Polymorphisms in Genes Involved in Osteoblast Differentiation and Function Are Associated with Anthropometric Phenotypes in Spanish Women
Abstract
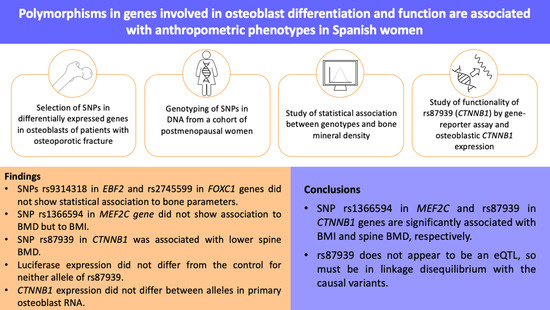
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Biochemical Assays, and Anthropometric and Bone Mineral Density (BMD) Data
2.3. Genes, Single-Nucleotide Polymorphisms (SNPs), and Genotyping
2.4. Functionality of rs87939 in CTNNB1 by Gene-Reporter Assay
2.5. Osteoblastic CTNNB1 Expression
2.6. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kanis, J.A.; Cooper, C.; Rizzoli, R.; Reginster, J.-Y.Y.; McCloskey, E.V.; Johansson, H.; Cooper, C.; Rizzoli, R.; Reginster, J.-Y.Y.; McCloskey, E.V.; et al. Executive summary of the european guidance for the diagnosis and management of osteoporosis in postmenopausal women. Calcif. Tissue Int. 2019, 104, 235–238. [Google Scholar] [CrossRef]
- Saad, F.A. Novel insights into the complex architecture of osteoporosis molecular genetics. Ann. N. Y. Acad. Sci. 2020, 1462, 37–52. [Google Scholar] [CrossRef]
- Hsu, Y.H.; Kiel, D.P. Genome-wide association studies of skeletal phenotypes: What we have learned and where we are headed. J. Clin. Endocrinol. Metab. 2012, 97, 1958–1977. [Google Scholar] [CrossRef] [Green Version]
- Morris, J.A.; Kemp, J.P.; Youlten, S.E.; Laurent, L.; Logan, J.G.; Chai, R.C.; Vulpescu, N.A.; Forgetta, V.; Kleinman, A.; Mohanty, S.T.; et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019, 51, 258–266. [Google Scholar] [CrossRef]
- Zheng, H.F.; Forgetta, V.; Hsu, Y.-H.; Estrada, K.; Rosello-Diez, A.; Leo, P.J.; Dahia, C.L.; Park-Min, K.H.; Tobias, J.H.; Kooperberg, C.; et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature 2015, 526, 112–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Daily, J.W.; Song, M.Y.; Kwon, H.-K. Gene-gene and gene-lifestyle interactions of AKAP11, KCNMA1, PUM1, SPTBN1, and EPDR1 on osteoporosis risk in middle-aged adults. Nutrition 2020, 79, 110859. [Google Scholar] [CrossRef] [PubMed]
- Costantini, A.; Skarp, S.; Kämpe, A.; Mäkitie, R.E.; Pettersson, M.; Männikkö, M.; Jiao, H.; Taylan, F.; Lindstrand, A.; Mäkitie, O. Rare copy number variants in array-based comparative genomic hybridization in early-onset skeletal fragility. Front. Endocrinol. 2018, 9, 380. [Google Scholar] [CrossRef] [PubMed]
- Pineda, B.; Serna, E.; Laguna-Fernández, A.; Noguera, I.; Panach, L.; Hermenegildo, C.; Tarín, J.J.J.; Cano, A.; García-Pérez, M.Á.; Garcia-Perez, M. Gene expression profile induced by ovariectomy in bone marrow of mice: A functional approach to identify new candidate genes associated to osteoporosis risk in women. Bone 2014, 65, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Panach, L.; Serna, E.; Tarín, J.J.; Cano, A.; García-Pérez, M.Á. A translational approach from an animal model identifies CD80 as a candidate gene for the study of bone phenotypes in postmenopausal women. Osteoporos. Int. 2017, 28, 2445–2455. [Google Scholar] [CrossRef]
- Panach, L.; Pertusa, C.; Martínez-Rojas, B.; Acebron, A.; Mifsut, D.; Tarín, J.J.; Cano, A.; Garcia-Perez, M.A. Comparative transcriptome analysus identifies CARM1 and DNMT3A as genes associated with osteoporosis. Sci. Rep. 2020, 10, 16298. [Google Scholar] [CrossRef]
