The Tomato Interspecific NB-LRR Gene Arsenal and Its Impact on Breeding Strategies
Abstract
:1. Introduction
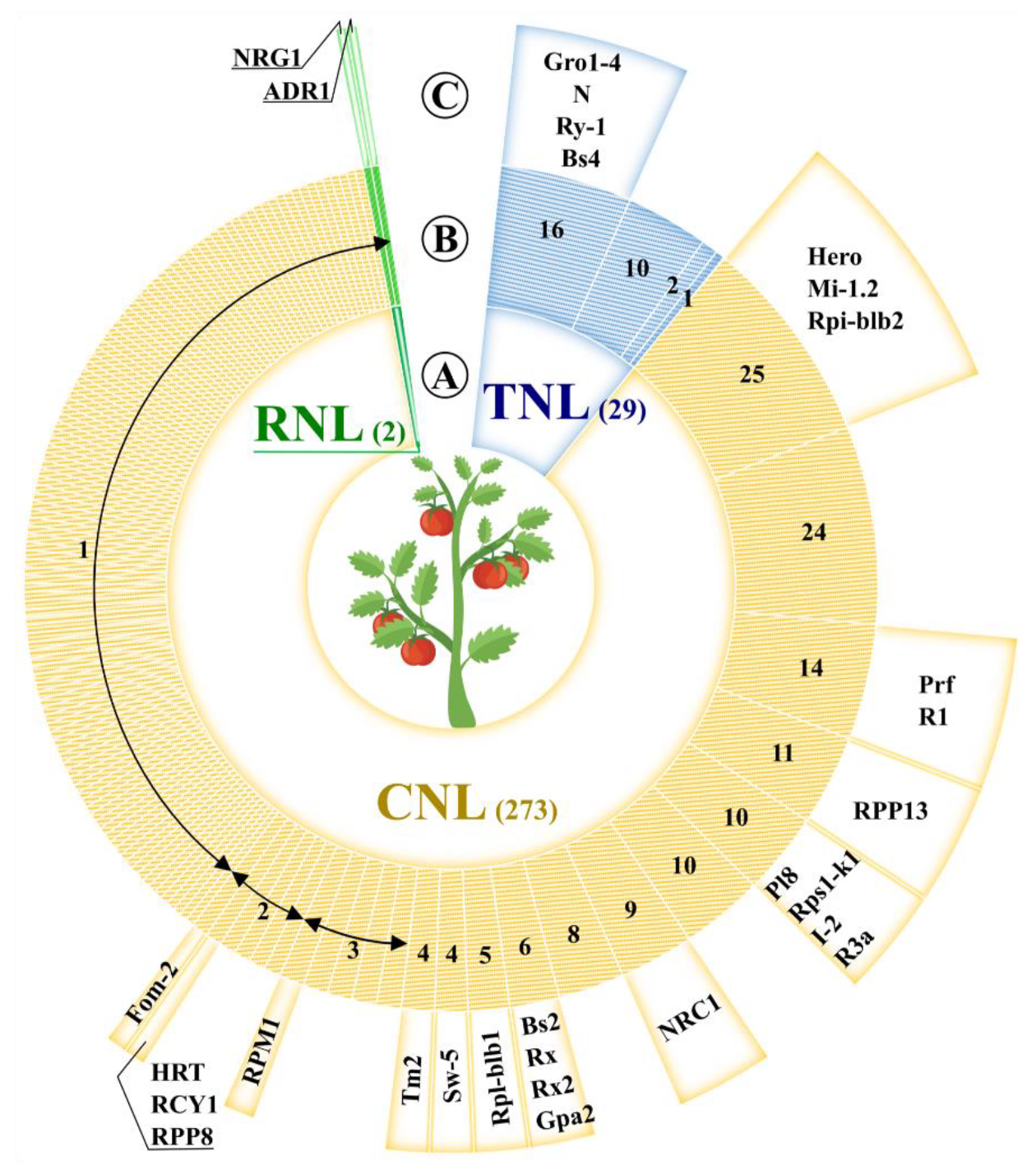
2. The Genome-Wide Arrangement of Tomato NB-LRR Genes
3. Resistance Sources in Wild Tomato Relatives
4. Evolution of R-Type Defense Genes within Solanaceae
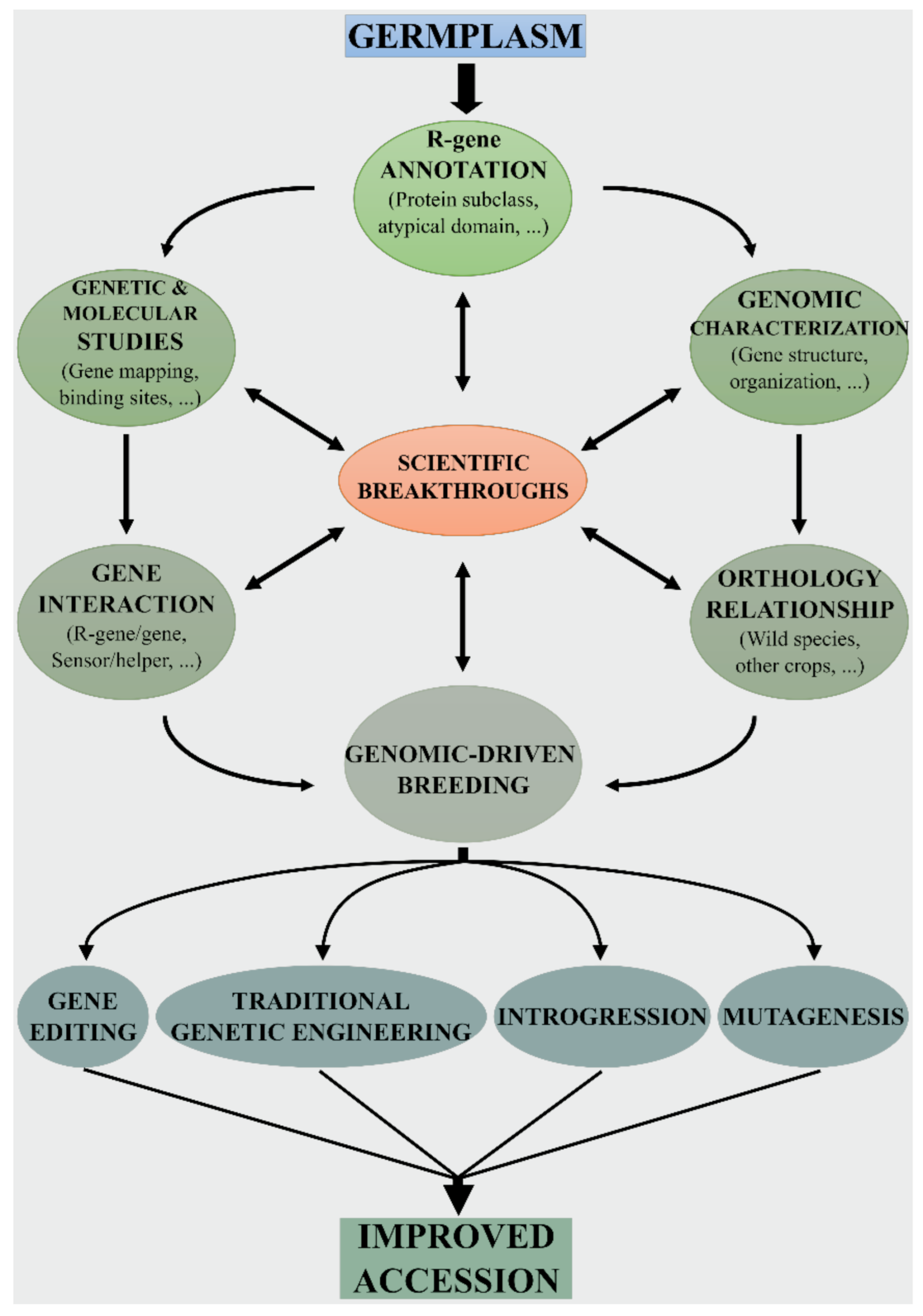
5. Genomic-Driven Breeding for Developing New Resistant Tomato Varieties
6. Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Peralta, I.E.; Spooner, D.M.; Knapp, S. Taxonomy of wild tomatoes and their relatives (Solanum sect. Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon; Solanaceae). Syst. Bot. Monogr. 2008, 84, 1–186. [Google Scholar]
- Arie, T.; Takahashi, H.; Kodama, M.; Teraoka, T. Tomato as a model plant for plant-pathogen interactions. Plant Biotechnol. 2007, 24, 135–147. [Google Scholar] [CrossRef] [Green Version]
- Andolfo, G.; Jupe, F.; Witek, K.; Etherington, G.J.; Ercolano, M.R.; Jones, J.D.G. Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biol. 2014, 14, 120. [Google Scholar] [CrossRef] [Green Version]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Andolfo, G.; Sanseverino, W.; Rombauts, S.; Van de Peer, Y.; Bradeen, J.M.; Carputo, D.; Frusciante, L.; Ercolano, M.R. Overview of tomato (Solanum lycopersicum) candidate pathogen recognition genes reveals important Solanum R locus dynamics. New Phytol. 2013, 197, 223–237. [Google Scholar] [CrossRef]
- Andolfo, G.; Sanseverino, W.; Aversano, R.; Frusciante, L.; Ercolano, M.R. Genome-wide identification and analysis of candidate genes for disease resistance in tomato. Mol. Breed. 2014, 33, 227–233. [Google Scholar] [CrossRef] [Green Version]
- Pease, J.B.; Haak, D.C.; Hahn, M.W.; Moyle, L.C. Phylogenomics Reveals Three Sources of Adaptive Variation during a Rapid Radiation. PLoS Biol. 2016, 14, e1002379. [Google Scholar] [CrossRef]
- Razifard, H.; Ramos, A.; Della Valle, A.L.; Bodary, C.; Goetz, E.; Manser, E.J.; Li, X.; Zhang, L.; Visa, S.; Tieman, D.; et al. Genomic Evidence for Complex Domestication History of the Cultivated Tomato in Latin America. Mol. Biol. Evol. 2020, 37, 1118–1132. [Google Scholar] [CrossRef]
- Tamburino, R.; Sannino, L.; Cafasso, D.; Cantarella, C.; Orrù, L.; Cardi, T.; Cozzolino, S.; D’Agostino, N.; Scotti, N. Cultivated Tomato (Solanum lycopersicum L.) Suffered a Severe Cytoplasmic Bottleneck during Domestication: Implications from Chloroplast Genomes. Plants 2020, 9, 1443. [Google Scholar] [CrossRef]
- Stam, R.; Silva-Arias, G.A.; Tellier, A. Subsets of NLR genes show differential signatures of adaptation during colonization of new habitats. New Phytol. 2019, 224, 367–379. [Google Scholar] [CrossRef]
