Molecular Characterization and Phylogenetic Analysis of MADS-Box Gene VroAGL11 Associated with Stenospermocarpic Seedlessness in Muscadine Grapes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Berry and Seed Characteristics
2.3. RNA Isolation and cDNA Synthesis
2.4. Detection and Confirmation of Muscadine Grape AGL11 Transcripts
2.5. Quantification of Relative Gene Expressions
2.6. DNA Isolation and Sequencing of Muscadine AGL11 Gene
2.7. Analysis of Deduced Protein Sequences
2.8. Phylogenetic Tree Constructions
2.9. Statistical Analysis
2.10. Accession Numbers
3. Results
3.1. Berry and Seed Characteristics
3.2. Identification and Analysis of Muscadine Grape AGL11 Gene
3.3. Detection and Confirmation of VroAGL11 Transcripts
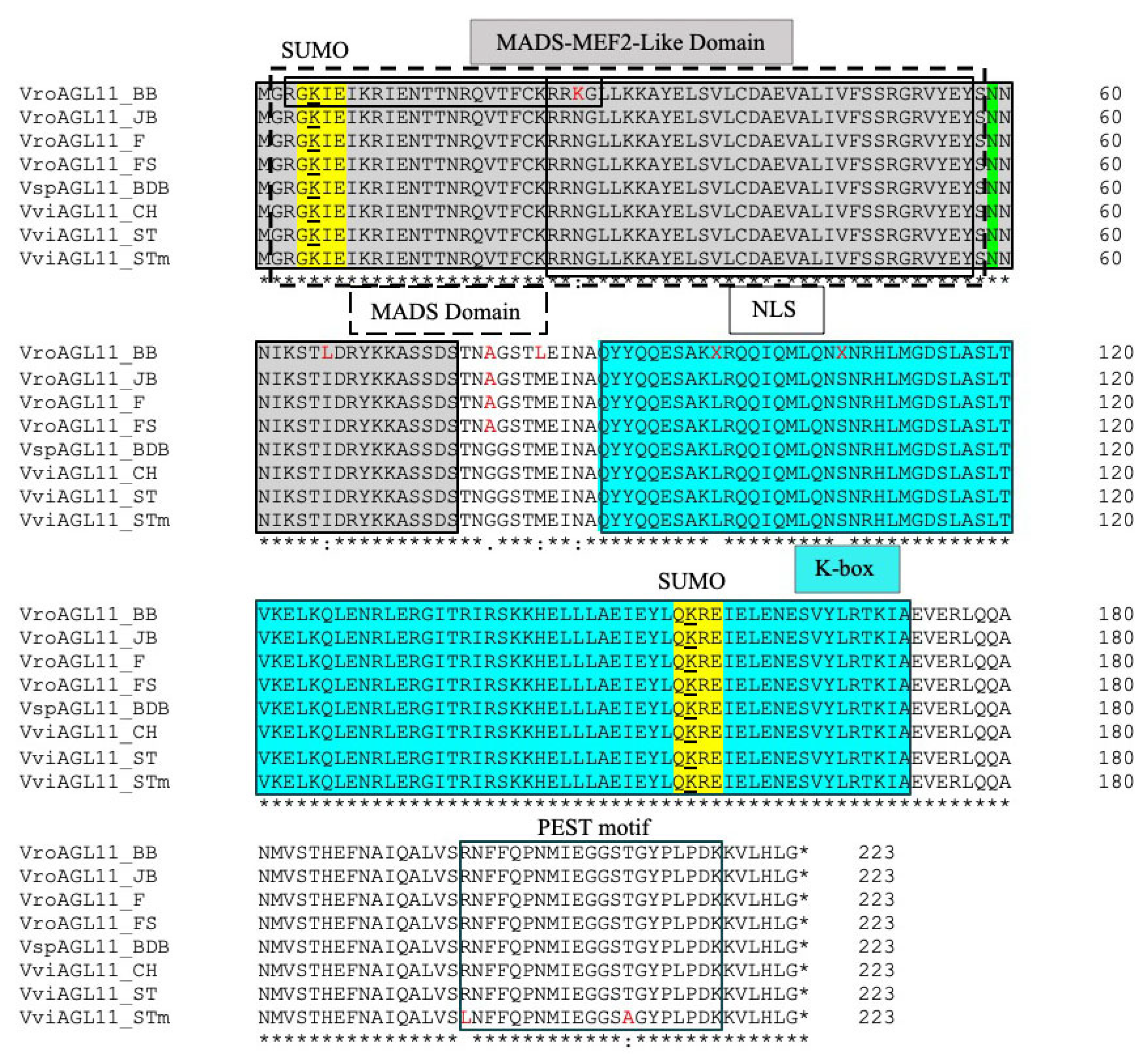
3.4. Analysis of Predicted AGL11 Protein Sequences
3.5. Relative Expressions of AGL11 Genes
3.6. Relationship between AGL11 Gene Expression Levels and Seed Characteristics
3.7. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FWGGA. Florida Wine and Grape Growers Association. Available online: https://www.fgga.org/ (accessed on 19 November 2020).
