm, enzymes) whose only common feature seems to be testis expression. If antagonistic selection in females plays a significant role in this process, we must conclude that it was present in the diverse set of genes that successfully moved to the Y (e.g., GK20609) and absent in those that failed (e.g., GK13044, GK13038, GK14208, GK13026, GK26902, GK28338; Figure 4).
Alternatively, chance might play a significant role in the “choice” of which gene copy (the original or the new Y-linked one) stays functional and which one becomes a pseudogene. Of course, chance does not imply 50% probability, and indeed in the CH964272 segmental duplication, only one out seven testis genes that duplicated to the Y eventually became established there. Given that Y chromosomes have few genes and the autosomes and X have many, this mechanism by itself would create gene traffic of male genes from the X and autosomes to the Y chromosome. While we cannot at this moment prove that this hypothesis is correct, it seems advisable to consider it, along with natural selection-based models, as possible explanations for the male gene traffic to the Y chromosome. Finally, it seems to us that the above discussion does not apply to the X-autosome gene traffic problem: unlike the Y chromosome, the X is neither restricted to one sex nor extremely gene-poor.
4.5. Concluding Remarks and Future Directions
There is much we do not understand about Drosophila Y chromosomes, and few of these chromosomes have been studied in detail. As a consequence, each newly studied species brings new aspects to light. The case of D. willistoni highlighted the role of segmental duplications in the origin of new genes in the Y chromosome and possibly also in other chromosomes. Besides this, the segmental duplications create replicas to study the evolutionary fate of genes that are duplicated to the Y, which opens a window to investigate the roles of natural selection and chance in the gene traffic to the Y. Further studies are likely to bring additional surprises, particularly in areas that have been barely touched, such as Drosophila outgroups [17] (e.g., how far in the phylogeny the pattern of “more gains than losses” goes) and non-coding RNA [62] (which may help explain the manifold effects of Drosophila Y chromosomes in phenotypes such as longevity [63], gene expression [64], and hybrid sterility [65]).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12111815/s1, Figure S1: Phylogeny of the Prosβ6 gene family in D. willistoni, Figure S2: Ancestral autosomal location of the Y-linked gene GK21220, Figure S3: Gene gains and losses in Y chromosomes of three Drosophila species, Table S1: Illumina datasets, Table S2: PCR primers used to test for Y-linkage in D. willistoni and other species, Table S3: Expression (in fpkm) of Prosβ6 gene family in D. willistoni, Table S4: Expression (in fpkm) of Y-linked genes and their close autosomal paralogs, Table S5: D. willistoni Y-linked pseudogenes, Table S6: Time of acquisition of genes that retained the autosomal or X-linked original copy, Table S7: D. willistoni Y-linked genes as represented in successive annotation efforts, Table S8: Gene gains and losses in the Y chromosomes of three Drosophila species.
Author Contributions
Conceptualization, methodology, funding acquisition, A.B.C.; investigation, formal analysis, J.R. and A.B.C.; data curation, F.U. and A.B.C.; writing—original draft preparation, A.B.C.; writing—review and editing, F.U. and A.B.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by FAPERJ—Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, grant number CNE2018, CNPq—Conselho Nacional de Desenvolvimento Científico e Tecnológico, grant number INCT-EM, and Wellcome Trust, grant 207486/Z/17/Z.
Institutional Review Board Statement
Ethical review and approval were waived for this study according to the Brazilian law, because it did not involve vertebrates.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the Supplementary Material.
Acknowledgments
We thank present and part members of the Carvalho’s lab for help and suggestions during the annotation of the Y-linked genes, in particular E. Dupim, G. Goldstein, and L. Araripe. We also thank G. Goldstein and M. Vibranovski for help with the testis transcriptome, and two anonymous reviewers for valuable suggestions in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Appendix A
In several passages of this work, we called attention to the acquisition of new genes by the Drosophila Y as a major feature of its evolution, in contrast with the standard model of Y chromosome degeneration. While we stand for this interpretation, we agree that there is room for disagreement. We think that it is worth summarizing this here, starting with the canonical view.