- Barchi, L.; Pietrella, M.; Venturini, L.; Minio, A.; Toppino, L.; Acquadro, A.; Andolfo, G.; Aprea, G.; Avanzato, C.; Bassolino, L.; et al. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019, 9, 11769. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, A.; Andolfo, G.; Ferrarini, A.; Delledonne, M.; Ercolano, M.R.; Crease, T. Investigation of orthologous pathogen recognition gene-rich regions in solanaceous species. Genome 2017, 60, 850–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.-Z. Origin and evolution of the plant immune system. New Phytol. 2019, 222, 70–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vakhrusheva, O.A.; Nedospasov, S.A. System of innate immunity in plants. Mol. Biol. 2011, 45, 16–23. [Google Scholar] [CrossRef]
- Andolfo, G.; Ercolano, M.R. Plant Innate Immunity Multicomponent Model. Front. Plant Sci. 2015, 6, 987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andolfo, G.; Di Donato, A.; Chiaiese, P.; De Natale, A.; Pollio, A.; Jones, J.D.G.; Frusciante, L.; Ercolano, M.R. Alien Domains Shaped the Modular Structure of Plant NLR Proteins. Genome Biol. Evol. 2019, 11, 3466–3477. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Christopoulou, M.; Caldwell, K.S. Impacts of Resistance Gene Genetics, Function, and Evolution on a Durable Future. Annu. Rev. Phytopathol. 2013, 51, 291–319. [Google Scholar] [CrossRef]
- Munch, D.; Teh, O.-K.; Malinovsky, F.G.; Liu, Q.; Vetukuri, R.R.; El Kasmi, F.; Brodersen, P.; Hara-Nishimura, I.; Dangl, J.L.; Petersen, M.; et al. Retromer Contributes to Immunity-Associated Cell Death in Arabidopsis. Plant Cell 2015, 27, 463. [Google Scholar] [CrossRef]
- Lee, H.-A.; Yeom, S.-I. Plant NB-LRR proteins: Tightly regulated sensors in a complex manner. Brief. Funct. Genomics 2015, 14, 233–242. [Google Scholar] [CrossRef] [Green Version]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-Wide Analysis of NBS-LRR—Encoding Genes in Arabidopsis. Plant Cell 2003, 15, 809. [Google Scholar] [CrossRef] [Green Version]
- Jubic, L.M.; Saile, S.; Furzer, O.J.; El Kasmi, F.; Dangl, J.L. Help wanted: Helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 2019, 50, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Belhaj, K.; Bozkurt, T.O.; Birk, M.S.; Kamoun, S. Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for Pto-mediated cell death and resistance in Nicotiana benthamiana. New Phytol. 2016, 209, 1344–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.-H.; Derevnina, L.; Kamoun, S. Receptor networks underpin plant immunity. Science 2018, 360, 1300. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113. [Google Scholar] [CrossRef] [Green Version]
- Andolfo, G.; Villano, C.; Errico, A.; Frusciante, L.; Carputo, D.; Aversano, R.; Ercolano, M.R. Inferring RPW8-NLRs’s evolution patterns in seed plants: Case study in Vitis vinifera. Planta 2019, 251, 32. [Google Scholar] [CrossRef]