- Balasubramani, S.P.; Rahman, M.A.; Basha, S.M. Synergistic Action of Stilbenes in Muscadine Grape Berry Extract Shows Better Cytotoxic Potential Against Cancer Cells Than Resveratrol Alone. Biomedicines 2019, 7, 96. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Kim, M.H.; Sandhu, A.K.; Gao, C.; Gu, L. Muscadine Grape (Vitis rotundifolia) or Wine Phyto-chemicals Reduce Intestinal Inflammation in Mice with Dextran Sulfate Sodium-Induced Colitis. J. Agric. Food Chem. 2017, 65, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Mendonca, P.; Darwish, A.G.; Tsolova, V.; El-Sharkawy, I.; Soliman, K.F.A. The Anticancer and Antioxidant Effects of Muscadine Grape Extracts on Racially Different Triple-negative Breast Cancer Cells. Anticancer Res. 2019, 39, 4043–4053. [Google Scholar] [CrossRef] [Green Version]
- Kovaleva, L.V.; Smirnova, N.K.; Milyaeva, E.L. Seedlessness: Structure and Metabolic activity of Vitis vinifera L. female gametophyte (cv. Kishmish Chernyi). Russ. J. Plant Physiol. 1997, 44, 368–373. [Google Scholar]
- Ledbetter, C.A.; Ramming, D.W. Seedlessness in Grapes. In Horticultural Reviews; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1989; pp. 159–184. [Google Scholar] [CrossRef]
- Bouquet, A.; Danglot, Y. Inheritance of seedlessness in grapevine (Vitis vinifera L.). Vitis 1996, 35, 35–42. [Google Scholar]
- Mejía, N.; Soto, B.; Guerrero, M.; Casanueva, X.; Houel, C.; de los Ángeles Miccono, M.; Ramos, R.; Le Cunff, L.; Boursiquot, J.-M.; Hinrichsen, P.; et al. Molecular, genetic and transcriptional evidence for a role of VvAGL11 in stenospermocarpic seedlessness in grapevine. BMC Plant Biol. 2011, 11, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basiouny, F.M.; Himelrick, D.G. Muscadine Grapes; ASHS Press: Alexandria, VA, USA, 2001. [Google Scholar]
- Adam-Blondon, A.F.; Lahogue-Esnault, F.; Bouquet, A.; Boursiquot, J.M.; This, P. Usefulness of two SCAR markers for marker-assisted selection of seedless grapevine cultivars. Vitis 2001, 40, 147–155. [Google Scholar]
- Dangl, G.S.; Mendum, M.L.; Prins, B.H.; Walker, M.A.; Meredith, C.P.; Simon, C.J. Simple sequence repeat analysis of a clonally propagated species: A tool for managing a grape germplasm collection. Genome 2001, 44, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, J.; Vargas, A.M.; Palancar, M.; Borrego, J.; de Andrés, M.T. Genetic relationships among table-grape varieties. Am. J. Enol. Vitic. 2009, 60, 35–42. [Google Scholar]
- Ibáñez, J.; Carreño, J.; Yuste, J.; Martínez-Zapater, J.M. Grapevine breeding and clonal selection programmes in Spain. In Grapevine Breeding Programs for the Wine Industry; Reynolds, A.G., Ed.; Woodhead Publishing: Oxford, UK, 2015; pp. 183–209. [Google Scholar]
- Di Genova, A.; Almeida, A.M.; Munoz-Espinoza, C.; Vizoso, P.; Travisany, D.; Moraga, C.; Pinto, M.; Hinrichsen, P.; Orellana, A.; Maass, A. Whole genome comparison between table and wine grapes reveals a comprehensive catalog of structural variants. BMC Plant Biol. 2014, 14, 7. [Google Scholar] [CrossRef] [Green Version]
- Lahogue, F.; This, P.; Bouquet, A. Identification of a codominant scar marker linked to the seedlessness character in grapevine. Theor. Appl. Genet. 1998, 97, 950–959. [Google Scholar] [CrossRef]
- Cabezas, J.A.; Cervera, M.T.; Ruiz-García, L.; Carreño, J.; Martínez-Zapater, J.M. A genetic analysis of seed and berry weight in grapevine. Genome 2006, 49, 1572–1585. [Google Scholar] [CrossRef]
