Evolution of the IRF Family in Salmonids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phylogenetic and Gene Synteny Analysis
2.2. IRF Primer Design
2.3. Animal Work
2.4. RNA Extraction and Reverse Transcription
2.5. Quantitative Real-Time PCR
2.6. Statistical Analysis
3. Results
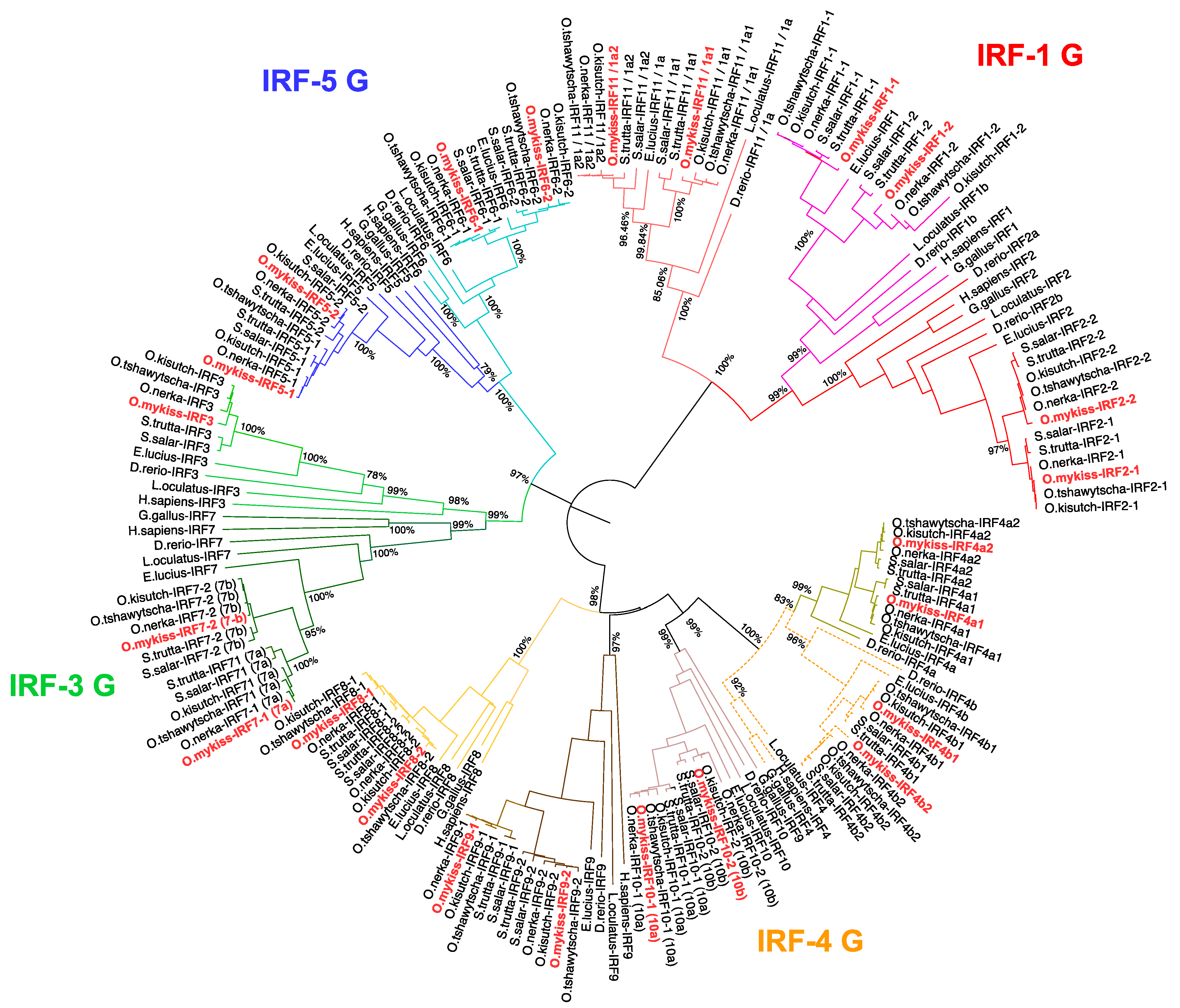
3.1. Phylogenetic Analysis of Salmonid IRF Family
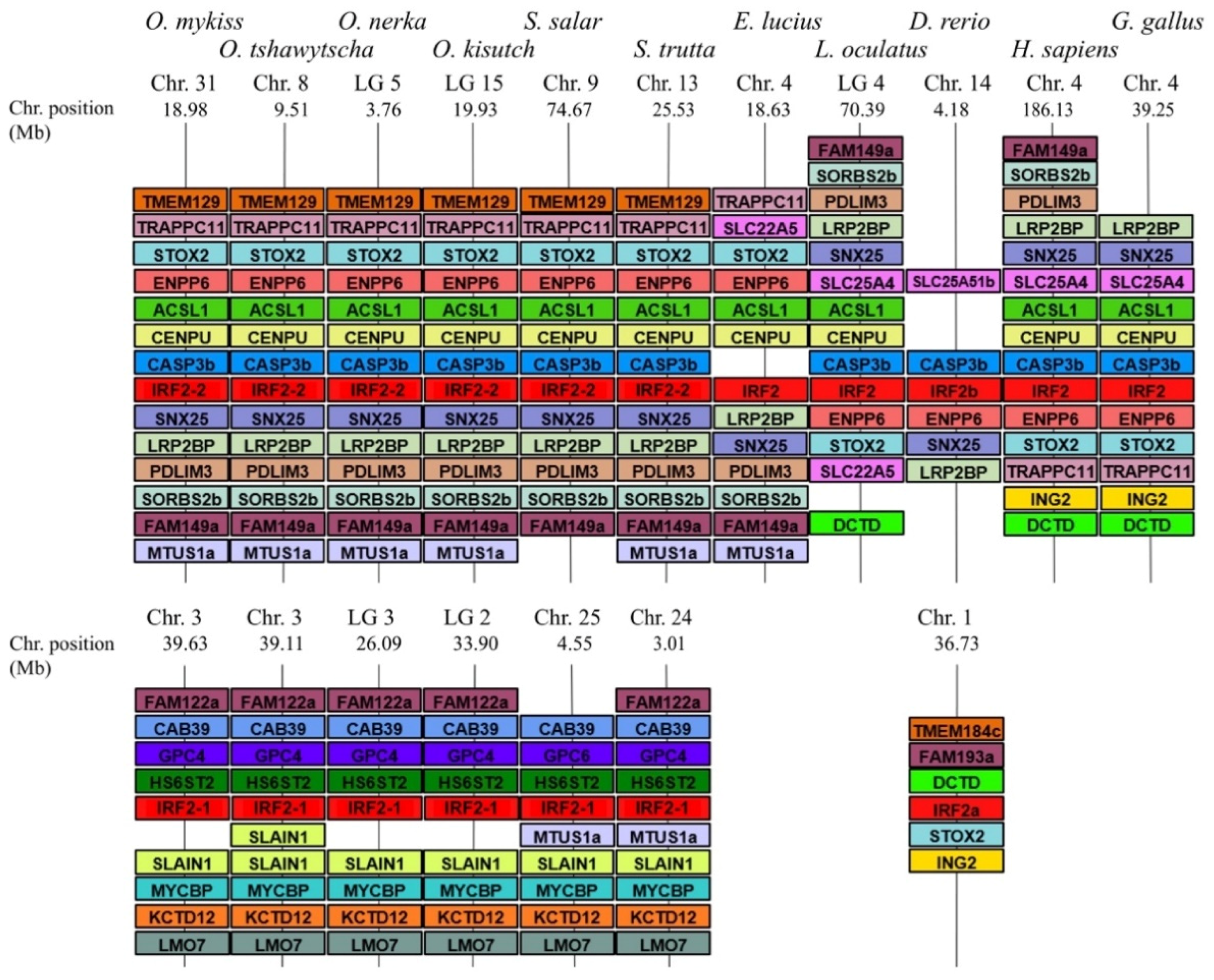
3.2. Comparative Phylogenetic and Synteny Analysis on the Case of Group 1 Salmonid Irfs
3.3. A Consistent Nomenclature for Salmonid IRF Family
3.4. Constitutive mRNA Expression Levels of the IRF Gene Family
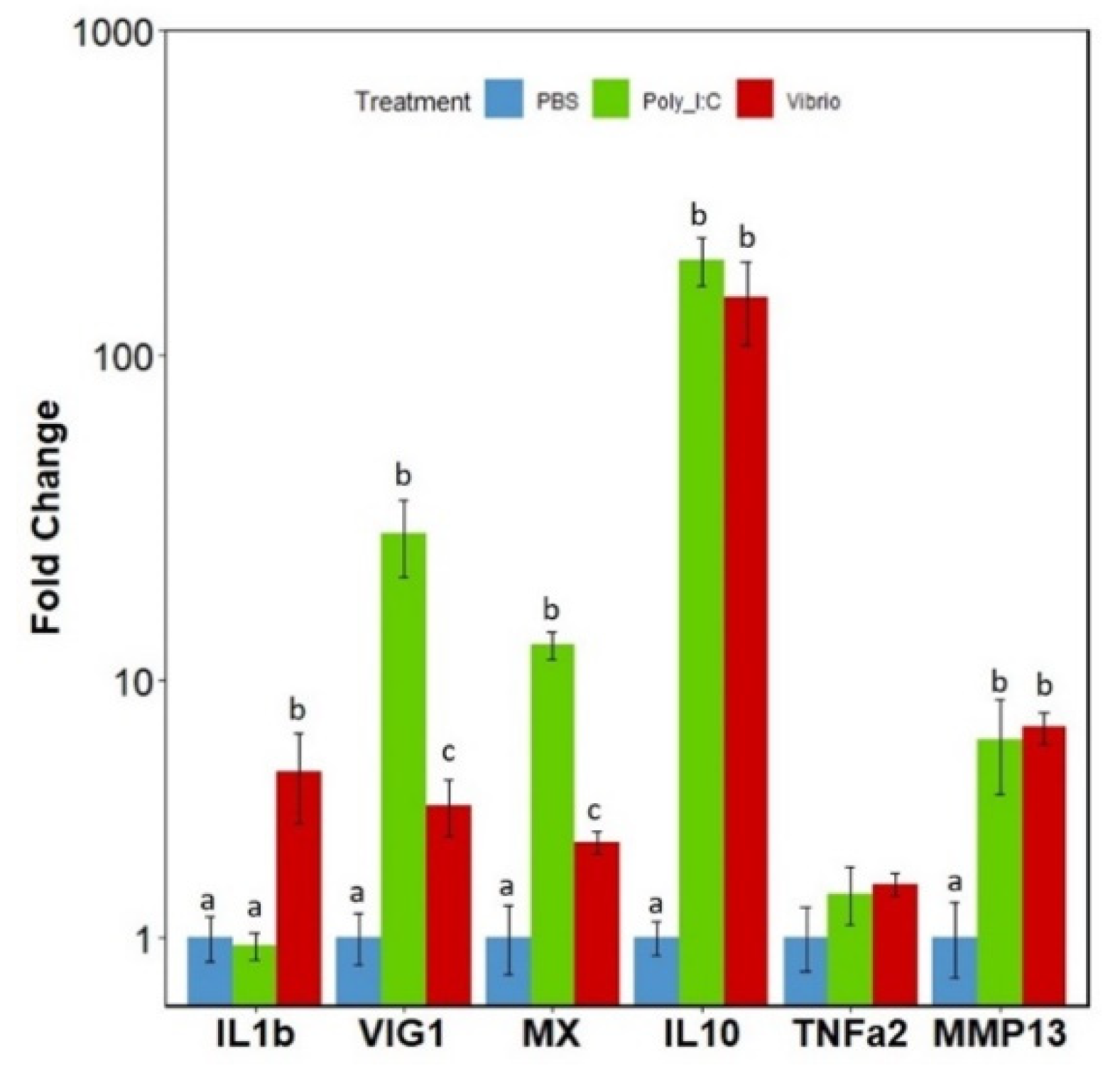
3.5. mRNA Expression Levels of the IRFs in Response to Poly I:C or Vibrio Extract
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nehyba, J.; Hrdličková, R.; Bose, H.R. Dynamic Evolution of Immune System Regulators: The History of the Interferon Regulatory Factor Family. Mol. Biol. Evol. 2009, 26, 2539–2550. [Google Scholar] [CrossRef]
- Yanai, H.; Negishi, H.; Taniguchi, T. The IRF Family of Transcription Factors Inception, Impact and Implications in Oncogenesis. OncoImmunology 2012, 1376–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Royer William, E.J.E. Structural Insights into Interferon Regulatory Factor Activation. Cell. Signal. 2010, 22, 883–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikushima, H.; Negishi, H.; Taniguchi, T. The IRF Family Transcription Factors at the Interface of Innate and Adaptive Immune Responses. Cold Spring Harb. Symp. Quant. Biol. 2013, 78, 105–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jefferies, C.A. Regulating IRFs in IFN Driven Disease. In Frontiers in Immunology; Frontiers Media: Lausanne, Switzerland, 2019; p. 325. [Google Scholar] [CrossRef] [Green Version]