- Chandraprakash, M.; Thomas, R. Genome wide identification of nucleotide-binding site leucine-rich repeat (NBS-LRR) gene encoding regions in Solanum lycopersicum resistant to environmental pathogens by computational methods. J. Environ. Biol. 2019, 40, 613–618. [Google Scholar] [CrossRef]
- Peart, J.R.; Mestre, P.; Lu, R.; Malcuit, I.; Baulcombe, D.C. NRG1, a CC-NB-LRR Protein, together with N, a TIR-NB-LRR Protein, Mediates Resistance against Tobacco Mosaic Virus. Curr. Biol. 2005, 15, 968–973. [Google Scholar] [CrossRef] [Green Version]
- Bonardi, V.; Tang, S.; Stallmann, A.; Roberts, M.; Cherkis, K.; Dangl, J.L. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA 2011, 108, 16463. [Google Scholar] [CrossRef] [Green Version]
- Marone, D.; Russo, M.A.; Laidò, G.; De Leonardis, A.M.; Mastrangelo, A.M. Plant Nucleotide Binding Site—Leucine-Rich Repeat (NBS-LRR) Genes: Active Guardians in Host Defense Responses. Int. J. Mol. Sci. 2013, 14. [Google Scholar] [CrossRef] [Green Version]
- Park, T.-H.; Gros, J.; Sikkema, A.; Vleeshouwers, V.G.A.A.; Muskens, M.; Allefs, S.; Jacobsen, E.; Visser, R.G.F.; van der Vossen, E.A.G. The Late Blight Resistance Locus Rpi-blb3 from Solanum bulbocastanum Belongs to a Major Late Blight R Gene Cluster on Chromosome 4 of Potato. Mol. Plant-Microbe Interact. 2005, 18, 722–729. [Google Scholar] [CrossRef] [Green Version]
- Osuna-Cruz, C.M.; Paytuvi-Gallart, A.; Di Donato, A.; Sundesha, V.; Andolfo, G.; Cigliano, R.A.; Sanseverino, W.; Ercolano, M.R. PRGdb 3.0: A comprehensive platform for prediction and analysis of plant disease resistance genes. Nucleic Acids Res. 2018, 46, D1197–D1201. [Google Scholar] [CrossRef] [PubMed]
- Lanfermeijer, F.C.; Jiang, G.; Ferwerda, M.A.; Dijkhuis, J.; de Haan, P.; Yang, R.; Hille, J. The durable resistance gene Tm-22 from tomato confers resistance against ToMV in tobacco and preserves its viral specificity. Plant Sci. 2004, 167, 687–692. [Google Scholar] [CrossRef]
- Lin, N.-C.; Martin, G.B. Pto- and Prf-Mediated Recognition of AvrPto and AvrPtoB Restricts the Ability of Diverse Pseudomonas syringae Pathovars to Infect Tomato. Mol. Plant-Microbe Interact. 2007, 20, 806–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulbert, S.H.; Webb, C.A.; Smith, S.M.; Sun, Q. Resitence Gene Complexes: Evolution and Utilization. Annu. Rev. Phytopathol. 2001, 39, 285–312. [Google Scholar] [CrossRef]
- Nieri, D.; Di Donato, A.; Ercolano, M.R. Analysis of tomato meiotic recombination profile reveals preferential chromosome positions for NB-LRR genes. Euphytica 2017, 213, 206. [Google Scholar] [CrossRef]
- Rossi, M.; Goggin, F.L.; Milligan, S.B.; Kaloshian, I.; Ullman, D.E.; Williamson, V.M. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 1998, 95, 9750–9754. [Google Scholar] [CrossRef] [Green Version]
