PUFA Treatment Affects C2C12 Myocyte Differentiation, Myogenesis Related Genes and Energy Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. PUFA Preparation for In Vitro Culture
2.3. Assessment of PUFA Effect on Cell Viability by xCELLigence Real-Time Assay
2.4. Cell Differentiation Experiment
2.5. Quantitative Real-Time PCR
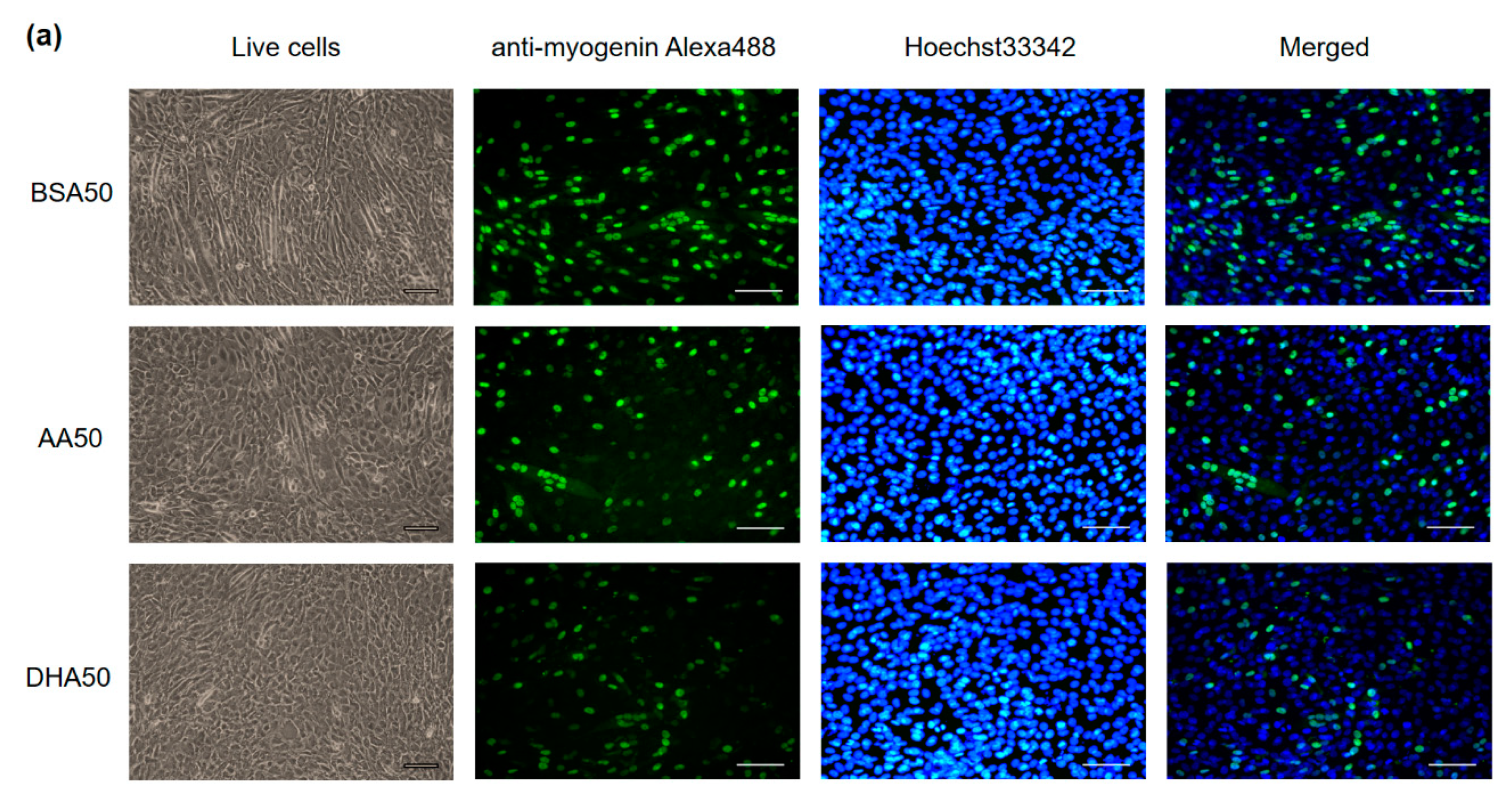
2.6. Immunofluorescence Assays
2.7. Bioenergetics Assay
2.8. Data Analysis
3. Results
3.1. Assessment of PUFA Effect on Cell Viability
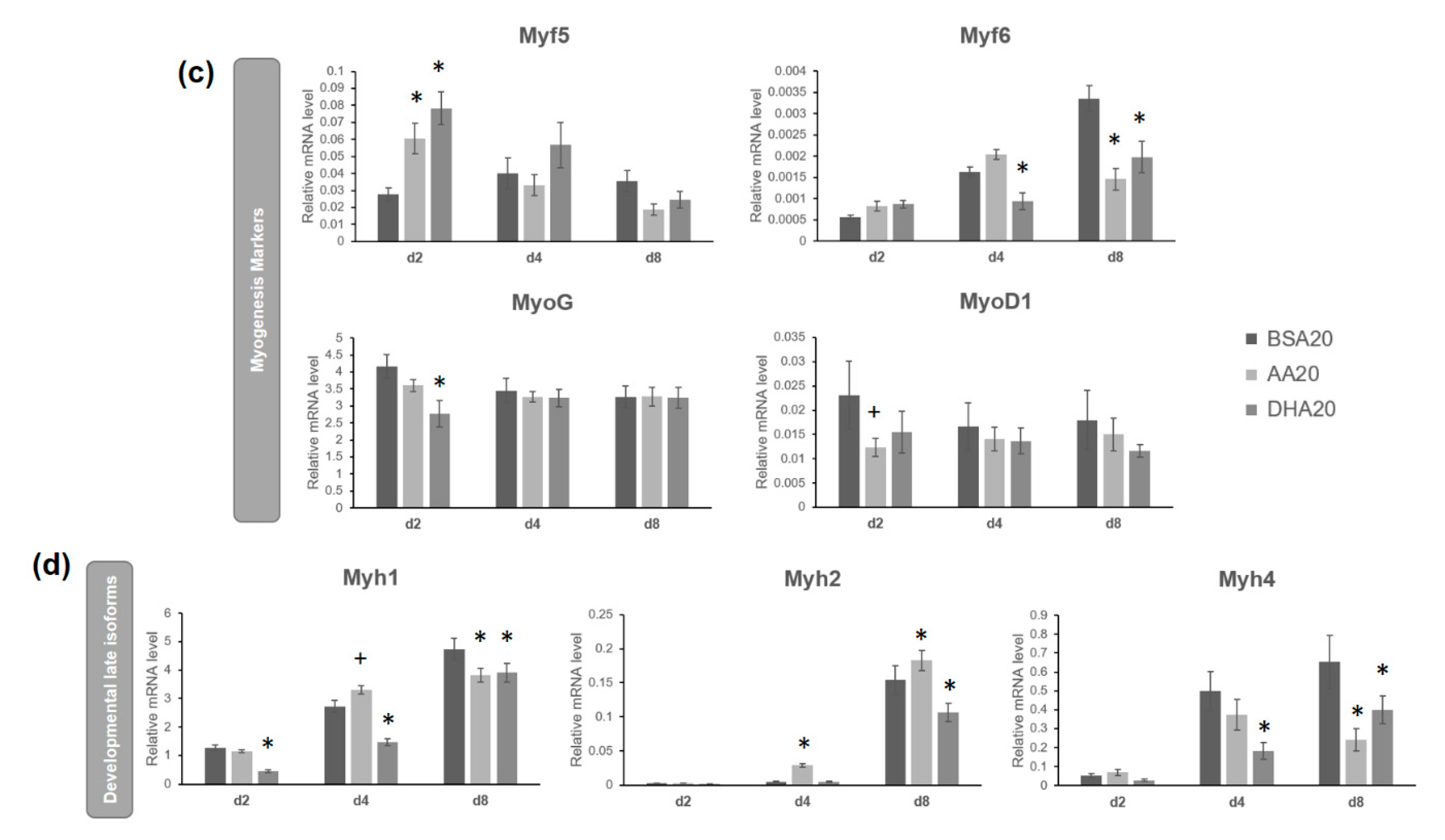
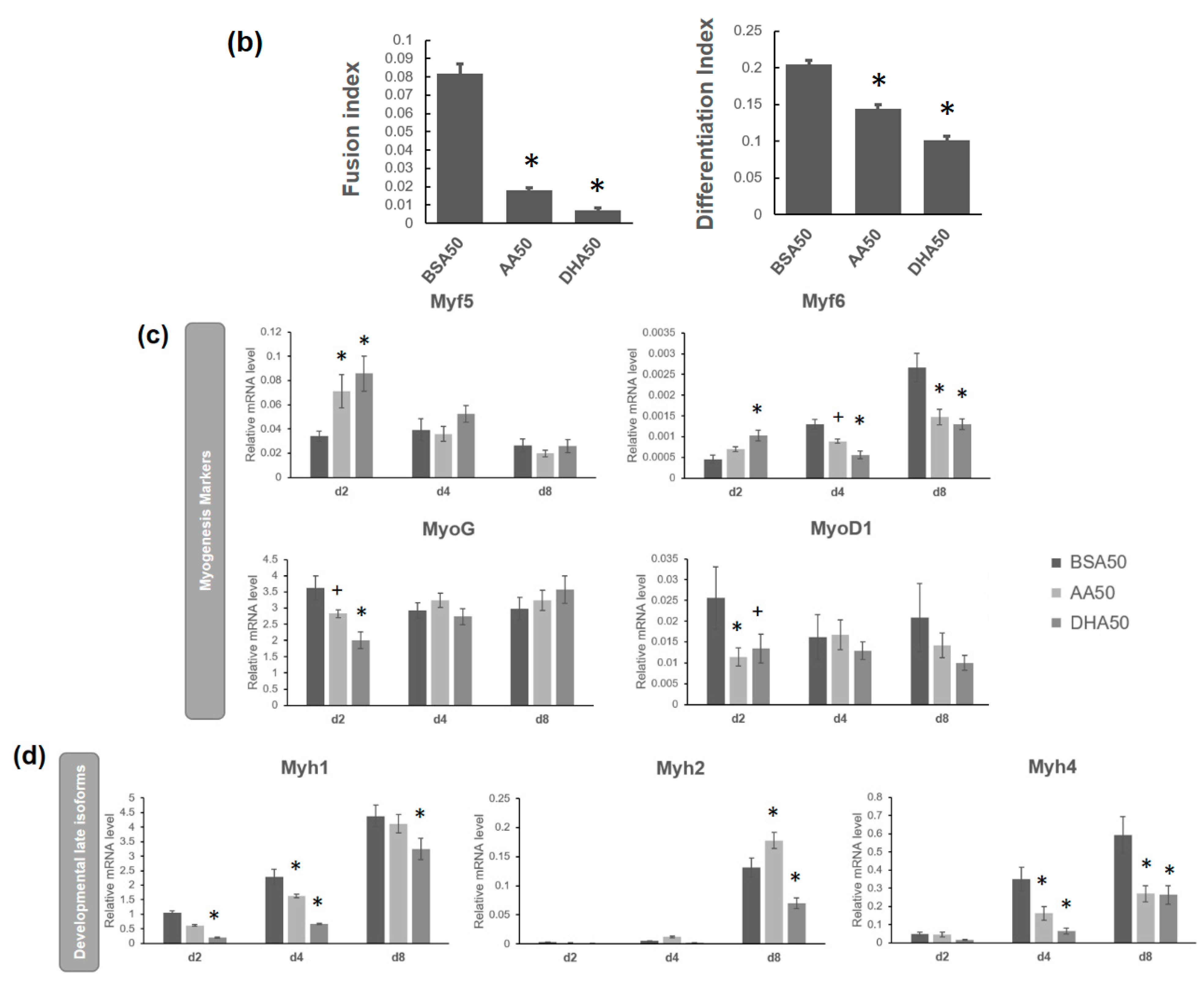
3.2. AA and DHA Reduced Myogenic Differentiation
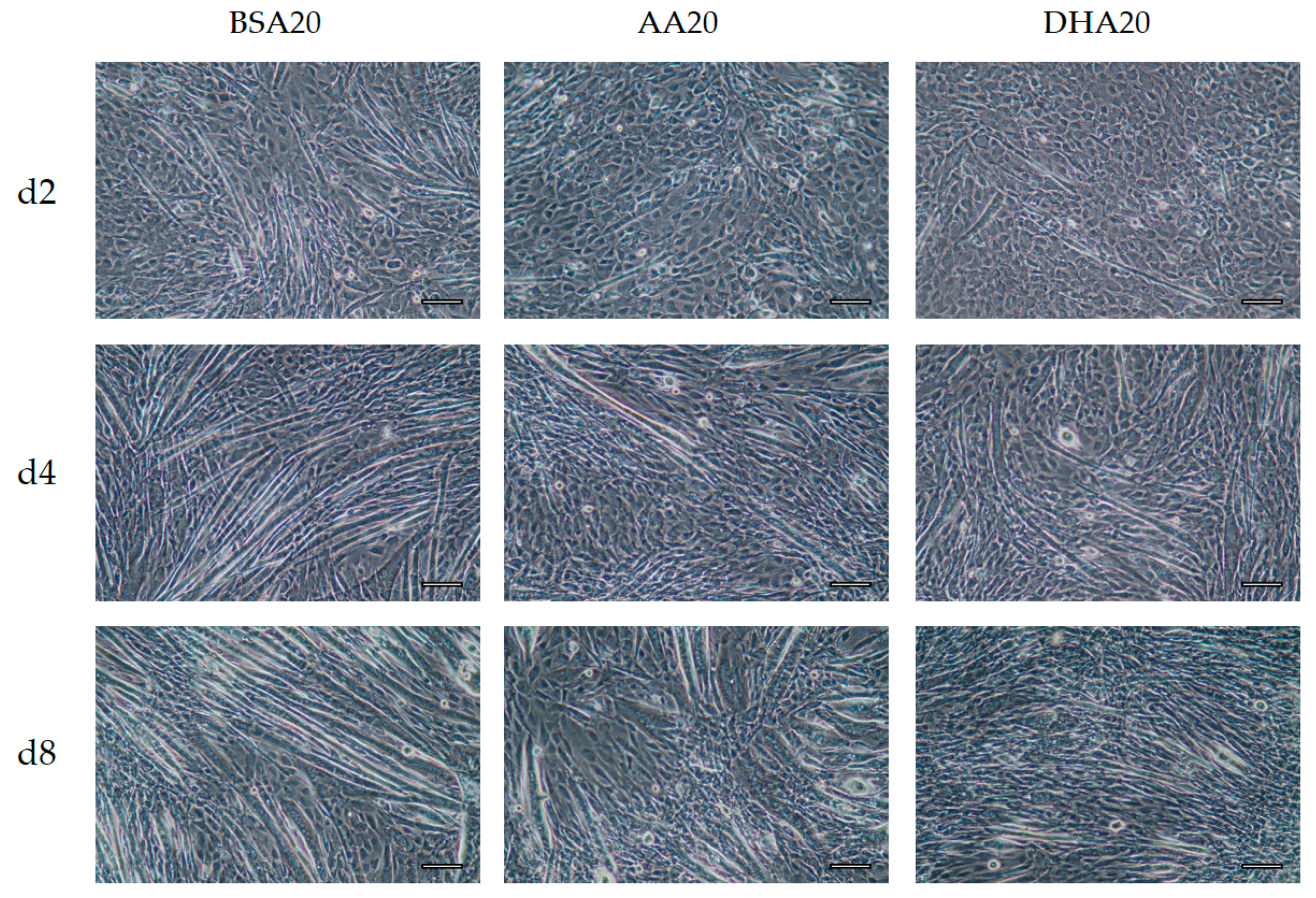
3.3. Morphological Changes after 6 Days Recovery
3.4. AA and DHA Affects the mRNA Abundance of Genes Involved in Wnt Signaling Pathway
3.5. AA and DHA Affects Myoblasts Energy Metabolism
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PUFAs | Polyunsaturated fatty acids |
| DHA | Docosahexaenoic acid |
| AA | Arachidonic acid |
| Myh1 | Myosin Heavy Chain 1 |
| Myh2 | Myosin Heavy Chain 2 |
| Myh4 | Myosin Heavy Chain 4 |
| Myf6 | Myogenic Factor 6 |
| Myf5 | Myogenic Factor 5 |
| MyoG | Myogenin |
| MyoD | Myogenic myogenic differentiation |
| Daam1 | Dishevelled Associated Activator of Morphogenesis 1 |
| Znrf3 | Zinc and Ring Finger 3 |
| Wif1 | Wnt Inhibitory Factor 1 |
| Rac1 | Rac Family Small GTPase 1 |
| PGC-1α | PPARG Coactivator 1 Alpha |
| Prkaa1 | Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 |
| Sirt1 | Sirtuin 1 |
References
- Friedrichs, M.; Wirsdöerfer, F.; Flohé, S.B.; Schneider, S.; Wuelling, M.; Vortkamp, A. BMP signaling balances proliferation and differentiation of muscle satellite cell descendants. BMC Cell Biol. 2011, 12, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straface, G.; Aprahamian, T.; Flex, A.; Gaetani, E.; Biscetti, F.; Smith, R.C.; Pecorini, G.; Pola, E.; Angelini, F.; Stigliano, E.; et al. Sonic hedgehog regulates angiogenesis and myogenesis during post-natal skeletal muscle regeneration. J. Cell Mol. Med. 2009, 13, 2424–2435. [Google Scholar] [CrossRef] [PubMed]