- Costantini, L.; Battilana, J.; Lamaj, F.; Fanizza, G.; Grando, M.S. Berry and phenology-related traits in grapevine (Vitis vinifera L.): From Quantitative Trait Loci to underlying genes. BMC Plant Biol. 2008, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Doligez, A.; Bouquet, A.; Danglot, Y.; Lahogue, F.; Riaz, S.; Meredith, C.; Edwards, K.; This, P. Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor. Appl. Genet. 2002, 105, 780–795. [Google Scholar] [CrossRef]
- Mejía, N.; Gebauer, M.; Muñoz, L.; Hewstone, N.; Muñoz, C.; Hinrichsen, P. Identification of QTLs for Seedlessness, Berry Size, and Ripening Date in a Seedless × Seedless Table Grape Progeny. Am. J. Enol. Vitic. 2007, 58, 499–507. [Google Scholar]
- Diaz-Riquelme, J.; Grimplet, J.; Martínez-Zapater, J.M.; Carmona, M.J. Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol. 2012, 12, 181. [Google Scholar] [CrossRef] [Green Version]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.; Kater, M.; Colobo, L. MADS-Box Protein Complexes Control Carpel and Ovule Development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef] [Green Version]
- Malabarba, J.; Buffon, V.; Mariath, J.E.A.; Gaeta, M.L.; Dornelas, M.C.; Margis-Pinheiro, M.; Pasquali, G.; Revers, L.F. The MADS-box gene Agamous-like 11 is essential for seed morphogenesis in grapevine. J. Exp. Bot. 2017, 68, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Mizzotti, C.; Ezquer, I.; Paolo, D.; Rueda-Romero, P.; Guerra, R.F.; Battaglia, R.; Rogachev, I.; Aharoni, A.; Kater, M.M.; Caporali, E.; et al. SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat. PLoS Genet. 2014, 10, e1004856. [Google Scholar] [CrossRef] [PubMed]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Ocarez, N.; Mejía, N. Suppression of the D-class MADS-box AGL11 gene triggers seedlessness in fleshy fruits. Plant Cell Rep. 2016, 35, 239–254. [Google Scholar] [CrossRef] [PubMed]
- Royo, C.; Torres-Pérez, R.; Mauri, N.; Diestro, N.; Cabezas, J.A.; Marchal, C.; Lacombe, T.; Ibáñez, J.; Tornel, M.; Carreño, J.; et al. The Major Origin of Seedless Grapes Is Associated with a Missense Mutation in the MADS-Box Gene VviAGL11. Plant Physiol. 2018, 177, 1234–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malabarba, J.; Buffon, V.; Mariath, J.E.A.; Maraschin, F.S.; Margis-Pinheiro, M.; Pasquali, G.; Revers, L.F. Manipulation of VviAGL11 Expression Changes the Seed Content in Grapevine (Vitis vinifera L.). Plant Sci. 2018, 269, 126–135. [Google Scholar] [CrossRef] [Green Version]
- Coombe, B.G. Growth Stages of the Grapevine: Adoption of a System for Identifying Grapevine Growth Stages. Aust. J. Grape Wine Res. 1995, 1, 104–110. [Google Scholar] [CrossRef]
- Rahman, M.A.; Moody, M.A.; Nassuth, A. Grape contains 4 ICE genes whose expression includes alternative polyadenylation, leading to transcripts encoding at least 7 different ICE proteins. Environ. Exp. Bot. 2014, 106, 70–78. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.S.; Lee, C.H.; Shin, J.S.; Chung, Y.S.; Hyung, N.I. A Simple and Rapid Method for Isolation of High Quality Genomic DNA from Fruit Trees and Conifers Using PVP. Nucleic Acids Res. 1997, 25, 1085–1086. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic Identification of Cell Cycle-Dependent Yeast Nucleocytoplasmic Shuttling Proteins by Prediction of Composite Motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [Green Version]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 20 October 2020).