- Hasani-Ranjbar, S.; Jafari-Adli, S.; Payab, M.; Qorbani, M.; Ahanjideh, F.; Keshtkar, A.; Larijani, B. Association of osteoporosis with anthropometric measures in a representative sample of iranian adults: The iranian multicenter osteoporosis study. Int. J. Prev. Med. 2019, 10, 157. [Google Scholar] [CrossRef]
- Armstrong, M.E.; Kirichek, O.; Cairns, B.J.; Green, J.; Reeves, G.K. Relationship of height to site-specific fracture risk in postmenopausal women. J. Bone Miner. Res. 2016, 31, 725–731. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Li, C.; Pan, J.; Zhang, S.; Dong, H.; Wu, Y.; Lv, J. Causal associations of anthropometric measurements with fracture risk and bone mineral density: A mendelian randomization study. J. Bone Miner. Res. 2021, 36, 1281–1287. [Google Scholar] [CrossRef]
- Pineda, B.; Pertusa, C.; Panach, L.; Tarín, J.J.; Cano, A.; García-Pérez, M.Á. Polymorphisms in genes involved in T-cell co-stimulation are associated with blood pressure in women. Gene 2020, 754, 144838. [Google Scholar] [CrossRef] [PubMed]
- Zolfaroli, I.; Ortiz, E.; García-Pérez, M.Á.; Hidalgo-Mora, J.J.; Tarín, J.J.; Cano, A. Positive association of high-density lipoprotein cholesterol with lumbar and femoral neck bone mineral density in postmenopausal women. Maturitas 2021, 147, 41–46. [Google Scholar] [CrossRef]
- Wein, M.N.; Spatz, J.; Nishimori, S.; Doench, J.; Root, D.; Babij, P.; Nagano, K.; Baron, R.; Brooks, D.; Bouxsein, M.; et al. HDAC5 controls MEF2C-driven sclerostin expression in osteocytes. J. Bone Miner. Res. 2015, 30, 400–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieslinger, M.; Folberth, S.; Dobreva, G.; Dorn, T.; Croci, L.; Erben, R.; Consalez, G.G.; Grosschedl, R. EBF2 regulates osteoblast-dependent differentiation of osteoclasts. Dev. Cell 2005, 9, 757–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirzayans, F.; Lavy, R.; Penner-Chea, J.; Berry, F.B. Initiation of early osteoblast differentiation events through the direct transcriptional regulation of Msx2 by FOXC1. PLoS ONE 2012, 7, e49095. [Google Scholar] [CrossRef]
- Day, T.F.; Guo, X.; Garrett-Beal, L.; Yang, Y. Wnt/Beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell 2005, 8, 739–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivadeneira, F.; Styrkársdottir, U.; Estrada, K.; Halldórsson, B.V.; Hsu, Y.-H.; Richards, J.B.; Zillikens, M.C.; Kavvoura, F.K.; Amin, N.; Aulchenko, Y.S.; et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet. 2009, 41, 1199–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.-F.; Duncan, E.L.; Yerges-Armstrong, L.M.; Eriksson, J.; Bergström, U.; Leo, P.J.; Leslie, W.D.; Goltzman, D.; Blangero, J.; Hanley, D.A.; et al. Meta-analysis of genome-wide studies identifies MEF2C SNPs associated with bone mineral density at forearm. J. Med. Genet. 2013, 50, 473–478. [Google Scholar] [CrossRef] [Green Version]