- Moncaut, N.; Rigby, P.W.; Carvajal, J.J. Dial M(RF) for myogenesis. FEBS J. 2013, 280, 3980–3990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ott, M.O.; Bober, E.; Lyons, G.; Arnold, H.; Buckingham, M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development 1991, 111, 1097. [Google Scholar] [PubMed]
- Knight, J.D.R.; Kothary, R. The myogenic kinome: Protein kinases critical to mammalian skeletal myogenesis. Skelet. Muscle 2011, 1, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buonanno, A.; Apone, L.; Morasso, M.I.; Beers, R.; Brenner, H.R.; Eftimie, R. The MyoD family of myogenic factors is regulated by electrical activity: Isolation and characterization of a mouse Myf-5 cDNA. Nucleic Acids Res. 1992, 20, 539–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demonbreun, A.R.; Biersmith, B.H.; McNally, E.M. Membrane fusion in muscle development and repair. Semin. Cell Dev. Biol. 2015, 45, 48–56. [Google Scholar] [CrossRef] [Green Version]
- Simons, K.; Ikonen, E. Functional rafts in cell membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef]
- Calder, P.C. n-3 polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am. J. Clin. Nutr. 2006, 83, 1505s–1519s. [Google Scholar] [CrossRef]
- Calder, P.C. Polyunsaturated fatty acids, inflammation, and immunity. Lipids 2001, 36, 1007–1024. [Google Scholar] [CrossRef]
- Fournier, N.; Benoist, J.-F.; Allaoui, F.; Nowak, M.; Dakroub, H.; Vedie, B.; Paul, J.-L. Contrasting effects of membrane enrichment with polyunsaturated fatty acids on phospholipid composition and cholesterol efflux from cholesterol-loaded J774 mouse or primary human macrophages. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158536. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Tachibana, H.; Morinaga, Y.; Fujimura, Y.; Yamada, K. Modulation of proliferation and differentiation of C2C12 skeletal muscle cells by fatty acids. Life Sci. 2009, 84, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Hurley, M.S.; Flux, C.; Salter, A.M.; Brameld, J.M. Effects of fatty