Circulating Transcriptional Profile Modulation in Response to Metabolic Unbalance Due to Long-Term Exercise in Equine Athletes: A Pilot Study
Abstract
1. Introduction
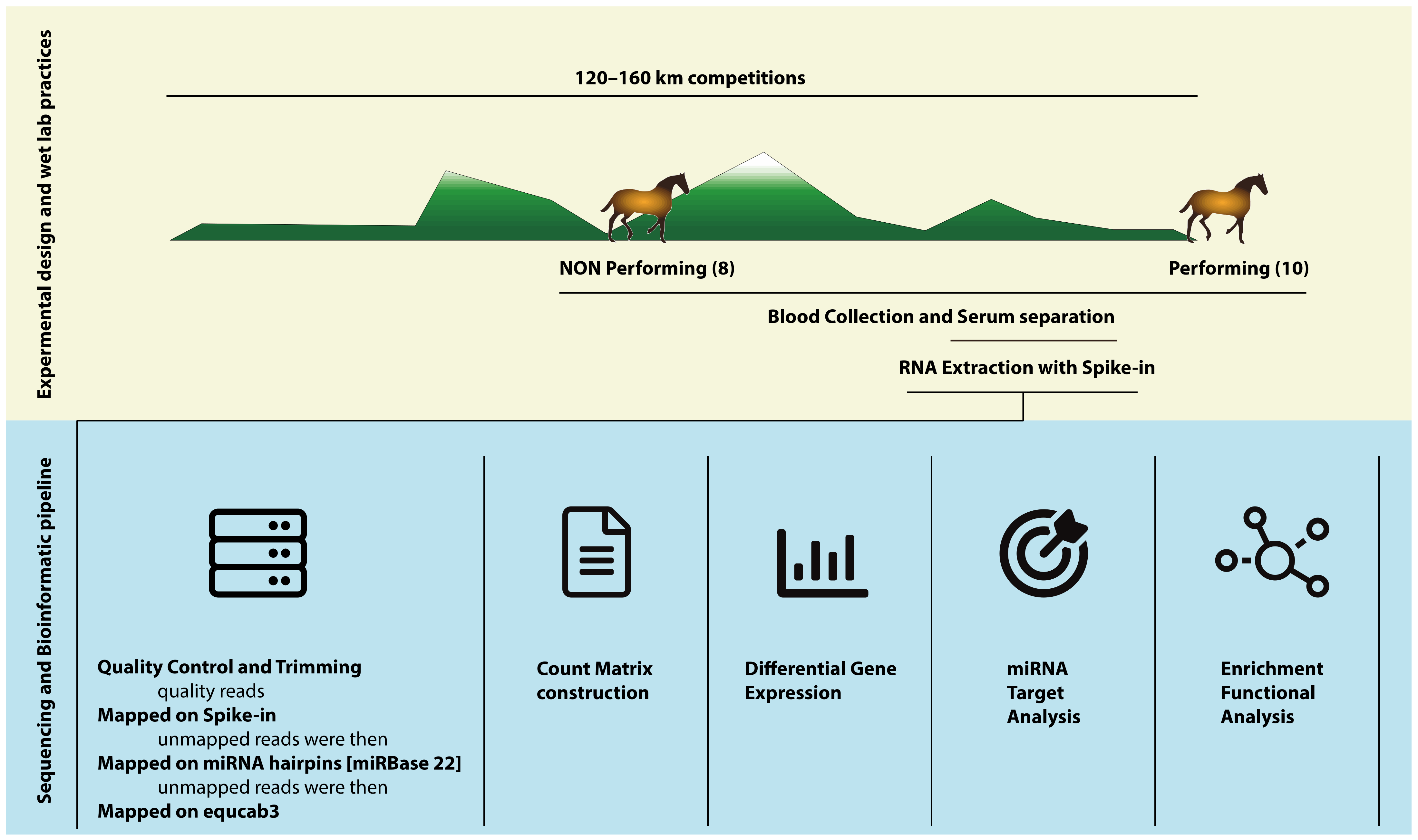
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
2.3. Blood Count and Biochemical Analysis
2.4. RNA Extraction and Library Preparation
2.5. Bioinformatic Analysis
Target Genes and Enrichment Analysis
3. Results
3.1. Hematology and Clinical Chemistry Analyses
3.2. Sequencing Statistics
3.3. Differential Expression Analysis of miRNA
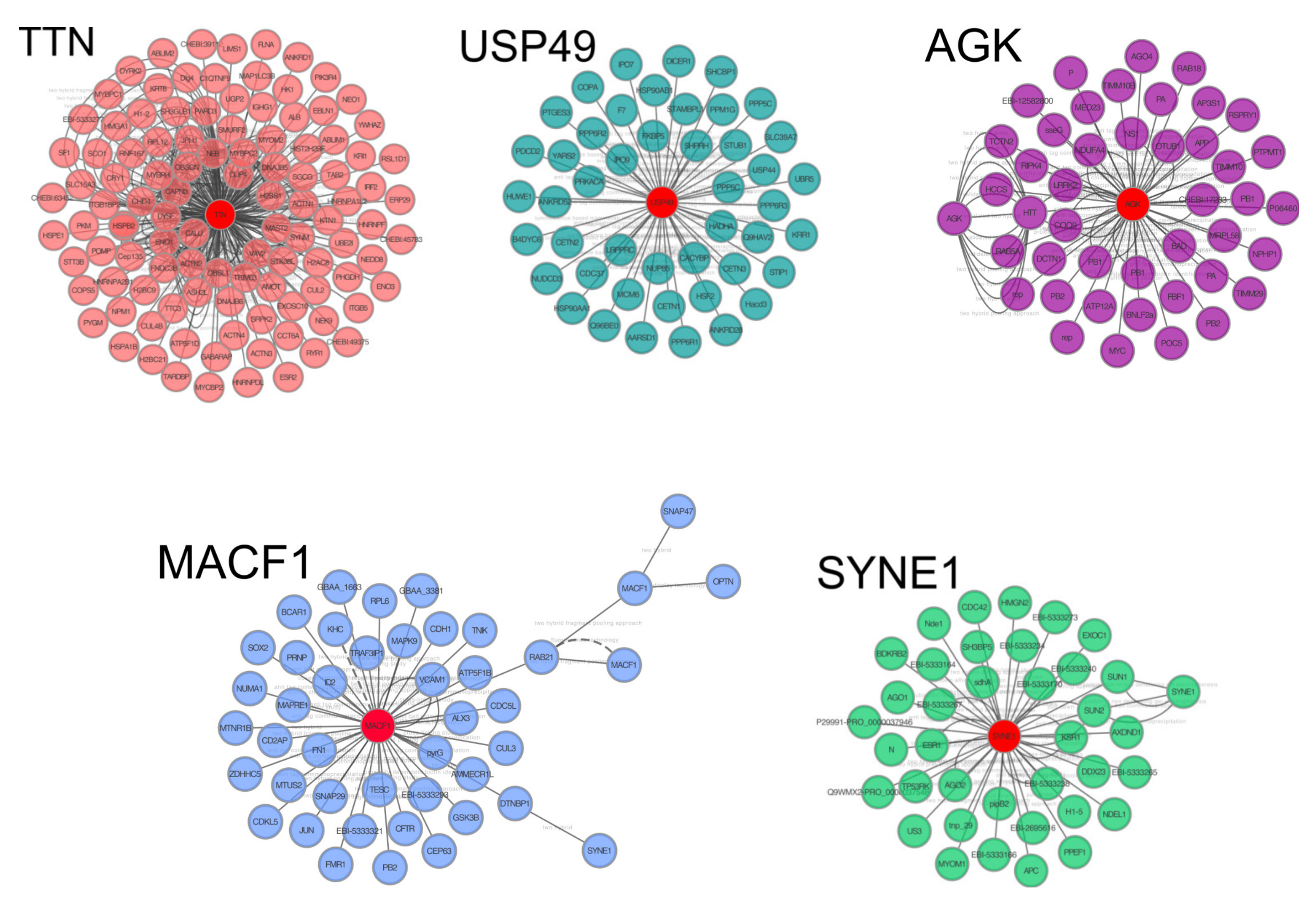
3.4. miRNA-Target Evaluation and Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dhabhar, F.S. Effects of stress on immune function: The good, the bad, and the beautiful. Immunol. Res. 2014, 58, 193–210. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.P.; Kayani, A.C.; McArdle, A.; Drust, B. The Exercise-Induced stress response of skeletal muscle, with specific emphasis on humans. Sports Med. 2009, 39, 643–662. [Google Scholar] [CrossRef]
- Cappelli, K.; Mecocci, S.; Gioiosa, S.; Giontella, A.; Silvestrelli, M.; Cherchi, R.; Valentini, A.; Chillemi, G.; Capomaccio, S. Gallop racing shifts mature mRNA towards introns: Does exercise-induced stress enhance genome plasticity? Genes 2020, 11, 410. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, K.; Amadori, M.; Mecocci, S.; Miglio, A.; Antognoni, M.T.; Razzuoli, E. Immune response in young thoroughbred racehorses under training. Animals 2020, 10, 1809. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, K.; Felicetti, M.; Capomaccio, S.; Nocelli, C.; Silvestrelli, M.; Verini-Supplizi, A. Effect of training status on immune defence related gene expression in Thoroughbred: Are genes ready for the sprint? Vet. J. 2013, 195, 373–376. [Google Scholar] [CrossRef]
- Noakes, T.D. Physiological models to understand exercise fatigue and the adaptations that predict or enhance athletic performance. Scand. J. Med. Sci. Sports 2000, 10, 123–145. [Google Scholar] [CrossRef] [PubMed]
- Nagy, A.; Dyson, S.J.; Murray, J.K. A veterinary review of endurance riding as an international competitive sport. Vet. J. 2012, 194, 288–293. [Google Scholar] [CrossRef]
- Amory, H.; Votion, D.M.; Fraipont, A.; Goachet, A.G.; Robert, C.; Farnir, F.; Van Erck, E. Altered systolic left ventricular function in horses completing a long distance endurance race. Equine Vet. J. 2010, 42, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Scoppetta, F.; Tartaglia, M.; Renzone, G.; Avellini, L.; Gaiti, A.; Scaloni, A.; Chiaradia, E. Plasma protein changes in horse after prolonged physical exercise: A proteomic study. J. Proteom. 2012, 75, 4494–4504. [Google Scholar] [CrossRef]