- Antonczyk, A.; Krist, B.; Sajek, M.; Michalska, A.; Piaszyk-Borychowska, A.; Plens-Galaska, M.; Wesoly, J.; Bluyssen, H.A.R. Direct Inhibition of IRF-Dependent Transcriptional Regulatory Mechanisms Associated with Disease. Front. Immunol. 2019, 10, 1176. [Google Scholar] [CrossRef] [Green Version]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I Inteferon Gene Induction by the Interferon Regulatory Factor Family of Transcription Factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Paun, A.; Pitha, P.M. The IRF Family, Revisited. Biochimie 2007, 89, 744–753. [Google Scholar] [CrossRef]
- Eguchi, J.; Yan, Q.W.; Schones, D.E.; Kamal, M.; Hsu, C.H.; Zhang, M.Q.; Crawford, G.E.; Rosen, E.D. Interferon Regulatory Factors Are Transcriptional Regulators of Adipogenesis. Cell Metab. 2008, 7, 86–94. [Google Scholar] [CrossRef] [Green Version]
- Gabriele, L.; Ozato, K. The Role of the Interferon Regulatory Factor (IRF) Family in Dendritic Cell Development and Function. Cytokine Growth Factor Rev. 2007, 18, 503–510. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, K.; Peng, L.; Xiong, H. Regulation of T Helper Cell Differentiation by Interferon Regulatory Factor Family Members. Immunol. Res. 2012, 54, 169–176. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Myasoedova, V.A.; Revin, V.V.; Orekhov, A.N.; Bobryshev, Y.V. The Impact of Interferon-Regulatory Factors to Macrophage Differentiation and Polarization into M1 and M2. Immunobiology 2018, 223, 101–111. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C. The Toll Receptor Family and Microbial Recognition. Trends Microbiol. 2000, 8, 452–456. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA Helicase RIG-I Has an Essential Function in Double-Stranded RNA-Induced Innate Antiviral Responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Langer, J.A.; Cutrone, E.C.; Kotenko, S. The Class II Cytokine Receptor (CRF2) Family: Overview and Patterns of Receptor-Ligand Interactions. Cytokine Growth Factor Rev. 2004, 15, 33–48. [Google Scholar] [CrossRef]
- Pestka, S.; Krause, C.D.; Walter, M.R. Interferons, Interferon-like Cytokines, and Their Receptors. Immunol. Rev. Immunol. Rev. Dec. 2004, 202, 8–32. [Google Scholar] [CrossRef]