acids on skeletal muscle cell differentiation in vitro. Br. J. Nutr. 2006, 95, 623–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Xu, X.; Liu, Y.; Zhang, L.; Odle, J.; Lin, X.; Zhu, H.; Wang, X.; Liu, Y. EPA and DHA Inhibit Myogenesis and Downregulate the Expression of Muscle-related Genes in C2C12 Myoblasts. Genes 2019, 10, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, A.; Nalsen, C.; Tengblad, S.; Vessby, B. Fatty acid composition of skeletal muscle reflects dietary fat composition in humans. Am. J. Clin. Nutr. 2002, 76, 1222–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philipsen, M.H.; Sämfors, S.; Malmberg, P.; Ewing, A.G. Relative quantification of deuterated omega-3 and -6 fatty acids and their lipid turnover in PC12 cell membranes using TOF-SIMS. J. Lipid Res. 2018, 59, 2098–2107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaikh, S.R.; Kinnun, J.J.; Leng, X.; Williams, J.A.; Wassall, S.R. How polyunsaturated fatty acids modify molecular organization in membranes: Insight from NMR studies of model systems. Biochim. Biophys. Acta 2015, 1848, 211–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glatz, J.F.C.; Luiken, J.J.F.P. Fatty acids in cell signaling: Historical perspective and future outlook. Prostaglandins Leukot. Essent. Fat. Acids 2015, 92, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Cossu, G.; Borello, U. Wnt signaling and the activation of myogenesis in mammals. EMBO J. 1999, 18, 6867–6872. [Google Scholar] [CrossRef] [Green Version]
- Linker, C.; Lesbros, C.; Gros, J.; Burrus, L.W.; Rawls, A.; Marcelle, C. Beta-Catenin-dependent Wnt signalling controls the epithelial organisation of somites through the activation of paraxis. Development 2005, 132, 3895–3905. [Google Scholar] [CrossRef] [Green Version]
- Otto, A.; Schmidt, C.; Luke, G.; Allen, S.; Valasek, P.; Muntoni, F.; Lawrence-Watt, D.; Patel, K. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J. Cell Sci. 2008, 121, 2939–2950. [Google Scholar] [CrossRef] [Green Version]