- Simons, G.; Groenendijk, J.; Wijbrandi, J.; Reijans, M.; Groenen, J.; Diergaarde, P.; Van der Lee, T.; Bleeker, M.; Onstenk, J.; de Both, M.; et al. Dissection of the Fusarium I2 Gene Cluster in Tomato Reveals Six Homologs and One Active Gene Copy. Plant Cell 1998, 10, 1055. [Google Scholar] [CrossRef] [Green Version]
- Ballvora, A.; Pierre, M.; van den Ackerveken, G.; Schornack, S.; Rossier, O.; Ganal, M.; Lahaye, T.; Bonas, U. Genetic Mapping and Functional Analysis of the Tomato Bs4 Locus Governing Recognition of the Xanthomonas campestris pv. vesicatoria AvrBs4 Protein. Mol. Plant-Microbe Interact. 2001, 14, 629–638. [Google Scholar] [CrossRef] [Green Version]
- Ernst, K.; Kumar, A.; Kriseleit, D.; Kloos, D.-U.; Phillips, M.S.; Ganal, M.W. The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. Plant J. 2002, 31, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Mahfouze, S.; Saxena, S.; Mahfouze, H.; Rajam, M. Characterization of Mi1.2 whitefly (Bemisia tabaci) resistance gene. Online J. Biol. Sci. 2017, 17, 323–334. [Google Scholar] [CrossRef]
- Jablonska, B.; Ammiraju, J.S.S.; Bhattarai, K.K.; Mantelin, S.; de Ilarduya, O.M.; Roberts, P.A.; Kaloshian, I. The Mi-9 Gene from Solanum arcanum Conferring Heat-Stable Resistance to Root-Knot Nematodes Is a Homolog of Mi-1. Plant Physiol. 2007, 143, 1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmeron, J.M.; Oldroyd, G.E.D.; Rommens, C.M.T.; Scofield, S.R.; Kim, H.-S.; Lavelle, D.T.; Dahlbeck, D.; Staskawicz, B.J. Tomato Prf Is a Member of the Leucine-Rich Repeat Class of Plant Disease Resistance Genes and Lies Embedded within the Pto Kinase Gene Cluster. Cell 1996, 86, 123–133. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Liu, L.; Wang, X.; Vossen, J.; Li, G.; Li, T.; Zheng, Z.; Gao, J.; Guo, Y.; Visser, R.G.F.; et al. The Ph-3 gene from Solanum pimpinellifolium encodes CC-NBS-LRR protein conferring resistance to Phytophthora infestans. Theor. Appl. Genet. 2014, 127, 1353–1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brommonschenkel, S.H.; Frary, A.; Frary, A.; Tanksley, S.D. The Broad-Spectrum Tospovirus Resistance Gene Sw-5 of Tomato Is a Homolog of the Root-Knot Nematode Resistance Gene Mi. Mol. Plant-Microbe Interact. 2000, 13, 1130–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishibashi, K.; Masuda, K.; Naito, S.; Meshi, T.; Ishikawa, M. An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc. Natl. Acad. Sci. USA 2007, 104, 13833. [Google Scholar] [CrossRef] [Green Version]
- Lanfermeijer, F.C.; Warmink, J.; Hille, J. The products of the broken Tm-2 and the durable Tm-22 resistance genes from tomato differ in four amino acids. J. Exp. Bot. 2005, 56, 2925–2933. [Google Scholar] [CrossRef] [Green Version]