- Levraud, J.-P.; Boudinot, P.; Colin, I.; Benmansour, A.; Peyrieras, N.; Herbomel, P.; Lutfalla, G. Identification of the Zebrafish IFN Receptor: Implications for the Origin of the Vertebrate IFN System. J. Immunol. 2007, 178, 4385–4394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, O.; Akira, S. Innate Immunity to Virus Infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H. IRF and STAT Transcription Factors—From Basic Biology to Roles in Infection, Protective Immunity, and Primary Immunodeficiencies. Front. Immunol. 2019, 9, 3047. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E.; Kerr, I.M.; Stark, G.R. Jak-STAT Pathways and Transcriptional Activation in Response to IFNs and Other Extracellular Signaling Proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bluyssen, H.A.R.; Durbin, J.E.; Levy, D.E. ISGF3γ P48, a Specificity Switch for Interferon Activated Transcription Factors. Cytokine Growth Factor Rev. 1996, 7, 11–17. [Google Scholar] [CrossRef]
- Taniguchi, T.; Ogasawara, K.; Takaoka, A.; Tanaka, N. IRF Family of Transcription Factors as Regulators of Host Defense. Annu. Rev. Immunol. 2001, 19, 623–655. [Google Scholar] [CrossRef] [PubMed]
- Collet, B. Innate Immune Responses of Salmonid Fish to Viral Infections. Dev. Comp. Immunol. 2014, 43, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Qi, Z.T.; Xu, Z.; Nie, P. Global Characterization of Interferon Regulatory Factor (IRF) Genes in Vertebrates: Glimpse of the Diversification in Evolution. BMC Immunol. 2010, 5, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The Atlantic Salmon Genome Provides Insights into Rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef] [Green Version]
- Bergan, V.; Kileng, Ø.; Sun, B.; Robertsen, B. Regulation and Function of Interferon Regulatory Factors of Atlantic Salmon. Mol. Immunol. 2010, 47, 2005–2014. [Google Scholar] [CrossRef]
- Xu, Q.; Jiang, Y.; Wangkahart, E.; Zou, J.; Chang, M.; Yang, D.; Secombes, C.J.; Nie, P.; Wang, T. Sequence and Expression Analysis of Interferon Regulatory Factor 10 (IRF10) in Three Diverse Teleost Fish Reveals Its Role in Antiviral Defense. PLoS ONE 2016, 11, e0147181. [Google Scholar] [CrossRef] [Green Version]