- Sokol, S.Y. Maintaining embryonic stem cell pluripotency with Wnt signaling. Development 2011, 138, 4341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Cavaliere, G.; Trinchese, G.; Bergamo, P.; De Filippo, C.; Mattace Raso, G.; Gifuni, G.; Putti, R.; Moni, B.H.; Canani, R.B.; Meli, R.; et al. Polyunsaturated Fatty Acids Attenuate Diet Induced Obesity and Insulin Resistance, Modulating Mitochondrial Respiratory Uncoupling in Rat Skeletal Muscle. PLoS ONE 2016, 11, e0149033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva, E.P., Jr.; Nachbar, R.T.; Levada-Pires, A.C.; Hirabara, S.M.; Lambertucci, R.H. Omega-3 fatty acids differentially modulate enzymatic anti-oxidant systems in skeletal muscle cells. Cell Stress Chaperones 2016, 21, 87–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tachtsis, B.; Camera, D.; Lacham-Kaplan, O. Potential Roles of n-3 PUFAs during Skeletal Muscle Growth and Regeneration. Nutrients 2018, 10, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kho, D.; MacDonald, C.; Johnson, R.; Unsworth, C.P.; O’Carroll, S.J.; du Mez, E.; Angel, C.E.; Graham, E.S. Application of xCELLigence RTCA Biosensor Technology for Revealing the Profile and Window of Drug Responsiveness in Real Time. Biosensors 2015, 5, 199–222. [Google Scholar] [CrossRef] [Green Version]
- Sajjanar, B.; Siengdee, P.; Trakooljul, N.; Liu, X.; Kalbe, C.; Wimmers, K.; Ponsuksili, S. Cross-talk between energy metabolism and epigenetics during temperature stress response in C2C12 myoblasts. Int. J. Hyperth. Off. J. Eur. Soc. Hyperthermic Oncol. North Am. Hyperth. Group 2019, 36, 776–784. [Google Scholar] [CrossRef] [Green Version]
- Abdelhamid, A.; Hooper, L.; Sivakaran, R.; Hayhoe, R.P.G.; Welch, A. The Relationship Between Omega-3, Omega-6 and Total Polyunsaturated Fat and Musculoskeletal Health and Functional Status in Adults: A Systematic Review and Meta-analysis of RCTs. Calcif. Tissue Int. 2019, 105, 353–372. [Google Scholar] [CrossRef] [Green Version]
- Jeromson, S.; Gallagher, I.J.; Galloway, S.D.R.; Hamilton, D.L. Omega-3 Fatty Acids and Skeletal Muscle Health. Mar. Drugs 2015, 13, 6977–7004. [Google Scholar] [CrossRef]