Sex-chromosome evolution provides a compelling blend of empirical data and theory (reviewed in [8]). Briefly, the canonical theory states that X–Y chromosomes evolve from a common autosomal ancestor, after one homolog acquires one or more strong sex-determining genes. Gradual suppression of X–Y recombination ensues, accompanied by gene losses in the Y. The above theory is very general, being valid for plant and mammalian Y chromosomes (e.g., Refs. [3,66]). Furthermore, it applies to X0 species, which would be the last step in the Y degeneration process [67]; note that it is difficult to imagine an alternative route for the evolution of X chromosomes. Given the above commented generality and the similarities between the Drosophila Y and other systems (e.g., low gene content, accumulation of repetitive DNA), it most likely is a degenerated X chromosome. Hence, it must have lost a huge number of genes, in fact all its ancestral protein coding genes and the original sex-determining gene. Drosophila X and Y share rRNA genes [53], which is evidence of their common origin. The nearly complete loss of X-Y homology in Drosophila strongly suggests that it is an old system. Indeed, the origin of the Drosophila Y predates the split of the Drosophila and Sophophora subgenera (30–60 Mya; [14,32,33]). Gene loss is quite fast in Drosophila when a genome segment becomes male-restricted: the young neo-Y of D. miranda (~1.5 Myr old) is already degenerating and ~13 Myr-old neo-Y of D. pseudoobscura has completely degenerated [48,68,69]. Hence, the lack or paucity of X–Y homology is expected. The complete loss of several thousand genes from a chromosome is a much more relevant biological phenomenon than the acquisition of a few genes from other chromosomes, as reported here for the D. willistoni Y.
Our view may be summarized as follows (see [16,19] for details). Around the origin of the Drosophilidae family, the original X and Y chromosomes evolved, probably as described by the canonical theory, and it is possible that at some point this lineage became X0 (Figure 1 in Ref. [19]). The actual history is complex and largely unknown, since there were numerous replacements of the X chromosome within Diptera [70,71]). Regarding the current Drosophila Y, there is no solid evidence that it is a degenerated X chromosome and there are alternative explanations for its origin. For example, Hackstein et al. [72] noticed similarities between the Drosophila Y and B-chromosomes and suggested that the former originated from the latter. B-chromosomes are parasitic genetic elements that employ a variety of mechanisms that ensure their propagation, including the ability to pair with the X in X0 species [73]. In some Homoptera species, the B-X segregation is nearly perfect (99.5% correct); these B-chromosomes are now called Y chromosomes [74,75,76]. A B-chromosome origin of the Drosophila Y nicely explains its lack of a sex-determining gene and shared genes with the X. More recently, Fraïsse et al. [77] suggested a similar non-canonical origin for the lepidopteran W. Interestingly, B-chromosomes frequently contain rRNA genes, which confirms that repetitive genes are poor indicators of common ancestry (e.g., [78]). So we think that we cannot assume that the current Drosophila Y is a degenerated X (and that it suffered massive gene loss). However, even if it is, there is something unexpected in its evolution. The canonical theory predicts gradual gene loss, which would deaccelerate as the number of remaining genes diminish [8,79], possibly approaching a gain-loss steady-state equilibrium, but it would not explain a net gain of genes, for which there is direct evidence in D. melanogaster [14], D. virilis [13], and now in D. willistoni. The number of gene gains is small, but in the last 30–60 Myr, which is the period we currently have good data for, it has been the dominating force in the evolution of the Drosophila Y. It is worth noting here that demasculinization of the X chromosomes, which is a major biological phenomenon, is also manifested by a small number of gene movements (e.g., Table 3 of [80] and Table S3 of [10]).
References
- Bridges, C.B. Non-disjunction as proof of the chromosome theory of heredity. Genetics 1916, 1, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J. Evolution of Sex Determining Mechanisms; Benjamin/Cummings Pub. Co. Advanced Book Program: Menlo Park, CA, USA, 1983; p. xx, 316. [Google Scholar]