- Panach, L.; Mifsut, D.; Tarín, J.J.; Cano, A.; García-Pérez, M.Á. Replication study of three functional polymorphisms associated with bone mineral density in a cohort of spanish women. J. Bone Miner. Metab. 2014, 32, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Sole, X.; Guino, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Schwarz, J.; Bucana, C.; Olson, E.N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 1997, 276, 1404–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heard-Costa, N.L.; Carola Zillikens, M.; Monda, K.L.; Johansson, Å.; Harris, T.B.; Fu, M.; Haritunians, T.; Feitosa, M.F.; Aspelund, T.; Eiriksdottir, G.; et al. NRXN3 is a novel locus for waist circumference: A genome-wide association study from the CHARGE consortium. PLoS Genet. 2009, 5, e1000539. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Chromosome | Gene | Fold Change 1 | SNP | Position (GRCh38.p12) | Location | Major Allele | Minor Allele | MAF | p-HWE 2 |
|---|---|---|---|---|---|---|---|---|---|
| 3 | CTNNB1 | 1.3 | rs87939 | 41096353 | Intergenic | A | G | 0.47 | 0.70 |
| 8 | EBF2 | −1.79 | rs9314318 | 26046872 | 2 KB Upstream Variant | G | A | 0.25 | 0.21 |
| 6 | FOXC1 | 1.64 | rs2745599 | 1613451 | 3′UTR Variant | A | G | 0.45 | 0.14 |
| 5 | MEF2C | 1.55 | rs1366594 | 89080244 | Intronic | A | C | 0.49 | 0.95 |
| Values | |
|---|---|
| Age (y) | 55.7 ± 8.7 |
| Weight (kg) | 66.5 ± 10.3 |
| Height (cm) | 157.5 ± 6.0 |
| BMI (kg/m2) | 26.8 ± 4.1 |
| Waist circumference (cm) | 85.9 ± 10.4 |
| Hip circumference (cm) | 102.4 ± 8.2 |
| Postmenopausal women (%) | 97.3 |
| Antiresorptive therapy user (%) | 25.3 |
| FN-sBMD (g/cm2) | 0.795 ± 0.117 |
| FN T-score | −1.006 ± 0.994 |
| FN Z-score | −0.081 ± 0.926 |
| LS-sBMD (g/cm2) | 0.989 ± 0.150 |
| LS T-score | −1.207 ± 1.331 |
| LS Z-score | −1.121 ± 1.227 |
| Gene SNP (rs) | Genotype (N) | FN-sBMD | Genotype (N) | LS-sBMD | ||
|---|---|---|---|---|---|---|
| Unadjusted | Adjusted | Unadjusted (N) | Adjusted (N) | |||
| CTNNB1 rs87939 | A/A (284) | 0.807 ± 0.123 | 0.807 ± 0.007 | A/A (284) | 1.014 ± 0.162 | 1.049 ± 0.017 |
| A/G–G/G (716) | 0.791 ± 0.114 | 0.789 ± 0.004 | A/G–G/G (716) | 0.979 ± 0.144 | 1.015 ± 0.015 | |
| p-value | 0.047 | 0.020 | p-value | 0.0009 | 0.0017 | |
| EBF2 rs9314318 | A/A (55) | 0.823 ± 0.108 | 0.813 ± 0.016 | A/G (391) | 0.996 ± 0.150 | 1.034 ± 0.016 |
| A/G–G/G (939) | 0.794 ± 0.117 | 0.793 ± 0.004 | A/A–G/G (603) | 0.984 ± 0.150 | 1.017 ± 0.015 | |
| p-value | 0.073 | 0.210 | p-value | 0.211 | 0.079 | |
| FOXC1 rs2745599 | G/G (218) | 0.808 ± 0.113 | 0.805 ± 0.008 | G/G (218) | 1.009 ± 0.149 | 1.025 ± 0.019 |
| A/G–A/A (779) | 0.792 ± 0.118 | 0.791 ± 0.004 | A/G–A/A (779) | 0.983 ± 0.149 | 1.013 ± 0.016 | |
| p-value | 0.066 | 0.114 | p-value | 0.023 | 0.407 | |
| MEF2C rs1366594 | A/A (260) | 0.808 ± 0.127 | 0.876 ± 0.028 | A/A (260) | 0.998 ± 0.153 | 1.103 ± 0.037 |
| A/C–C/C (729) | 0.791 ± 0.113 | 0.798 ± 0.012 | A/C–C/C (729) | 0.986 ± 0.149 | 1.009 ± 0.016 | |
| p-value | 0.042 | 0.009 | p-value | 0.252 | 0.020 | |
| Gene SNP (rs) | Genotype (N) | Waist Circumference | Hip Circumference | BMI | |||
|---|---|---|---|---|---|---|---|
| Unadjusted | Adjusted | Unadjusted | Adjusted | Unadjusted | Adjusted | ||
| MEF2C rs1366594 | A/A (260) | 84.1 ± 9.7 | 85.6 ± 0.4 | 101.0 ± 8.0 | 102.1 ± 0.3 | 26.1 ± 3.9 | 26.1 ± 0.2 |
| A/C–C/C (729) | 86.5 ± 10.6 | 85.9 ± 0.2 | 102.9 ± 8.3 | 102.5 ± 0.2 | 27.1 ± 4.1 | 27.1 ± 0.1 | |
| p-value | 0.0018 | 0.386 | 0.0014 | 0.386 | 0.0011 | 0.0010 | |
| rs87939 (CTNNB1) | p | rs1366594 (MEF2C) | p | |||