- Lanfermeijer, F.C.; Dijkhuis, J.; Sturre, M.J.G.; de Haan, P.; Hille, J. Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-22 from Lycopersicon esculentum. Plant Mol. Biol. 2003, 52, 1039–1051. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Caro, M.; Hutton, S.F.; Scott, J.W.; Guo, Y.; Wang, X.; Rashid, M.H.; Szinay, D.; de Jong, H.; Visser, R.G.F.; et al. Fine mapping of the tomato yellow leaf curl virus resistance gene Ty-2 on chromosome 11 of tomato. Mol. Breed. 2014, 34, 749–760. [Google Scholar] [CrossRef] [Green Version]
- Stam, R.; Nosenko, T.; Hörger, A.C.; Stephan, W.; Seidel, M.; Kuhn, J.M.M.; Haberer, G.; Tellier, A. The de Novo Reference Genome and Transcriptome Assemblies of the Wild Tomato Species Solanum chilense Highlights Birth and Death of NLR Genes Between Tomato Species. G3 Genes | Genomes | Genet. 2019, 9, 3933. [Google Scholar] [CrossRef] [Green Version]
- Stam, R.; Scheikl, D.; Tellier, A. Pooled Enrichment Sequencing Identifies Diversity and Evolutionary Pressures at NLR Resistance Genes within a Wild Tomato Population. Genome Biol. Evol. 2016, 8, 1501–1515. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Liu, J.; Guo, Q.; Pan, L.; Chai, S.; Cheng, Y.; Ruan, M.; Ye, Q.; Wang, R.; Yao, Z. Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana. Evol. Bioinform. 2020, 16, 1176934320911055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melito, S.; Sanseverino, W.; Carli, P.; Monti, L.; Frusciante, L.; Ercolano Maria, R. Solanaceae Evolutionary Dynamics of the I2-NBS Domain. Am. J. Plant Sci. 2012, 3, 283–294. [Google Scholar]
- Hörger, A.C.; Ilyas, M.; Stephan, W.; Tellier, A.; van der Hoorn, R.A.L.; Rose, L.E. Balancing Selection at the Tomato RCR3 Guardee Gene Family Maintains Variation in Strength of Pathogen Defense. PLoS Genet. 2012, 8, e1002813. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, M.; Yamamoto-Katou, A.; Katou, S.; Hirai, K.; Meshi, T.; Ohashi, Y.; Mitsuhara, I. Identification of an amino acid residue required for differential recognition of a viral movement protein by the Tomato mosaic virus resistance gene Tm-22. J. Plant Physiol. 2011, 168, 1142–1145. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.; Seo, E.; Witek, K.; Li, M.; Staskawicz, B. Evolution of NLR resistance genes with noncanonical N-terminal domains in wild tomato species. New Phytol. 2020, 227, 1530–1543. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.-H.; Zhou, G.-C.; Sun, X.-Q.; Lei, Z.; Zhang, Y.-M.; Xue, J.-Y.; Hang, Y.-Y. Distinct Patterns of Gene Gain and Loss: Diverse Evolutionary Modes of NBS-Encoding Genes in Three Solanaceae Crop Species. G3 Genes | Genomes | Genet. 2017, 7, 1577. [Google Scholar] [CrossRef] [Green Version]
- Destefanis, M.; Nagy, I.; Rigney, B.; Bryan, G.J.; McLean, K.; Hein, I.; Griffin, D.; Milbourne, D. A disease resistance locus on potato and tomato chromosome 4 exhibits a conserved multipartite structure displaying different rates of evolution in different lineages. BMC Plant Biol. 2015, 15, 255. [Google Scholar] [CrossRef] [Green Version]