| Serial Number | Name of Cultivars | Seeded/Seedless 1 | Mean Berry Weight (=BW/BN) g | Mean Seed Number /Berry (=SN/BN) | Mean Seed Weight /Berry (=SW/BN) mg | Mean Seed Weight (=SW/SN) mg | Seed Length (mm) (Mean ± SE) | Seed Width (mm) (Mean ± SE) | Seed Content of Berry (%) =(SW/BW) * 100 |
|---|---|---|---|---|---|---|---|---|---|
| Muscadine grapes | |||||||||
| 1 | African Queen | S | 9.9 | 3.3 | 280.6 | 85.0 | 5.7 ± 0.18 | 3.9 ± 0.16 | 2.8 |
| 2 | Alachua | S | 8.9 | 4.1 | 349.8 | 85.3 | 6.3 ± 0.12 | 3.8 ± 0.12 | 3.9 |
| 3 | Albermale | S | 6.1 | 4.2 | 318.2 | 76.4 | 5.7 ± 0.10 | 3.0 ± 0.10 | 5.2 |
| 4 | Black Beauty | S | 15.9 | 3.8 | 332.8 | 86.8 | 6.0 ± 0.11 | 3.6 ± 0.10 | 2.1 |
| 5 | Black Fry | S | 10.5 | 3.5 | 307.8 | 88.0 | 6.2 ± 0.20 | 3.7 ± 0.17 | 2.9 |
| 6 | Carlos | S | 5.0 | 3.8 | 272.3 | 71.0 | 7.3 ± 0.06 | 4.8 ± 0.06 | 5.4 |
| 7 | Cowart | S | 7.1 | 4.0 | 366.7 | 91.7 | 6.0 ± 0.29 | 3.6 ± 0.11 | 5.2 |
| 8 | Darlene | S | 15.1 | 3.8 | 373.9 | 99.7 | 5.9 ± 0.10 | 3.8 ± 0.08 | 2.5 |
| 9 | Dixie | S | 5.1 | 3.7 | 251.8 | 68.7 | 6.7 ± 0.06 | 4.8 ± 0.15 | 5.0 |
| 10 | Dixie Red | S | 9.3 | 3.9 | 315.8 | 81.0 | 6.4 ± 0.09 | 3.8 ± 0.08 | 3.4 |
| 11 | Doreen | S | 10.7 | 3.5 | 354.5 | 101.3 | 6.2 ± 0.09 | 3.8 ± 0.08 | 3.3 |
| 12 | Farrer | S | 11.2 | 3.0 | 344.0 | 114.7 | 7.0 ± 0.13 | 3.9 ± 0.08 | 3.1 |
| 13 | Fry | S | 10.2 | 2.6 | 244.6 | 94.7 | 5.9 ± 0.08 | 3.8 ± 0.10 | 2.4 |
| 14 | Fry Seedless | SL | 2.5 | ||||||
| 15 | Granny Val | S | 11.7 | 3.1 | 298.3 | 96.8 | 7.0 ± 0.06 | 3.9 ± 0.18 | 2.6 |
| 16 | Higgins | S | 6.0 | 4.1 | 275.5 | 67.5 | 6.8 ± 0.18 | 4.6 ± 0.05 | 4.6 |
| 17 | Hunt | S | 5.5 | 4.2 | 268.5 | 63.9 | 5.5 ± 0.05 | 3.0 ± 0.06 | 4.9 |
| 18 | Ison | S | 6.7 | 3.8 | 332.0 | 86.6 | 7.9 ± 0.08 | 5.0 ± 0.07 | 5.0 |
| 19 | Jane Bell | S | 5.3 | 3.2 | 383.0 | 119.7 | 7.2 ± 0.08 | 4.1 ± 0.11 | 7.2 |
| 20 | Janet | S | 15.5 | 4.9 | 578.9 | 118.1 | 6.8 ± 0.06 | 3.9 ± 0.10 | 3.7 |
| 21 | Jumbo | S | 10.5 | 4.4 | 409.3 | 92.7 | 6.4 ± 0.07 | 3.4 ± 0.08 | 3.9 |
| 22 | Late Fry | S | 14.7 | 3.8 | 415.9 | 108.5 | 7.9 ± 0.06 | 5.6 ± 0.14 | 2.8 |
| 23 | Loomis | S | 10.2 | 3.4 | 319.1 | 93.4 | 7.4 ± 0.07 | 5.2 ± 0.05 | 3.1 |
| 24 | Magnolia | S | 7.8 | 3.7 | 219.8 | 59.9 | 7.5 ± 0.11 | 4.9 ± 0.09 | 2.8 |
| 25 | Nesbitt | S | 10.1 | 3.5 | 327.2 | 93.5 | 6.3 ± 0.08 | 3.7 ± 0.12 | 3.2 |
| 26 | Noble | S | 4.7 | 4.0 | 235.3 | 58.8 | 6.4 ± 0.05 | 4.6 ± 0.04 | 5.0 |