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T.; et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003, 423, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Meccariello, A.; Salvemini, M.; Primo, P.; Hall, B.; Koskinioti, P.; Dalikova, M.; Gravina, A.; Gucciardino, M.A.; Forlenza, F.; Gregoriou, M.E.; et al. Maleness-on-the-Y (MoY) orchestrates male sex determination in major agricultural fruit fly pests. Science 2019, 365, 1457–1460. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.; Celniker, S.; Holt, R.; Evans, C.; Gocayne, J.; Amanatides, P.; Scherer, S.; Li, P.; Hoskins, R.; Galle, R. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef]
- Muller, H.J. A gene for the fourth chromosome of Drosophila. J. Exp. Zool 1914, 17, 325–336. [Google Scholar] [CrossRef]
- Rice, W.R. Evolution of the Y sex chromosome in animals. Bioscience 1996, 46, 331–343. [Google Scholar] [CrossRef]
- Charlesworth, B.; Charlesworth, D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1563–1572. [Google Scholar] [CrossRef]
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef]
- Sturgill, D.; Zhang, Y.; Parisi, M.; Oliver, B. Demasculinization of X chromosomes in the Drosophila genus. Nature 2007, 450, 238–241. [Google Scholar] [CrossRef]
- Parisi, M.; Nuttall, R.; Naiman, D.; Bouffard, G.; Malley, J.; Andrews, J.; Eastman, S.; Oliver, B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 2003, 299, 697–700. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.F.; Rozen, S. Genomics and genetics of human and primate Y chromosomes. Annu. Rev. Genom. Hum. Genet. 2012, 13, 83–108. [Google Scholar] [CrossRef]
- Carvalho, A.B.; Clark, A.G. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 2013, 23, 1894–1907. [Google Scholar] [CrossRef]
- Koerich, L.B.; Wang, X.; Clark, A.G.; Carvalho, A.B. Low conservation of gene content in the Drosophila Y chromosome. Nature 2008, 456, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Vicoso, B.; Russo, C.A.; Swenor, B.; Clark, A.G. Birth of a new gene on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2015, 112, 12450–12455. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Koerich, L.B.; Clark, A.G. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009, 25, 270–277. [Google Scholar] [CrossRef]
- Mahajan, S.; Bachtrog, D. Convergent evolution of Y chromosome gene content in flies. Nat. Commun. 2017, 8, 785. [Google Scholar] [CrossRef] [PubMed]
- Gepner, J.; Hays, T.S. A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc. Natl. Acad. Sci. USA 1993, 90, 11132–11136. [Google Scholar] [CrossRef]
- Carvalho, A.B. Origin and evolution of the Drosophila Y chromosome. Curr. Opin. Genet. Dev. 2002, 12, 664–668. [Google Scholar] [CrossRef]
- Hall, A.B.; Qi, Y.; Timoshevskiy, V.; Sharakhova, M.V.; Sharakhov, I.V.; Tu, Z. Six novel Y chromosome genes in Anopheles mosquitoes discovered by independently sequencing males and females. BMC Genom. 2013, 14, 273. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Dobo, B.A.; Vibranovski, M.D.; Clark, A.G. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2001, 98, 13225–13230. [Google Scholar] [CrossRef]
- Long, M.; VanKuren, N.W.; Chen, S.; Vibranovski, M.D. New gene evolution: Little did we know. Annu. Rev. Genet. 2013, 47, 307–333. [Google Scholar] [CrossRef]
- Bhutkar, A.; Russo, S.M.; Smith, T.F.; Gelbart, W.M. Genome-scale analysis of positionally relocated genes. Genome Res. 2007, 17, 1880–1887. [Google Scholar] [CrossRef] [PubMed]
- Reugels, A.M.; Kurek, R.; Lammermann, U.; Bunemann, H. Mega-introns in the dynein gene DhDhc7(Y) on the heterochromatic Y chromosome give rise to the giant Threads loops in primary spermatocytes of Drosophila hydei. Genetics 2000, 154, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.D.; Koerich, L.B.; Carvalho, A.B.; Clark, A.G. Positive and purifying selection on the Drosophila Y chromosome. Mol. Biol. Evol. 2014, 31, 2612–2623. [Google Scholar] [CrossRef]
- Carvalho, A.B.; Clark, A.G. Intron size and natural selection. Nature 1999, 401, 344. [Google Scholar] [CrossRef]
- Diaz-Castillo, C.; Golic, K.G. Evolution of gene sequence in response to chromosomal location. Genetics 2007, 177, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Vibranovski, M.D.; Zhang, Y.; Long, M. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009, 19, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Lazzaro, B.P.; Clark, A.G. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc. Natl. Acad. Sci. USA 2000, 97, 13239–13244. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. The evolution of dominance. Biol. Rev. 1931, 6, 345–368. [Google Scholar] [CrossRef]