- Macqueen, D.J.; Johnston, I.A. A Well-Constrained Estimate for the Timing of the Salmonid Whole Genome Duplication Reveals Major Decoupling from Species Diversification. Proc. R. Soc. B Biol. Sci. 2014, 281, 1778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hernández, P.P.; Strzelecka, P.M.; Athanasiadis, E.I.; Hall, D.; Robalo, A.F.; Collins, C.M.; Boudinot, P.; Levraud, J.P.; Cvejic, A. Single-Cell Transcriptional Analysis Reveals ILC-like Cells in Zebrafish. Sci. Immunol. 2018, 3, eaau5265. [Google Scholar] [CrossRef] [Green Version]
- Yano, A.; Guyomard, R.; Nicol, B.; Jouanno, E.; Quillet, E.; Klopp, C.; Cabau, C.; Bouchez, O.; Fostier, A.; Guiguen, Y. An Immune-Related Gene Evolved into the Master Sex-Determining Gene in Rainbow Trout, Oncorhynchus Mykiss. Curr. Biol. 2012, 22, 1423–1428. [Google Scholar] [CrossRef] [Green Version]
- Inkpen, S.M.; Solbakken, M.H.; Jentoft, S.; Eslamloo, K.; Rise, M.L. Full Characterization and Transcript Expression Profiling of the Interferon Regulatory Factor (IRF) Gene Family in Atlantic Cod (Gadus Morhua). Dev. Comp. Immunol. 2019, 98, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Qi, C.; Shan, S.; Zhang, F.; Li, H.; An, L.; Yang, G. Characterization of Common Carp (Cyprinus Carpio L.) Interferon Regulatory Factor 5 (IRF5) and Its Expression in Response to Viral and Bacterial Challenges. BMC Vet. Res 2016, 12, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, R.; Takizawa, F.; Chaara, W.; Lunazzi, A.; Dang, T.H.; Koellner, B.; Quillet, E.; Six, A.; Fischer, U.; Boudinot, P. Contrasted TCRβ Diversity of CD8+ and CD8− T Cells in Rainbow Trout. PLoS ONE 2013, 8, e60175. [Google Scholar] [CrossRef] [Green Version]
- Langevin, C.; Blanco, M.; Martin, S.A.M.; Jouneau, L.; Bernardet, J.-F.; Houel, A.; Lunazzi, A.; Duchaud, E.; Michel, C.; Quillet, E.; et al. Transcriptional Responses of Resistant and Susceptible Fish Clones to the Bacterial Pathogen Flavobacterium Psychrophilum. PLoS ONE 2012, 7, e39126. [Google Scholar] [CrossRef] [Green Version]