- Mooren, F.C.; Viereck, J.; Krüger, K.; Thum, T. Circulating micrornas as potential biomarkers of aerobic exercise capacity. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H557–H563. [Google Scholar] [CrossRef] [PubMed]
- Polakovičová, M.; Musil, P.; Laczo, E.; Hamar, D.; Kyselovič, J. Circulating MicroRNAs as potential biomarkers of exercise response. Int. J. Mol. Sci. 2016, 17, 1553. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Liu, Q.; Yao, J.; Dai, Y.; Wang, H.; Xiao, J. Circulating microRNAs in response to exercise. Scand. J. Med. Sci. Sports 2015, 25, e149–e154. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, G.; Perego, S.; Sansoni, V.; Banfi, G. Circulating miRNA as fine regulators of the physiological responses to physical activity: Pre-analytical warnings for a novel class of biomarkers. Clin. Biochem. 2016, 49, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Makarova, J.A.; Maltseva, D.V.; Galatenko, V.V.; Abbasi, A.; Maximenko, D.G.; Grigoriev, A.I.; Tonevitsky, A.G.; Northoff, H. Exercise immunology meets MiRNAs. Exerc. Immunol. Meets MiRNAs 2014, 20, 135–164. [Google Scholar]
- Cappelli, K.; Capomaccio, S.; Viglino, A.; Silvestrelli, M.; Beccati, F.; Moscati, L.; Chiaradia, E. Circulating miRNAs as putative biomarkers of exercise adaptation in endurance horses. Front. Physiol. 2018, 9, 429. [Google Scholar] [CrossRef] [PubMed]
- Head, S.R.; Kiyomi Komori, H.; LaMere, S.A.; Whisenant, T.; Van Nieuwerburgh, F.; Salomon, D.R.; Ordoukhanian, P. Library construction for next-generation sequencing: Overviews and challenges. Biotechniques 2014, 56, 61–77. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kalbfleisch, T.S.; Rice, E.S.; DePriest, M.S.; Walenz, B.P.; Hestand, M.S.; Vermeesch, J.R.; O’Connell, B.L.; Fiddes, I.T.; Vershinina, A.O.; Saremi, N.F.; et al. Improved reference genome for the domestic horse increases assembly contiguity and composition. Commun. Biol. 2018, 1, 1–8. [Google Scholar] [CrossRef]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. ClusterMaker: A multi-algorithm clustering plugin for Cytoscape. BMC Bioinform. 2011, 12, 436. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef]