- Markworth, J.F.; Cameron-Smith, D. Arachidonic acid supplementation enhances in vitro skeletal muscle cell growth via a COX-2-dependent pathway. Am. J. Physiol.-Cell Physiol. 2012, 304, C56–C67. [Google Scholar] [CrossRef] [Green Version]
- Bhullar, A.S.; Putman, C.T.; Mazurak, V.C. Potential Role of Omega-3 Fatty Acids on the Myogenic Program of Satellite Cells. Nutr. Metab. Insights 2016, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsueh, T.-Y.; Baum, J.I.; Huang, Y. Effect of Eicosapentaenoic Acid and Docosahexaenoic Acid on Myogenesis and Mitochondrial Biosynthesis during Murine Skeletal Muscle Cell Differentiation. Front. Nutr. 2018, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briolay, A.; Jaafar, R.; Nemoz, G.; Bessueille, L. Myogenic differentiation and lipid-raft composition of L6 skeletal muscle cells are modulated by PUFAs. Biochim. Biophys. Acta 2013, 1828, 602–613. [Google Scholar] [CrossRef] [PubMed]
- Alsabeeh, N.; Chausse, B.; Kakimoto, P.A.; Kowaltowski, A.J.; Shirihai, O. Cell culture models of fatty acid overload: Problems and solutions. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Relaix, F.; Marcelle, C. Muscle stem cells. Curr. Opin. Cell Biol. 2009, 21, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Gil, A. Polyunsaturated fatty acids and inflammatory diseases. Biomed. Pharmacother. 2002, 56, 388–396. [Google Scholar] [CrossRef]
- Panda, A.C.; Abdelmohsen, K.; Martindale, J.L.; Di Germanio, C.; Yang, X.; Grammatikakis, I.; Noh, J.H.; Zhang, Y.; Lehrmann, E.; Dudekula, D.B.; et al. Novel RNA-binding activity of MYF5 enhances Ccnd1/Cyclin D1 mRNA translation during myogenesis. Nucleic Acids Res. 2016, 44, 2393–2408. [Google Scholar] [CrossRef] [Green Version]
- Kitzmann, M.; Carnac, G.; Vandromme, M.; Primig, M.; Lamb, N.J.; Fernandez, A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J. Cell Biol. 1998, 142, 1447–1459. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Zheng, Y.; Zhang, Y.; Zhao, J.; Chang, F.; Lu, T.; Zhang, R.; Li, Q.; Hu, X.; Li, N. Different effects of omega-3 fatty acids on the cell cycle in C2C12 myoblast proliferation. Mol. Cell Biochem. 2012, 367, 165–173. [Google Scholar] [CrossRef]