- Castro, R.; Jouneau, L.; Tacchi, L.; Macqueen, D.J.; Alzaid, A.; Secombes, C.J.; Martin, S.A.M.; Boudinot, P. Disparate Developmental Patterns of Immune Responses to Bacterial and Viral Infections in Fish. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Boudinot, P.; Bird, S.; Du Pasquier, L.; Collet, B. The Repertoire of Vertebrate STAT Transcription Factors: Origin and Variations in Fish. Dev. Comp. Immunol. 2021, 116, 103929. [Google Scholar] [CrossRef]
- Froschauer, A.; Braasch, I.; Volff, J. Fish Genomes, Comparative Genomics and Vertebrate Evolution. Curr. Genom. 2006, 7, 43–57. [Google Scholar] [CrossRef]
- Jiang, Y.; Gao, X.; Liu, S.; Zhang, Y.; Liu, H.; Sun, F.; Bao, L.; Waldbieser, G.; Liu, Z. Whole Genome Comparative Analysis of Channel Catfish (Ictalurus Punctatus) with Four Model Fish Species. BMC Genom. 2013, 14, 780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briolat, V.; Jouneau, L.; Carvalho, R.; Palha, N.; Langevin, C.; Herbomel, P.; Schwartz, O.; Spaink, H.P.; Levraud, J.-P.; Boudinot, P. Contrasted Innate Responses to Two Viruses in Zebrafish: Insights into the Ancestral Repertoire of Vertebrate IFN-Stimulated Genes. J. Immunol. 2014, 192, 4328–4341. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant Shark Genome Provides Unique Insights into Gnathostome Evolution. Nature 2014, 505, 174–179. [Google Scholar] [CrossRef] [Green Version]
- Langham, R.J.; Walsh, J.; Dunn, M.; Ko, C.; Goff, S.A.; Freeling, M. Genomic Duplication, Fractionation and the Origin of Regulatory Novelty. Genetics 2004, 166, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noël, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The Rainbow Trout Genome Provides Novel Insights into Evolution after Whole-Genome Duplication in Vertebrates. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glasauer, S.M.K.; Neuhauss, S.C.F. Whole-Genome Duplication in Teleost Fishes and Its Evolutionary Consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [Green Version]
- Pasquier, J.; Cabau, C.; Nguyen, T.; Jouanno, E.; Severac, D.; Braasch, I.; Journot, L.; Pontarotti, P.; Klopp, C.; Postlethwait, J.H.; et al. Gene Evolution and Gene Expression after Whole Genome Duplication in Fish: The PhyloFish Database. BMC Genom. 2016, 17, 368. [Google Scholar] [CrossRef]