- Fielding, C.L.; Magdesian, K.G.; Rhodes, D.M.; Meier, C.A.; Higgins, J.C. Clinical and biochemical abnormalities in endurance horses eliminated from competition for medical complications and requiring emergency medical treatment: 30 cases (2005–2006): Retrospective study. J. Vet. Emerg. Crit. Care 2009, 19, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.B.; Konhilas, J.P.; Kelly, D.P.; Leinwand, L.A. Molecular Mechanisms Underlying Cardiac Adaptation to Exercise. Cell Metab. 2017, 25, 1012–1026. [Google Scholar] [CrossRef]
- Garciarena, C.D.; Pinilla, O.A.; Nolly, M.B.; Laguens, R.P.; Escudero, E.M.; Cingolani, H.E.; Ennis, I.L. Endurance training in the spontaneously hypertensive rat conversion of pathological into physiological cardiac hypertrophy. Hypertension 2009, 53, 708–714. [Google Scholar] [CrossRef]
- Feng, H.J.; Ouyang, W.; Liu, J.H.; Sun, Y.G.; Hu, R.; Huang, L.H.; Xian, J.L.; Jing, C.F.; Zhou, M.J. Global microRNA profiles and signaling pathways in the development of cardiac hypertrophy. Braz. J. Med. Biol. Res. 2014, 47, 361–368. [Google Scholar] [CrossRef][Green Version]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef]
- Das, A.; Samidurai, A.; Salloum, F.N. Deciphering Non-coding RNAs in Cardiovascular Health and Disease. Front. Cardiovasc. Med. 2018, 5, 73. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.; Vollaard, N.B.J.; Gustafsson, T.; Gallagher, I.J.; Sundberg, C.J.; Rankinen, T.; Britton, S.L.; Bouchard, C.; Koch, L.G.; Timmons, J.A. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J. Appl. Physiol. 2011, 110, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Solich, J.; Kuśmider, M.; Faron-Górecka, A.; Pabian, P.; Kolasa, M.; Zemła, B.; Dziedzicka-Wasylewska, M. Serum Level of miR-1 and miR-155 as Potential Biomarkers of Stress-Resilience of NET-KO and SWR/J Mice. Cells 2020, 9, 917. [Google Scholar] [CrossRef] [PubMed]
- Håkansson, K.E.J.; Sollie, O.; Simons, K.H.; Quax, P.H.A.; Jensen, J.; Nossent, A.Y. Circulating Small Non-coding RNAs as Biomarkers for Recovery After Exhaustive or Repetitive Exercise. Front. Physiol. 2018, 9, 1136. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.; Åkerström, T.; Rinnov, A.; Yfanti, C.; Scheele, C.; Pedersen, B.K.; Laye, M.J. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS ONE 2014, 9, e87308. [Google Scholar] [CrossRef]
- Brown, M.D.; Hudlicka, O. Modulation of physiological angiogenesis in skeletal muscle by mechanical forces: Involvement of VEGF and metalloproteinases. Angiogenesis 2003, 6, 1–14. [Google Scholar] [CrossRef]
- Baggish, A.L.; Hale, A.; Weiner, R.B.; Lewis, G.D.; Systrom, D.; Wang, F.; Wang, T.J.; Chan, S.Y. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J. Physiol. 2011, 589, 3983–3994. [Google Scholar] [CrossRef]