- Lindon, C.; Montarras, D.; Pinset, C. Cell cycle-regulated expression of the muscle determination factor Myf5 in proliferating myoblasts. J. Cell Biol. 1998, 140, 111–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, A.; Lee, E.J.; Jan, A.T.; Ahmad, S.; Cho, K.-H.; Kim, J.; Choi, I. Network Analysis for the Identification of Differentially Expressed Hub Genes Using Myogenin Knock-down Muscle Satellite Cells. PLoS ONE 2015, 10, e0133597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, S.J.; Hansen, J.M.; Merrell, A.J.; Murphy, M.M.; Lawson, J.A.; Hutcheson, D.A.; Hansen, M.S.; Angus-Hill, M.; Kardon, G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 2011, 138, 371–384. [Google Scholar] [CrossRef] [Green Version]
- von Maltzahn, J.; Chang, N.C.; Bentzinger, C.F.; Rudnicki, M.A. Wnt signaling in myogenesis. Trends Cell Biol. 2012, 22, 602–609. [Google Scholar] [CrossRef] [Green Version]
- Huraskin, D.; Eiber, N.; Reichel, M.; Zidek, L.M.; Kravic, B.; Bernkopf, D.; von Maltzahn, J.; Behrens, J.; Hashemolhosseini, S. Wnt/β-catenin signaling via Axin2 is required for myogenesis and, together with YAP/Taz and Tead1, active in IIa/IIx muscle fibers. Development 2016, 143, 3128–3142. [Google Scholar] [CrossRef] [Green Version]
- Hall, A. Rho GTPases and the actin cytoskeleton. Science 1998, 279, 509–514. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Li, H. RAC1 overexpression promotes the proliferation, migration and epithelial-mesenchymal transition of lens epithelial cells. Int. J. Clin. Exp. Pathol. 2015, 8, 10760–10767. [Google Scholar]
- Li, D.; Hallett, M.A.; Zhu, W.; Rubart, M.; Liu, Y.; Yang, Z.; Chen, H.; Haneline, L.S.; Chan, R.J.; Schwartz, R.J.; et al. Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development 2011, 138, 303–315. [Google Scholar] [CrossRef] [Green Version]