|---|---|---|---|---|---|---|
| AA (N = 284) | AG/GG (N = 716) | AA (N = 260) | AC/CC (N = 729) | |||
| Age (y) | 55.7 ± 9.0 | 55.6 ± 8.5 | 0.915 | 55.7 ± 9.1 | 55.7 ± 8.5 | 0.998 |
| BMI (kg/m2) | 27.0 ± 4.2 | 26.8 ± 4.1 | 0.495 | 26.1 ± 3.9 | 27.1 ± 4.1 | 0.0011 |
| FN-BMD (g/cm2) | 0.807 ± 0.124 | 0.791 ± 0.114 | 0.047 | 0.808 ± 0.127 | 0.791 ± 0.113 | 0.042 |
| FN Z-score | 0.040 ± 0.946 | −0.130 ± 0.914 | 0.009 | 0.038 ± 0.953 | −0.121 ± 0.916 | 0.018 |
| LS-BMD (g/cm2) | 1.014 ± 0.162 | 0.979 ± 0.144 | 0.0009 | 0.998 ± 0.153 | 0.986 ± 0.149 | 0.252 |
| LS Z-score | 0.094 ± 1.279 | −0.209 ± 1.193 | 0.0004 | −0.023 ± 1.250 | −0.153 ± 1.219 | 0.142 |
| Estradiol (pg/mL) | 16.5 ± 25.2 | 14.7 ± 8.4 | 0.089 | 17.1 ± 26.2 | 14.5 ± 8.4 | 0.021 |
| FSH (U/mL) | 69.7 ± 29.1 | 73.8 ± 27.1 | 0.039 | 71.3 ± 30.7 | 73.0 ± 26.8 | 0.410 |
| 25(OH)D3 (ng/mL) | 26.1 ± 9.4 | 25.6 ± 9.3 | 0.479 | 25.7 ± 8.5 | 25.8 ± 9.6 | 0.877 |
| CTx (ng/mL) | 0.436 ± 0.205 | 0.439 ± 0.190 | 0.793 | 0.448 ± 0.203 | 0.434 ± 0.192 | 0.346 |
| Total ALP (U/L) | 168.9 ± 52.0 | 165.8 ± 46.3 | 0.364 | 167.7 ± 48.5 | 166.6 ± 47.7 | 0.755 |
| Calcium (mg/dL) | 9.6 ± 0.4 | 9.6 ± 0.4 | 0.326 | 9.6 ± 0.4 | 9.6 ± 0.4 | 0.475 |
| Phosphate (mg/dL) | 3.6 ± 0.5 | 3.6 ± 0.5 | 0.722 | 3.6 ± 0.5 | 3.6 ± 0.5 | 0.425 |
| Triglycerides (mg/dL) | 102.9 ± 39.6 | 100.7 ± 39.4 | 0.430 | 99.7 ± 37.3 | 101.7 ± 40.4 | 0.496 |
| Total cholesterol (mg/dL) | 214.8 ± 34.0 | 215.8 ± 35.1 | 0.691 | 216.4 ± 33.7 | 215.1 ± 35.3 | 0.613 |
| HDL-cholesterol (mg/dL) | 64.3 ± 14.0 | 64.2 ± 14.1 | 0.946 | 64.5 ± 14.0 | 64.2 ± 14.2 | 0.747 |
| LDL-cholesterol (mg/dL) | 129.9 ± 29.1 | 131.8 ± 29.1 | 0.330 | 132.2 ± 27.3 | 130.8 ± 29.8 | 0.502 |
| Glucose (mg/dL) | 100.9 ± 11.7 | 99.9 ± 11.5 | 0.209 | 99.5 ± 10.7 | 100.3 ± 11.9 | 0.304 |
| Insulin (μU/mL) | 8.4 ± 4.7 | 8.5 ± 5.2 | 0.762 | 8.2 ± 4.2 | 8.5 ± 5.4 | 0.359 |
| HOMA-IR index | 2.1 ± 1.4 | 2.2 ± 1.6 | 0.878 | 2.1 ± 1.2 | 2.2 ± 1.6 | 0.279 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pertusa, C.; Ruzo, S.P.; Panach, L.; Mifsut, D.; Tarín, J.J.; Cano, A.; García-Pérez, M.Á. Polymorphisms in Genes Involved in Osteoblast Differentiation and Function Are Associated with Anthropometric Phenotypes in Spanish Women. Genes 2021, 12, 2012. https://doi.org/10.3390/genes12122012
Pertusa C, Ruzo SP, Panach L, Mifsut D, Tarín JJ, Cano A, García-Pérez MÁ. Polymorphisms in Genes Involved in Osteoblast Differentiation and Function Are Associated with Anthropometric Phenotypes in Spanish Women. Genes. 2021; 12(12):2012. https://doi.org/10.3390/genes12122012
Chicago/Turabian StylePertusa, Clara, Sofía P. Ruzo, Layla Panach, Damián Mifsut, Juan J. Tarín, Antonio Cano, and Miguel Ángel García-Pérez. 2021. "Polymorphisms in Genes Involved in Osteoblast Differentiation and Function Are Associated with Anthropometric Phenotypes in Spanish Women" Genes 12, no. 12: 2012. https://doi.org/10.3390/genes12122012
APA StylePertusa, C., Ruzo, S. P., Panach, L., Mifsut, D., Tarín, J. J., Cano, A., & García-Pérez, M. Á. (2021). Polymorphisms in Genes Involved in Osteoblast Differentiation and Function Are Associated with Anthropometric Phenotypes in Spanish Women. Genes, 12(12), 2012. https://doi.org/10.3390/genes12122012