- Grube, R.C.; Blauth, J.R.; Andrés, M.S.A.; Caranta, C.; Jahn, M.K. Identification and comparative mapping of a dominant potyvirus resistance gene cluster in Capsicum. Theor. Appl. Genet. 2000, 101, 852–859. [Google Scholar] [CrossRef]
- Djian-Caporalino, C.; Fazari, A.; Arguel, M.J.; Vernie, T.; VandeCasteele, C.; Faure, I.; Brunoud, G.; Pijarowski, L.; Palloix, A.; Lefebvre, V.; et al. Root-knot nematode (Meloidogyne spp.) Me resistance genes in pepper (Capsicum annuum L.) are clustered on the P9 chromosome. Theor. Appl. Genet. 2007, 114, 473–486. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Park, J.; Yeom, S.-I.; Kim, Y.-M.; Seo, E.; Kim, K.-T.; Kim, M.-S.; Lee, J.M.; Cheong, K.; Shin, H.-S.; et al. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 2017, 18, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aversano, R.; Contaldi, F.; Ercolano, M.R.; Grosso, V.; Iorizzo, M.; Tatino, F.; Xumerle, L.; Dal Molin, A.; Avanzato, C.; Ferrarini, A.; et al. The Solanum commersonii Genome Sequence Provides Insights into Adaptation to Stress Conditions and Genome Evolution of Wild Potato Relatives. Plant Cell 2015, 27, 954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Z.-Q.; Xue, J.-Y.; Wu, P.; Zhang, Y.-M.; Wu, Y.; Hang, Y.-Y.; Wang, B.; Chen, J.-Q. Large-Scale Analyses of Angiosperm Nucleotide-Binding Site-Leucine-Rich Repeat Genes Reveal Three Anciently Diverged Classes with Distinct Evolutionary Patterns. Plant Physiol. 2016, 170, 2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catanzariti, A.-M.; Do, H.T.T.; Bru, P.; de Sain, M.; Thatcher, L.F.; Rep, M.; Jones, D.A. The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J. 2017, 89, 1195–1209. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, H.; Ohnishi, J.; Saito, A.; Ohyama, A.; Nunome, T.; Miyatake, K.; Fukuoka, H. An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 2018, 131, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Ercolano, M.R.; Sanseverino, W.; Carli, P.; Ferriello, F.; Frusciante, L. Genetic and genomic approaches for R-gene mediated disease resistance in tomato: Retrospects and prospects. Plant Cell Rep. 2012, 31, 973–985. [Google Scholar] [CrossRef] [Green Version]
- Faino, L.; Carli, P.; Testa, A.; Cristinzio, G.; Frusciante, L.; Ercolano, M.R. Potato R1 resistance gene confers resistance against Phytophthora infestans in transgenic tomato plants. Eur. J. Plant Pathol. 2010, 128, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Tai, T.H.; Dahlbeck, D.; Clark, E.T.; Gajiwala, P.; Pasion, R.; Whalen, M.C.; Stall, R.E.; Staskawicz, B.J. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 1999, 96, 14153. [Google Scholar] [CrossRef] [Green Version]