| 27 | Pam | S | 14.5 | 3.3 | 326.6 | 98.0 | 6.2 ± 0.16 | 3.8 ± 0.11 | 2.3 |
| 28 | Pineapple | S | 12.2 | 3.5 | 369.2 | 105.5 | 6.2 ± 0.08 | 3.9 ± 0.12 | 3.0 |
| 29 | Pride | S | 4.3 | 3.8 | 202.9 | 54.1 | 4.8 ± 0.04 | 3.1 ± 0.03 | 4.7 |
| 30 | Regale | S | 5.1 | 3.8 | 259.8 | 69.3 | 5.5 ± 0.05 | 3.2 ± 0.07 | 5.1 |
| 31 | Scarlet | S | 12.1 | 2.7 | 261.2 | 96.7 | 6.7 ± 0.14 | 4.0 ± 0.15 | 2.2 |
| 32 | Scuppernong | S | 10.2 | 2.3 | 215.7 | 92.4 | 6.2 ± 0.08 | 3.4 ± 0.15 | 2.1 |
| 33 | Southern Home | S | 6.3 | 3.4 | 204.2 | 59.8 | 5.9 ± 0.12 | 2.7 ± 0.04 | 3.3 |
| 34 | Southern Land | S | 6.8 | 3.7 | 280.8 | 76.6 | 6.0 ± 0.07 | 3.5 ± 0.11 | 4.2 |
| 35 | Sterling | S | 8.8 | 3.9 | 333.2 | 85.1 | 6.1 ± 0.11 | 3.7 ± 0.08 | 3.8 |
| 36 | Sugar Pop | S | 10.5 | 3.9 | 391.3 | 99.9 | 6.2 ± 0.11 | 3.8 ± 0.06 | 3.7 |
| 37 | Summit | S | 10.7 | 3.0 | 242.8 | 80.9 | 6.1 ± 0.06 | 3.2 ± 0.08 | 2.3 |
| 38 | Supreme | S | 15.5 | 2.8 | 300.0 | 105.9 | 6.3 ± 0.07 | 4.0 ± 0.09 | 1.9 |
| 39 | Sweet Jenny | S | 11.8 | 4.1 | 379.8 | 93.0 | 7.5 ± 0.10 | 5.2 ± 0.13 | 3.2 |
| 40 | Triumph | S | 9.8 | 3.8 | 249.3 | 66.5 | 6.0 ± 0.07 | 3.5 ± 0.09 | 2.5 |
| 41 | Watergate | S | 8.7 | 3.2 | 229.9 | 72.6 | 7.3 ± 0.15 | 4.9 ± 0.10 | 2.6 |
| 42 | Welder | S | 3.3 | 3.3 | 217.2 | 65.2 | 4.8 ± 0.04 | 3.5 ± 0.10 | 6.5 |
| Florida hybrid bunch | |||||||||
| 43 | Blanc du Bois | S | 1.7 | 1.9 | 55.1 | 28.3 | 3.2 | ||
| 44 | Stover | S | 2.0 | 1.3 | 61.9 | 48.5 | 5.1 ± 0.12 | 2.7 ± 0.06 | 3.1 |
| Bunch grape | |||||||||
| 45 | Riesling | S | 1.6 | 3.6 | 107.9 | 30.1 | 4.0 ± 0.13 | 2.0 ± 0.10 | 6.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.A.; Balasubramani, S.P.; Basha, S.M. Molecular Characterization and Phylogenetic Analysis of MADS-Box Gene VroAGL11 Associated with Stenospermocarpic Seedlessness in Muscadine Grapes. Genes 2021, 12, 232. https://doi.org/10.3390/genes12020232
Rahman MA, Balasubramani SP, Basha SM. Molecular Characterization and Phylogenetic Analysis of MADS-Box Gene VroAGL11 Associated with Stenospermocarpic Seedlessness in Muscadine Grapes. Genes. 2021; 12(2):232. https://doi.org/10.3390/genes12020232
Chicago/Turabian StyleRahman, M Atikur, Subramani P Balasubramani, and Sheikh M Basha. 2021. "Molecular Characterization and Phylogenetic Analysis of MADS-Box Gene VroAGL11 Associated with Stenospermocarpic Seedlessness in Muscadine Grapes" Genes 12, no. 2: 232. https://doi.org/10.3390/genes12020232