- Bowie, A.G.; Unterholzner, L. Viral Evasion and Subversion of Pattern-Recognition Receptor Signalling. Nat. Rev. Immunol. 2008, 8, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.L.; Puri, M.; Horvath, C.M.; Sen, G.C. Select Paramyxoviral V Proteins Inhibit IRF3 Activation by Acting as Alternative Substrates for Inhibitor of ΚB Kinase ε (IKKe)/TBK1. J. Biol. Chem. 2008, 283, 14269–14276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehler, C.E.; Lester, K.; Della Pelle, G.; Jouneau, L.; Houel, A.; Collins, C.; Dovgan, T.; Machat, R.; Zou, J.; Boudinot, P.; et al. Viral Resistance and IFN Signaling in STAT2 Knockout Fish Cells. J. Immunol. 2019, 203, 465–475. [Google Scholar] [CrossRef] [Green Version]
- Tamura, T.; Yanai, H.; Savitsky, D.; Taniguchi, T. The IRF Family Transcription Factors in Immunity and Oncogenesis. Annu. Rev. Immunol. 2008, 26, 535–584. [Google Scholar] [CrossRef]
- Negishi, H.; Taniguchi, T.; Yanai, H. The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harb. Perspect. Biol. 2018, 10, a028423. [Google Scholar] [CrossRef]
- Taniguchi, T.; Takaoka, A. The Interferon-α/β System in Antiviral Responses: A Multimodal Machinery of Gene Regulation by the IRF Family of Transcription Factors. Curr. Opin. Immunol. 2002, 14, 111–116. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; McWhirter, S.M.; Faia, K.L.; Rowe, D.C.; Latz, E.; Golenbock, D.T.; Coyle, A.J.; Liao, S.M.; Maniatis, T. IKKE and TBKI Are Essential Components of the IRF3 Signalling Pathway. Nat. Immunol. 2003, 4, 491–496. [Google Scholar] [CrossRef]
- Seth, R.B.; Sun, L.; Chen, Z.J. Antiviral Innate Immunity Pathways. Cell Res. 2006, 16, 141–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forero, A.; Ozarkar, S.; Li, H.; Lee, C.H.; Hemann, E.A.; Nadjsombati, M.S.; Hendricks, M.R.; So, L.; Green, R.; Roy, C.N.; et al. Differential Activation of the Transcription Factor IRF1 Underlies the Distinct Immune Responses