- Chang, F.Z.; Wang, Q.; Zhang, Q.; Chang, L.L.; Li, W. Omega-3 polyunsaturated fatty acid inhibits the malignant progression of hepatocarcinoma by inhibiting the Wnt/beta-catenin pathway. Eur Rev. Med. Pharmacol. Sci. 2018, 22, 4500–4508. [Google Scholar] [CrossRef]
- Rahmani, B.; Hamedi Asl, D.; Naserpour Farivar, T.; Azad, M.; Sahmani, M.; Gheibi, N. Omega-3 PUFA Alters the Expression Level but Not the Methylation Pattern of the WIF1 Gene Promoter in a Pancreatic Cancer Cell Line (MIA PaCa-2). Biochem. Genet 2019, 57, 477–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Go, S.; Kramer, T.T.; Verhoeven, A.J.; Oude Elferink, R.P.J.; Chang, J.C. The extracellular lactate-to-pyruvate ratio modulates the sensitivity to oxidative stress-induced apoptosis via the cytosolic NADH/NAD(+) redox state. Apoptosis 2020. [Google Scholar] [CrossRef] [PubMed]
- Herbst, E.A.F.; Paglialunga, S.; Gerling, C.; Whitfield, J.; Mukai, K.; Chabowski, A.; Heigenhauser, G.J.F.; Spriet, L.L.; Holloway, G.P. Omega-3 supplementation alters mitochondrial membrane composition and respiration kinetics in human skeletal muscle. J. Physiol. 2014, 592, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Ventura-Clapier, R.; Garnier, A.; Veksler, V. Transcriptional control of mitochondrial biogenesis: The central role of PGC-1alpha. Cardiovasc. Res. 2008, 79, 208–217. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Trakooljul, N.; Hadlich, F.; Murani, E.; Wimmers, K.; Ponsuksili, S. Mitochondrial-nuclear crosstalk, haplotype and copy number variation distinct in muscle fiber type, mitochondrial respiratory and metabolic enzyme activities. Sci. Rep. 2017, 7, 14024. [Google Scholar] [CrossRef]
- Chang, W.-H.; Ting, H.-C.; Chen, W.-W.; Chan, J.-F.; Hsu, Y.-H.H. Omega-3 and omega-6 fatty acid differentially impact cardiolipin remodeling in activated macrophage. Lipids Health Dis. 2018, 17, 201. [Google Scholar] [CrossRef] [Green Version]
- Xue, B.; Yang, Z.; Wang, X.; Shi, H. Omega-3 polyunsaturated fatty acids antagonize macrophage inflammation via activation of AMPK/SIRT1 pathway. PLoS ONE 2012, 7, e45990. [Google Scholar] [CrossRef]