- Lynch, M.; Katju, V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004, 20, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Obbard, D.J.; Maclennan, J.; Kim, K.W.; Rambaut, A.; O’Grady, P.M.; Jiggins, F.M. Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol. Biol. Evol. 2012, 29, 3459–3473. [Google Scholar] [CrossRef]
- Tamura, K.; Subramanian, S.; Kumar, S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 2004, 21, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.G.; Eisen, M.B.; Smith, D.R.; Bergman, C.M.; Oliver, B.; Markow, T.A.; Kaufman, T.C.; Kellis, M.; Gelbart, W.; Iyer, V.N.; et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature 2007, 450, 203–218. [Google Scholar] [CrossRef]
- Mardiros, X.B.; Park, R.; Clifton, B.; Grewal, G.; Khizar, A.K.; Markow, T.A.; Ranz, J.M.; Civetta, A. Postmating reproductive isolation between strains of Drosophila willistoni. Fly 2016, 10, 162–171. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, B.Y.; Wang, J.R.; Miller, D.E.; Barmina, O.; Delaney, E.; Thompson, A.; Comeault, A.A.; Peede, D.; D’Agostino, E.R.; Pelaez, J.; et al. Highly contiguous assemblies of 101 drosophilid genomes. Elife 2021, 10, e66405. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.D.; Shu, S.Q.; Mungall, C.J.; Karpen, G.H. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science 2007, 316, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, R.A.; Smith, C.D.; Carlson, J.W.; Carvalho, A.B.; Halpern, A.; Kaminker, J.S.; Kennedy, C.; Mungall, C.J.; Sullivan, B.A.; Sutton, G.G.; et al. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 2002, 3, 1–16. [Google Scholar] [CrossRef]
- Dupim, E.G.; Goldstein, G.; Vanderlinde, T.; Vaz, S.C.; Krsticevic, F.; Bastos, A.; Pinhao, T.; Torres, M.; David, J.R.; Vilela, C.R.; et al. An investigation of Y chromosome incorporations in 400 species of Drosophila and related genera. PLoS Genet. 2018, 14, e1007770. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11.12.1–11.12.34. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Jaime, M.; Polihronakis, M.; Kanegawa, K.; Markow, T.; Kaneshiro, K.; Oliver, B. Re-annotation of eight Drosophila genomes. Life Sci. Alliance 2018, 1, e201800156. [Google Scholar] [CrossRef] [PubMed]
- Betran, E. The “life histories” of genes. J. Mol. Evol. 2015, 80, 186–188. [Google Scholar] [CrossRef] [PubMed]
- Tobler, R.; Nolte, V.; Schlotterer, C. High rate of translocation-based gene birth on the Drosophila Y chromosome. Proc. Natl. Acad. Sci. USA 2017, 114, 11721–11726. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Clark, A.G. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 2005, 307, 108–110. [Google Scholar] [CrossRef]
- Bailey, J.A.; Gu, Z.; Clark, R.A.; Reinert, K.; Samonte, R.V.; Schwartz, S.; Adams, M.D.; Myers, E.W.; Li, P.W.; Eichler, E.E. Recent segmental duplications in the human genome. Science 2002, 297, 1003–1007. [Google Scholar] [CrossRef]
- Coulombe-Huntington, J.; Majewski, J. Intron loss and gain in Drosophila. Mol. Biol. Evol. 2007, 24, 2842–2850. [Google Scholar] [CrossRef]
- Finet, C.; Kassner, V.A.; Carvalho, A.B.; Chung, H.; Day, J.P.; Day, S.; Delaney, E.K.; De Re, F.C.; Dufour, H.D.; Dupim, E.; et al. DrosoPhyla: Resources for Drosophilid Phylogeny and Systematics. Genome Biol. Evol. 2021, 13, evab179. [Google Scholar] [CrossRef]
- Ayala, F.J.; Tracey, M.L. Enzyme variability in the Drosophila willistoni group. VIII. Genetic differentiation and reproductive isolation between subespecies. J. Hered. 1973, 64, 120–124. [Google Scholar] [CrossRef]
- Larkin, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; Dos Santos, G.; Garapati, P.V.; Goodman, J.L.; Gramates, L.S.; Millburn, G.; Strelets, V.B.; et al. FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res 2021, 49, D899–D907. [Google Scholar] [CrossRef] [PubMed]
- Elgin, S.C.; Reuter, G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 2013, 5, a017780. [Google Scholar] [CrossRef] [PubMed]
- Zanini, R.; Muller, M.J.; Vieira, G.C.; Valiati, V.H.; Depra, M.; Valente, V. Combining morphology and molecular data to improve Drosophila paulistorum (Diptera, Drosophilidae) taxonomic status. Fly 2018, 12, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Krsticevic, F.J.; Schrago, C.G.; Carvalho, A.B. Long-read single molecule sequencing to resolve tandem gene copies: The Mst77Y region on the Drosophila melanogaster Y chromosome. G3 2015, 5, 1145–1150. [Google Scholar] [CrossRef]