- Wang, D.; Wang, Y.; Ma, J.; Wang, W.; Sun, B.; Zheng, T.; Wei, M.; Sun, Y. MicroRNA-20a participates in the aerobic exercise-based prevention of coronary artery disease by targeting PTEN. Biomed. Pharmacother. 2017, 95, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.A.; Semus, H.M.; Montgomery, R.L.; Stack, C.; Latimer, P.A.; Lewton, S.M.; Lynch, J.M.; Hullinger, T.G.; Seto, A.G.; Van Rooij, E. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur. J. Heart Fail. 2013, 15, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Lv, Q.; Ye, W.; Wong, C.K.A.; Cai, G.; Gu, D.; Ji, Y.; Zhao, C.; Wang, J.; Yang, B.B.; et al. Mirna-directed regulation of VEGF and other angiogenic under hypoxia. PLoS ONE 2006, 1, e116. [Google Scholar] [CrossRef] [PubMed]
- Triozzi, P.L.; Achberger, S.; Aldrich, W.; Singh, A.D.; Grane, R.; Borden, E.C. The association of blood angioregulatory microRNA levels with circulating endothelial cells and angiogenic proteins in patients receiving dacarbazine and interferon. J. Transl. Med. 2012, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Caporali, A.; Emanueli, C. MicroRNA-503 and the Extended MicroRNA-16 Family in Angiogenesis. Trends Cardiovasc. Med. 2011, 21, 162–166. [Google Scholar] [CrossRef]
- Tijsen, A.J.; Van der Made, I.; Van den Hoogenhof, M.M.; Wijnen, W.J.; Van Deel, E.D.; de Groot, N.E.; Alekseev, S.; Fluiter, K.; Schroen, B.; Goumans, M.-J.; et al. The microRNA-15 family inhibits the TGFβ-pathway in the heart. Cardiovasc. Res. 2014, 104, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Edelman, J.J.B.; Kao, S.C.H.; Vallely, M.P.; Van Zandwijk, N.; Reid, G. The impact of hemolysis on cell-free microRNA biomarkers. Front. Genet. 2013, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Doss, J.F.; Corcoran, D.L.; Jima, D.D.; Telen, M.J.; Dave, S.S.; Chi, J.T. A comprehensive joint analysis of the long and short RNA transcriptomes of human erythrocytes. BMC Genom. 2015, 16, 952. [Google Scholar] [CrossRef]
- Davidsen, P.K.; Gallagher, I.J.; Hartman, J.W.; Tarnopolsky, M.A.; Dela, F.; Helge, J.W.; Timmons, J.A.; Phillips, S.M. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J. Appl. Physiol. 2011, 110, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, J.; Xu, N.; Han, G.; Geng, Q.; Song, J.; Li, S.; Zhao, J.; Chen, H. Signature of circulating MicroRNAs As potential biomarkers in vulnerable coronary artery disease. PLoS ONE 2013, 8, e80738. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C.A.; Gomez, C.G.; Novak, S.M.; Mi-Mi, L.; Gregorio, C.C. Overview of the muscle cytoskeleton. Compr. Physiol. 2017, 7, 891–944. [Google Scholar] [PubMed]