- Whitham, S.; McCormick, S.; Baker, B. The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proc. Natl. Acad. Sci. USA 1996, 93, 8776. [Google Scholar] [CrossRef] [Green Version]
- Prohens, J.; Gramazio, P.; Plazas, M.; Dempewolf, H.; Kilian, B.; Díez, M.J.; Fita, A.; Herraiz, F.J.; Rodríguez-Burruezo, A.; Soler, S. Introgressiomics: A new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica 2017, 213, 158. [Google Scholar] [CrossRef]
- Schouten, H.J.; Tikunov, Y.; Verkerke, W.; Finkers, R.; Bovy, A.; Bai, Y.; Visser, R.G.F. Breeding Has Increased the Diversity of Cultivated Tomato in The Netherlands. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andolfo, G.; Iovieno, P.; Frusciante, L.; Ercolano, M.R. Genome-Editing Technologies for Enhancing Plant Disease Resistance. Front. Plant Sci. 2016, 7, 1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharat, S.S.; Li, S.; Li, J.; Yan, L.; Xia, L. Base editing in plants: Current status and challenges. Crop J. 2020, 8, 384–395. [Google Scholar] [CrossRef]
- Giannakopoulou, A.; Steele, J.F.C.; Segretin, M.E.; Bozkurt, T.O.; Zhou, J.; Robatzek, S.; Banfield, M.J.; Pais, M.; Kamoun, S. Tomato I2 Immune Receptor Can Be Engineered to Confer Partial Resistance to the Oomycete Phytophthora infestans in Addition to the Fungus Fusarium oxysporum. Mol. Plant-Microbe Interact. 2015, 28, 1316–1329. [Google Scholar] [CrossRef] [Green Version]
- Segretin, M.E.; Pais, M.; Franceschetti, M.; Chaparro-Garcia, A.; Bos, J.I.B.; Banfield, M.J.; Kamoun, S. Single Amino Acid Mutations in the Potato Immune Receptor R3a Expand Response to Phytophthora Effectors. Mol. Plant-Microbe Interact. 2014, 27, 624–637. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Song, G.; Lal, N.K.; Nagalakshmi, U.; Li, Y.; Zheng, W.; Huang, P.-j.; Branon, T.C.; Ting, A.Y.; Walley, J.W.; et al. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat. Commun. 2019, 10, 3252. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xia, R.; Kuang, H.; Meyers, B.C. The Diversification of Plant NBS-LRR Defense Genes Directs the Evolution of MicroRNAs That Target Them. Mol. Biol. Evol. 2016, 33, 2692–2705. [Google Scholar] [CrossRef] [Green Version]
- Seo, E.; Kim, T.; Park, J.H.; Yeom, S.-I.; Kim, S.; Seo, M.-K.; Shin, C.; Choi, D. Genome-wide comparative analysis in Solanaceous species reveals evolution of microRNAs targeting defense genes in Capsicum spp. DNA Res. 2018, 25, 561–575. [Google Scholar] [CrossRef]
- Jiang, N.; Meng, J.; Cui, J.; Sun, G.; Luan, Y. Function identification of miR482b, a negative regulator during tomato resistance to Phytophthora infestans. Hortic. Res. 2018, 5, 9. [Google Scholar] [CrossRef] [Green Version]