Elicited by Type I and Type III Interferons. Immunity 2019, 51, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Zhong, Z.; Fang, C.; Dai, W.; Shen, Y.; Gan, X.; He, S. Ancient Duplications and Functional Divergence in the Interferon Regulatory Factors of Vertebrates Provide Insights into the Evolution of Vertebrate Immune Systems. Dev. Comp. Immunol. 2018, 81, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Hida, S.; Ogasawara, K.; Sato, K.; Abe, M.; Takayanagi, H.; Yokochi, T.; Sato, T.; Hirose, S.; Shirai, T.; Taki, S.; et al. CD8+ T Cell-Mediated Skin Disease in Mice Lacking IRF-2, the Transcriptional Attenuator of Interferon-α/β Signaling. Immunity 2000, 13, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Negishi, H.; Ohba, Y.; Yanai, H.; Takaoka, A.; Honma, K.; Yui, K.; Matsuyama, T.; Taniguchi, T.; Honda, K. Negative Regulation of Toll-like-Receptor Signaling by IRF-4. Proc. Natl. Acad. Sci. USA 2005, 102, 15989–15994. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Wang, T.; Petit, J.; Forlenza, M.; Chen, X.; Chen, L.; Zou, J.; Secombes, C.J. Evolution of IFN Subgroups in Bony Fish—2. Analysis of Subgroup Appearance and Expansion in Teleost Fish with a Focus on Salmonids. Fish Shellfish Immunol. 2020, 98, 564–573. [Google Scholar] [CrossRef]
- Aggad, D.; Mazel, M.; Boudinot, P.; Mogensen, K.E.; Hamming, O.J.; Hartmann, R.; Kotenko, S.; Herbomel, P.; Lutfalla, G.; Levraud, J.-P. The Two Groups of Zebrafish Virus-Induced Interferons Signal via Distinct Receptors with Specific and Shared Chains. J. Immunol. 2009, 183, 3924–3931. [Google Scholar] [CrossRef] [Green Version]
- De Weerd, N.A.; Nguyen, T. The Interferons and Their Receptors-Distribution and Regulation. Immunol. Cell Biol. 2012, 90, 483–491. [Google Scholar] [CrossRef]