- Anderson, B.R.; Granzier, H.L. Titin-based tension in the cardiac sarcomere: Molecular origin and physiological adaptations. Prog. Biophys. Mol. Biol. 2012, 110, 204–217. [Google Scholar] [CrossRef]
- Lewinter, M.M.; Granzier, H.L. Cardiac titin and heart disease. J. Cardiovasc. Pharmacol. 2014, 63, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Fassett, J.T.; Xu, X.; Kwak, D.; Wang, H.; Liu, X.; Hu, X.; Bache, R.J.; Chen, Y. Microtubule Actin Cross-Linking Factor 1 Regulates Cardiomyocyte Microtubule Distribution and Adaptation to Hemodynamic Overload. PLoS ONE 2013, 8, e73887. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.G.; Lamb, G.D.; Westerblad, H. Impaired calcium release during fatigue. J. Appl. Physiol. 2008, 104, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.; Hong, Z.; Zheng, X. Acylglycerol Kinase-Targeted Therapies in Oncology. Front. Cell Dev. Biol. 2021, 9, 1948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jones, A.; Joo, H.Y.; Zhou, D.; Cao, Y.; Chen, S.; Erdjument-Bromage, H.; Renfrow, M.; He, H.; Tempst, P.; et al. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev. 2013, 27, 1581–1595. [Google Scholar] [CrossRef] [PubMed]
- Todd, C.D.; Deniz, Ö.; Taylor, D.; Branco, M.R. Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. eLife 2019, 8, e44344. [Google Scholar] [CrossRef]
- Capomaccio, S.; Verini-Supplizi, A.; Galla, G.; Vitulo, N.; Barcaccia, G.; Felicetti, M.; Silvestrelli, M.; Cappelli, K. Transcription of LINE-derived sequences in exercise-induced stress in horses. Anim. Genet. 2010, 41, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Liu, X.; Li, P.; Akimoto, T.; Lee, S.Y.; Zhang, M.; Yan, Z. Transcriptional profiling in mouse skeletal muscle following a single bout of voluntary running: Evidence of increased cell proliferation. J. Appl. Physiol. 2005, 99, 2406–2415. [Google Scholar] [CrossRef][Green Version]
| miRNA | logFC | logCPM | FDR |
|---|---|---|---|
| eca-mir-211 | 4.9223101 | 0.1280119 | 0.0015486 |
| eca-mir-15b | 4.6239495 | 3.7887133 | 0.0027720 |
| eca-mir-451 | 2.1155572 | 9.8706133 | 0.0092029 |
| eca-mir-18a | 5.4076681 | 1.2325327 | 0.0092029 |
| eca-mir-20a | 3.2246991 | 2.7407456 | 0.0175767 |
| eca-mir-106b | 2.4277831 | 4.8328315 | 0.0213777 |
| eca-mir-101-1 | 2.0112158 | 7.7859690 | 0.0213777 |
| miRNA ID | Target 3′-UTR | Target 5′-UTR | Target CDS | Total Target |
|---|---|---|---|---|
| mir-101-3p | 315 | 43 | 322 | 680 |
| mir-106b-5p | 1442 | 263 | 1435 | 3140 |
| mir-15b-5p | 1102 | 285 | 1623 | 3010 |