- Reyes-Gordillo, K.; Shah, R.; Varatharajalu, R.; Garige, M.; Leckey, L.C.; Lakshman, M.R. Low-ω3 Fatty Acid and Soy Protein Attenuate Alcohol-Induced Fatty Liver and Injury by Regulating the Opposing Lipid Oxidation and Lipogenic Signaling Pathways. Oxid. Med. Cell Longev. 2016, 2016, 1840513. [Google Scholar] [CrossRef] [Green Version]



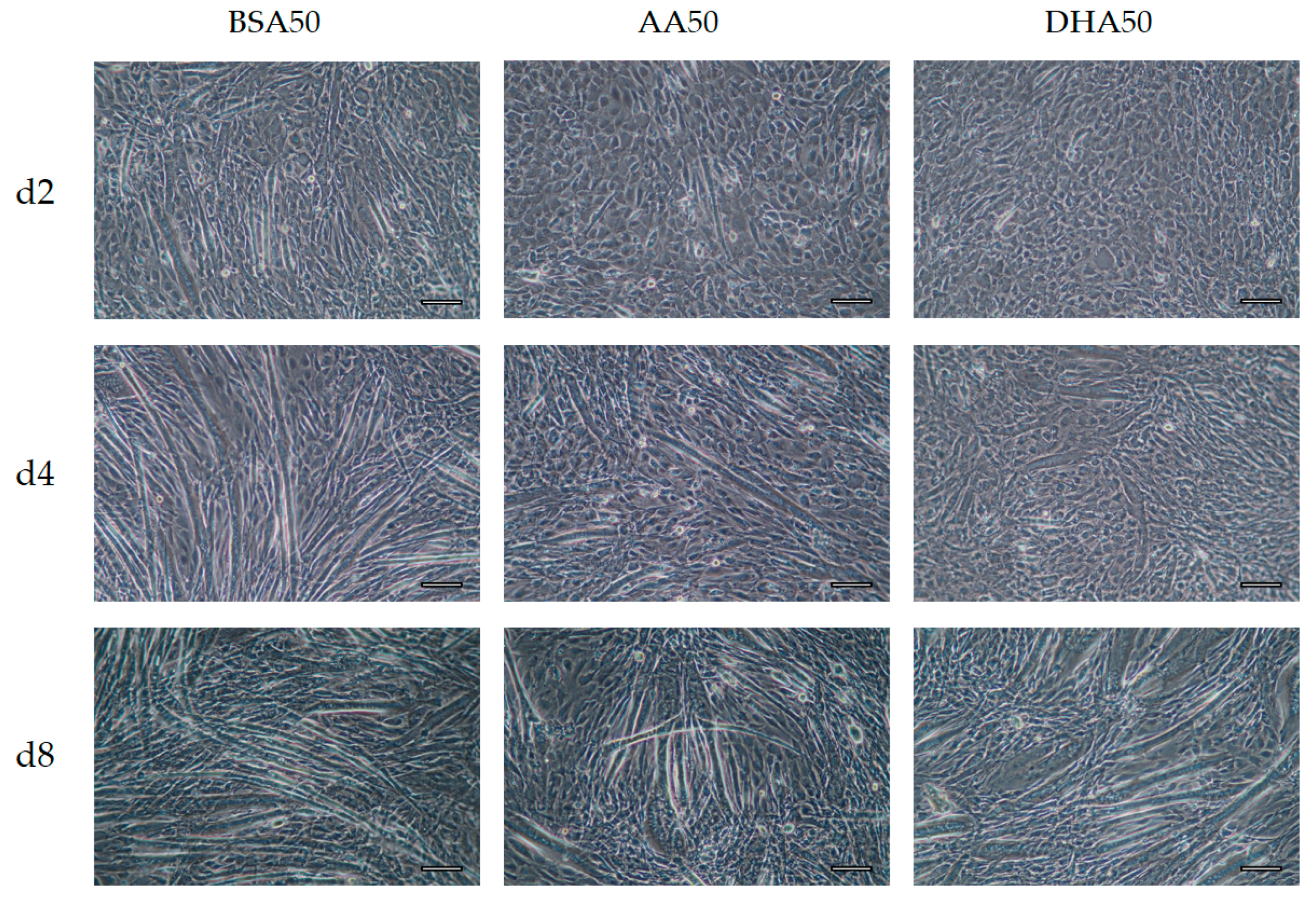
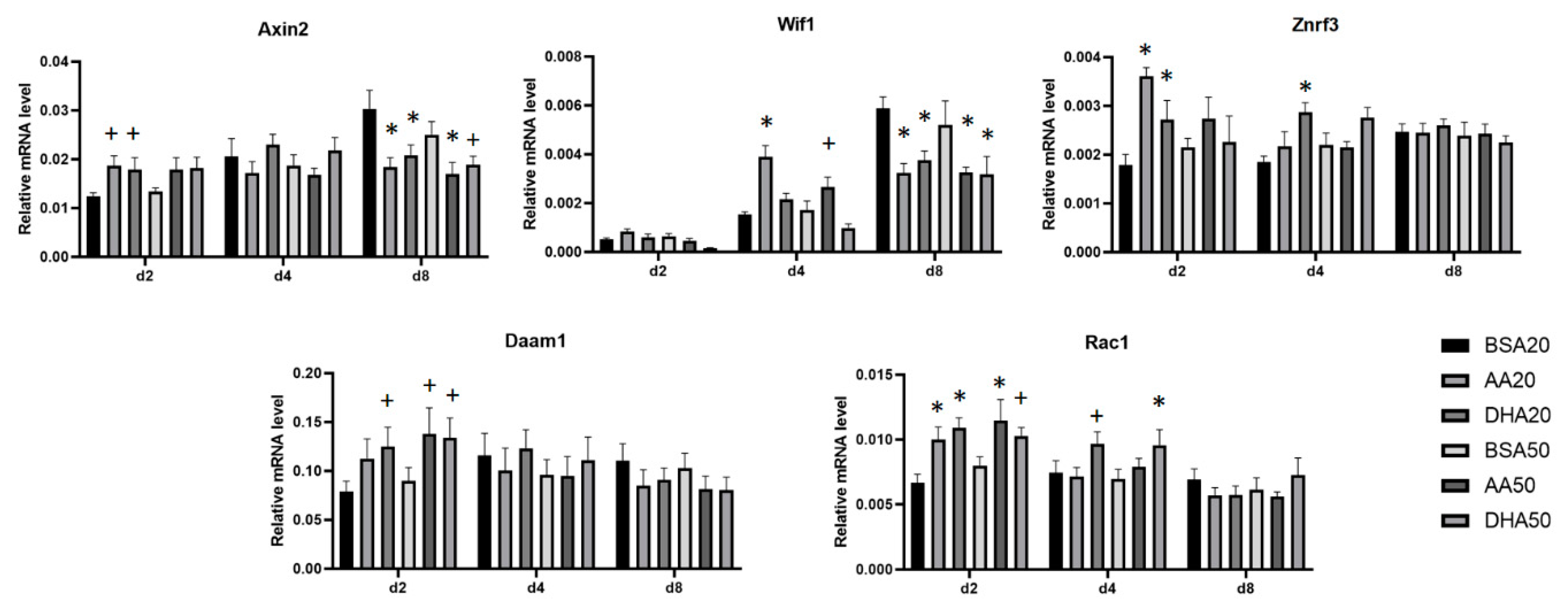
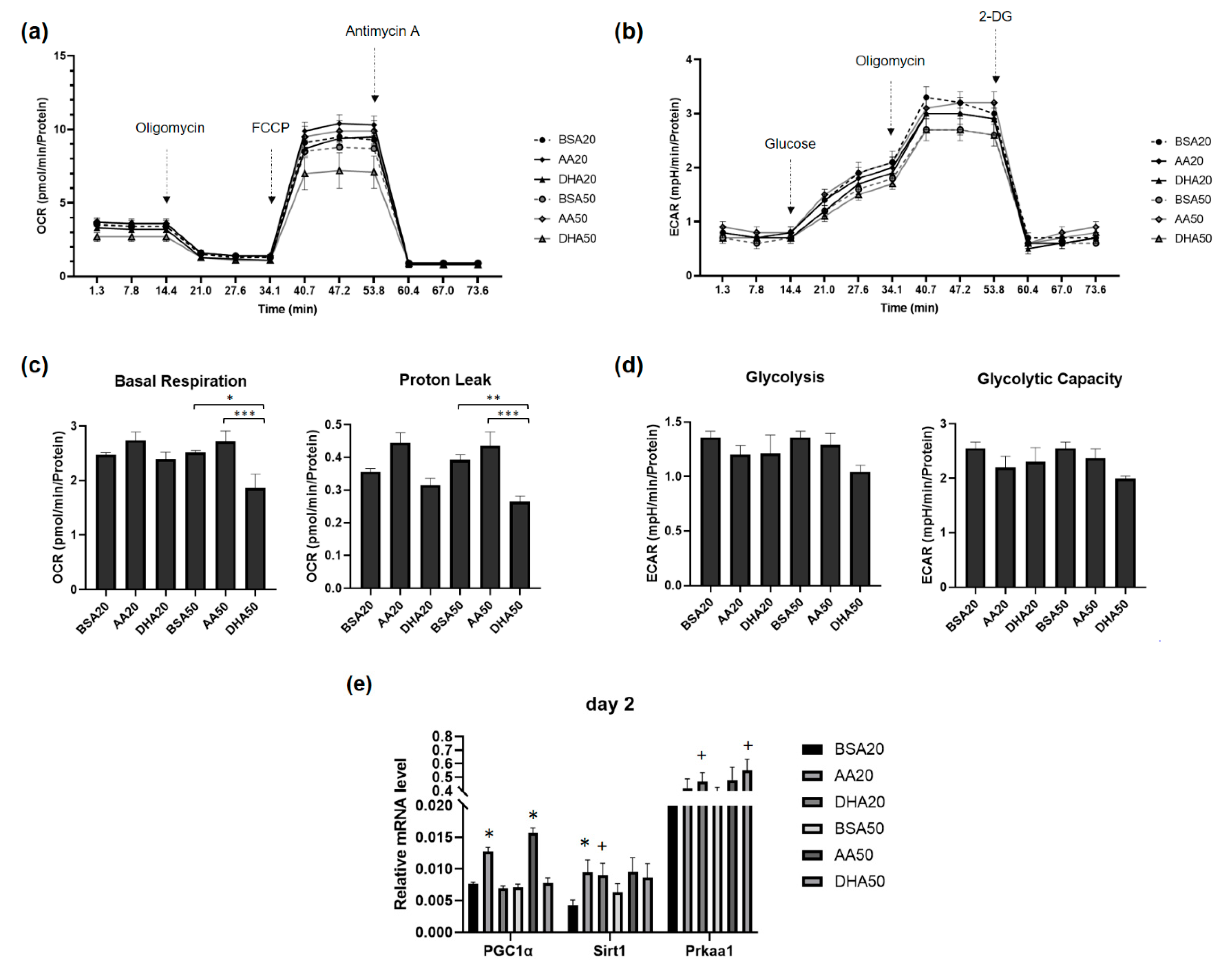
| Code | Treatment (Conditions) |
|---|---|
| AA20 | Arachidonic acid 20 μM + BSA 4.7 μM |
| AA30 | Arachidonic acid 30 μM + BSA 7.1 μM |
| AA50 | Arachidonic acid 50 μM + BSA 11.8 μM |
| AA100 | Arachidonic acid 100 μM + BSA 23.7 μM |
| DHA20 | Docosahexaenoic acid 20 μM + BSA 4.7 μM |
| DHA30 | Docosahexaenoic acid 30 μM + BSA 7.1 μM |
| DHA50 | Docosahexaenoic acid 50 μM + BSA 11.8 μM |
| DHA100 | Docosahexaenoic acid 100 μM + BSA 23.7 μM |
| BSA20 | Bovine Serum Albumin 4.7 μM |
| BSA30 | Bovine Serum Albumin 7.1 μM |
| BSAA50 | Bovine Serum Albumin 11.8 μM |
| BSA100 | Bovine Serum Albumin 23.7 μM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Risha, M.A.; Siengdee, P.; Dannenberger, D.; Wimmers, K.; Ponsuksili, S. PUFA Treatment Affects C2C12 Myocyte Differentiation, Myogenesis Related Genes and Energy Metabolism. Genes 2021, 12, 192. https://doi.org/10.3390/genes12020192
Risha MA, Siengdee P, Dannenberger D, Wimmers K, Ponsuksili S. PUFA Treatment Affects C2C12 Myocyte Differentiation, Myogenesis Related Genes and Energy Metabolism. Genes. 2021; 12(2):192. https://doi.org/10.3390/genes12020192
Chicago/Turabian StyleRisha, Marua Abu, Puntita Siengdee, Dirk Dannenberger, Klaus Wimmers, and Siriluck Ponsuksili. 2021. "PUFA Treatment Affects C2C12 Myocyte Differentiation, Myogenesis Related Genes and Energy Metabolism" Genes 12, no. 2: 192. https://doi.org/10.3390/genes12020192
APA StyleRisha, M. A., Siengdee, P., Dannenberger, D., Wimmers, K., & Ponsuksili, S. (2021). PUFA Treatment Affects C2C12 Myocyte Differentiation, Myogenesis Related Genes and Energy Metabolism. Genes, 12(2), 192. https://doi.org/10.3390/genes12020192