- Chapple, C.E.; Guigo, R. Relaxation of selective constraints causes independent selenoprotein extinction in insect genomes. PLoS ONE 2008, 3, e2968. [Google Scholar] [CrossRef]
- Coyne, J.A.; Orr, H.A. “Patterns of speciation in Drosophila”. Evolution 1997, 51, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Winge, H.; Cordeiro, A.R. Experimental hybrids between Drosophila willistoni Sturtevant and Drosophila paulistorum Dobzhansky and Pavan from southern marginal populations. Heredity 1963, 18, 215–222. [Google Scholar] [CrossRef]
- Masly, J.P.; Jones, D.; Noor, M.A.; Locke, J.; Orr, H.A. Gene transposition as a cause of hybrid sterility in Drosophila. Science 2006, 313, 1448–1450. [Google Scholar] [CrossRef]
- Gvozdev, V.A.; Kogan, G.L.; Usakin, L.A. The Y chromosome as a target for acquired and amplified genetic material in evolution. Bioessays 2005, 27, 1256–1262. [Google Scholar] [CrossRef]
- Chen, P.; Kotov, A.A.; Godneeva, B.K.; Bazylev, S.S.; Olenina, L.V.; Aravin, A.A. piRNA-mediated gene regulation and adaptation to sex-specific transposon expression in D. melanogaster male germline. Genes. Dev. 2021, 35, 914–935. [Google Scholar] [CrossRef]
- Brown, E.J.; Nguyen, A.H.; Bachtrog, D. The Y chromosome may contribute to sex-specific ageing in Drosophila. Nat. Ecol. Evol. 2020, 4, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Lemos, B.; Araripe, L.O.; Hartl, D.L. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 2008, 319, 91–93. [Google Scholar] [CrossRef]
- Lamnissou, K.; Loukas, M.; Zouros, E. Incompatibilities between Y chromosome and autosomes are responsible for male hybrid sterility in crosses between Drosophila virilis and Drosophila texana. Heredity 1996, 76, 603–609. [Google Scholar] [CrossRef]
- Marais, G.A.; Nicolas, M.; Bergero, R.; Chambrier, P.; Kejnovsky, E.; Moneger, F.; Hobza, R.; Widmer, A.; Charlesworth, D. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr. Biol. 2008, 18, 545–549. [Google Scholar] [CrossRef]
- White, M.J.D. Animal Cytology and Evolution, 3rd ed.; University Press: Cambridge, UK, 1973; 961p. [Google Scholar]
- Steinemann, M.; Steinemann, S. Degenerating Y-chromosome of Drosophila miranda-a trap for retrotransposons. Proc. Natl. Acad. Sci. USA 1992, 89, 7591–7595. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Wei, K.H.; Nalley, M.J.; Gibilisco, L.; Bachtrog, D. De novo assembly of a young Drosophila Y chromosome using single-molecule sequencing and chromatin conformation capture. PLoS Biol. 2018, 16, e2006348. [Google Scholar] [CrossRef]
- Vicoso, B.; Bachtrog, D. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 2015, 13, e1002078. [Google Scholar] [CrossRef]
- Vicoso, B.; Bachtrog, D. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 2013, 499, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Hackstein, J.H.P.; Hochstenbach, R.; Hauschteck-Jungen, E.; Beukeboom, L.W. Is the Y chromosome of Drosophila an evolved supernumerary chromosome? Bioessays 1996, 18, 317–323. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Sharbel, T.F.; Beukeboom, L.W. B-chromosome evolution. Philos. Trans. R. Soc. Lond. Ser. B-Biol. Sci. 2000, 355, 163–178. [Google Scholar] [CrossRef]
- Nokkala, S.; Nokkala, C. Interaction of B chromosomes with A or B chromosomes in segregation in insects. Cytogenet. Genome Res. 2004, 106, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Nokkala, S.; Grozeva, S.; Kuznetsova, V.; Maryanska-Nadachowska, A. The origin of the achiasmatic XY sex chromosome system in Cacopsylla peregrina (Frst.)(Psylloidea, Homoptera). Genetica 2003, 119, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Nokkala, S.; Kuznetsova, V.; Maryanska-Nadachowska, A. Achiasmate segregation of a B chromosome from the X chromosome in two species of psyllids (Psylloidea, Homoptera). Genetica 2000, 108, 181–189. [Google Scholar] [CrossRef]
- Fraisse, C.; Picard, M.A.L.; Vicoso, B. The deep conservation of the Lepidoptera Z chromosome suggests a non-canonical origin of the W. Nat. Commun. 2017, 8, 1486. [Google Scholar] [CrossRef] [PubMed]
- Steinemann, M.; Pinsker, W.; Sperlich, D. Chromosome homologies within the Drosophila obscura group probed by insitu hybridization. Chromosoma 1984, 91, 46–53. [Google Scholar] [CrossRef]
- Bachtrog, D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics 2008, 179, 1513–1525. [Google Scholar] [CrossRef]
- Betran, E.; Thornton, K.; Long, M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002, 12, 1854–1859. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).