| mir-18a-5p | 1550 | 356 | 2242 | 4148 |
| mir-20a-5p | 1167 | 183 | 1038 | 2388 |
| mir-211-5p | 2161 | 844 | 2626 | 5631 |
| mir-451-5p | 171 | 44 | 409 | 624 |
| Target Genes | miRNAs | # of miRNA |
|---|---|---|
| TTN | mir-101; mir-106b; mir-15b; mir-18a; mir-20a; mir-211 | 6 |
| MACF1 | mir-101; mir-106b; mir-15b; mir-18a; mir-20a | 5 |
| SYNE1 | mir-101; mir-15b; mir-18a; mir-20a; mir-211 | 5 |
| TDRD12 | mir-106b; mir-15b; mir-18a; mir-20a; mir-211 | 5 |
| USP49 | mir-101; mir-106b; mir-15b; mir-18a; mir-211 | 5 |
| ACTR8 | mir-101; mir-106b; mir-15b; mir-18a; mir-211 | 5 |
| AGK | mir-101; mir-106b; mir-15b; mir-20a; mir-211 | 5 |
| CACNA1B | mir-106b; mir-15b; mir-18a; mir-20a; mir-211 | 5 |
| COL6A5 | mir-101; mir-106b; mir-15b; mir-18a; mir-211 | 5 |
| FAM227A | mir-106b; mir-15b; mir-18a; mir-20a; mir-211 | 5 |
| HECW2 | mir-106b; mir-15b; mir-18a; mir-20a; mir-211 | 5 |
| PGM2L1 | mir-106b; mir-15b; mir-18a; mir-20a; mir-211 | 5 |
| PKD1L2 | mir-101; mir-106b; mir-18a; mir-20a; mir-211 | 5 |
| RIMBP2 | mir-106b; mir-15b; mir-18a; mir-20a; mir-211 | 5 |
| VPS13A | mir-101; mir-15b; mir-18a; mir-20a; mir-211 | 5 |
| ZBTB37 | mir-101; mir-106b; mir-18a; mir-20a; mir-211 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cappelli, K.; Mecocci, S.; Capomaccio, S.; Beccati, F.; Palumbo, A.R.; Tognoloni, A.; Pepe, M.; Chiaradia, E. Circulating Transcriptional Profile Modulation in Response to Metabolic Unbalance Due to Long-Term Exercise in Equine Athletes: A Pilot Study. Genes 2021, 12, 1965. https://doi.org/10.3390/genes12121965
Cappelli K, Mecocci S, Capomaccio S, Beccati F, Palumbo AR, Tognoloni A, Pepe M, Chiaradia E. Circulating Transcriptional Profile Modulation in Response to Metabolic Unbalance Due to Long-Term Exercise in Equine Athletes: A Pilot Study. Genes. 2021; 12(12):1965. https://doi.org/10.3390/genes12121965
Chicago/Turabian StyleCappelli, Katia, Samanta Mecocci, Stefano Capomaccio, Francesca Beccati, Andrea Rosario Palumbo, Alessia Tognoloni, Marco Pepe, and Elisabetta Chiaradia. 2021. "Circulating Transcriptional Profile Modulation in Response to Metabolic Unbalance Due to Long-Term Exercise in Equine Athletes: A Pilot Study" Genes 12, no. 12: 1965. https://doi.org/10.3390/genes12121965
APA StyleCappelli, K., Mecocci, S., Capomaccio, S., Beccati, F., Palumbo, A. R., Tognoloni, A., Pepe, M., & Chiaradia, E. (2021). Circulating Transcriptional Profile Modulation in Response to Metabolic Unbalance Due to Long-Term Exercise in Equine Athletes: A Pilot Study. Genes, 12(12), 1965. https://doi.org/10.3390/genes12121965