- de Vries, S.; Kloesges, T.; Rose, L.E. Evolutionarily Dynamic, but Robust, Targeting of Resistance Genes by the miR482/2118 Gene Family in the Solanaceae. Genome Biol. Evol. 2015, 7, 3307–3321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernie, A.R.; Yan, J. De Novo Domestication: An Alternative Route toward New Crops for the Future. Mol. Plant 2019, 12, 615–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courtier-Orgogozo, V.; Martin, A. The coding loci of evolution and domestication: Current knowledge and implications for bio-inspired genome editing. J. Exp. Biol. 2020, 223, jeb208934. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef]
- Burdett, H.; Bentham, A.R.; Williams, S.J.; Dodds, P.N.; Anderson, P.A.; Banfield, M.J.; Kobe, B. The Plant “Resistosome”: Structural Insights into Immune Signaling. Cell Host Microbe 2019, 26, 193–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Gene Name | Protein Class | Chromosome | Pathogen/Insect | Source of Resistance | Reference |
|---|---|---|---|---|---|
| Bs4 | TNL | 5 | Xanthomonas campestris pv. vesicatoria | S. lycopersicum | Ballvora et al. [38] |
| Hero | CNL | 4 | Globodera rostochiensis | S. pimpinellifolium | Ernst et al. [39] |
| I-2 | CNL | 11 | Fusarium oxysporum f. sp. Lycopersici | S. pimpinellifolium | Simons et al. [37] |
| Mi-1.2 | CNL | 6 | Melaydogyne spp.; Macrosiphum euphorbiae; Bemisia tabaci | S. peruvianum | Rossi et al. [36] Mahfouze et al. [40] |
| Mi-9 | CNL | 6 | Meloidogyne spp. | S. arcanum | Jablonska et al. [41] |
| Prf | CNL | 5 | Pseudomonas syringae | S. pimpinellifolium | Salmeron et al. [42] |
| Ph-3 | CNL | 9 | Phitophthora infestans | S. pimpinellifolium | Zhang et al. [43] |
| Sw-5 | CNL | 9 | Tomato spotted wilt virus | S. peruvianum | Brommonschenkel et al. [44] |
| Tm-1 | CNL | 2 | Tomato mosaic virus | S. habrochaites | Ishibashi et al. [45] |
| Tm-2 | CNL | 9 | Tobacco mosaic virus | S. habrochaites | Lanfermeijer et al. [46] |
| Tm-2a | CNL | 9 | Tobacco mosaic virus | S. peruvianum | Lanfermeijer et al. [47] |
| Ty 2 | CNL | 11 | Tomato leaf curly yellow virus | S. habrochaites | Yang et al. [48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andolfo, G.; D’Agostino, N.; Frusciante, L.; Ercolano, M.R. The Tomato Interspecific NB-LRR Gene Arsenal and Its Impact on Breeding Strategies. Genes 2021, 12, 184. https://doi.org/10.3390/genes12020184
Andolfo G, D’Agostino N, Frusciante L, Ercolano MR. The Tomato Interspecific NB-LRR Gene Arsenal and Its Impact on Breeding Strategies. Genes. 2021; 12(2):184. https://doi.org/10.3390/genes12020184
Chicago/Turabian StyleAndolfo, Giuseppe, Nunzio D’Agostino, Luigi Frusciante, and Maria Raffaella Ercolano. 2021. "The Tomato Interspecific NB-LRR Gene Arsenal and Its Impact on Breeding Strategies" Genes 12, no. 2: 184. https://doi.org/10.3390/genes12020184