- Sun, B.; Greiner-Tollersrud, L.; Koop, B.F.; Robertsen, B. Atlantic Salmon Possesses Two Clusters of Type I Interferon Receptor Genes on Different Chromosomes, Which Allows for a Larger Repertoire of Interferon Receptors than in Zebrafish and Mammals. Dev. Comp. Immunol. 2014, 47, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Laghari, Z.A.; Li, L.; Chen, S.N.; Huo, H.J.; Huang, B.; Zhou, Y.; Nie, P. Composition and Transcription of All Interferon Regulatory Factors (IRFs), IRF1‒11 in a Perciform Fish, the Mandarin Fish, Siniperca Chuatsi. Dev. Comp. Immunol. 2018, 81, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Gong, X.Y.; Li, Y.L.; Dan, C.; Gui, J.F.; Zhang, Y.B. Characterization of DNA Binding and Nuclear Retention Identifies Zebrafish IRF11 as a Positive Regulator of IFN Antiviral Response. J. Immunol. 2020, 205, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Sun, Y.; Xu, T. Molecular Characterization of Three IRF1 Subfamily Members Reveals Evolutionary Significance of IRF11 in Miiuy Croaker. Dev. Comp. Immunol. 2015, 53, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lu, L.-F.; Feng, H.; Wu, N.; Chen, D.-D.; Zhang, Y.-B.; Gui, J.-F.; Nie, P.; Zhang, Y.-A. IFN Regulatory Factor 10 Is a Negative Regulator of the IFN Responses in Fish. J. Immunol. 2014, 193, 1100–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Shan, S.; Zhao, H.; Liu, R.; Wang, H.; Chen, X.; Yang, G.; Li, H. Identification of an IRF10 Gene in Common Carp (Cyprinus Carpio L.) and Analysis of Its Function in the Antiviral and Antibacterial Immune Response. BMC Vet. Res. 2020, 16, 450. [Google Scholar] [CrossRef] [PubMed]



| Gene ID | Gene Name | Direction | Sequence | Annealing | Product Size (bps) | Accession |
|---|---|---|---|---|---|---|
| 100135845 | β actin 1 | Forward | GGTGGTAGGCCAGAGGC | 60 | 101 | NM_001124235.1 |
| Reverse | GGGAGAAGATGACCCAGATCATG | |||||
| 100136024 | IL1b 2 | Forward | GGAGAGGTTAAAGGGTGGCGA | 60 | 121 | XM_036979104 |
| Reverse | TGCCGACTCCAACTCCAACA | |||||
| 110494493 | MX3 | Forward | CCTCCTGAAATCAGCGAAGAC | 60 | 364 | XM_021569609.2 |
| Reverse | GAGTCTGAAGCATCTCCCTCCTG | |||||
| 100136835 | IL10 2 | Forward | CGACTTTAAATCTCCCATCGAC | 60 | 70 | NM_001245099.1 |
| Reverse | GCATTGGACGATCTCTTTCTTC | |||||
| 100135876 | VIG1 3 | Forward | GGCAACTCCAAGCAGTGTCAA | 60 | 187 | XM_021582972.2 |
| Reverse | GTCGTGTATGAAAGGCTCTCCG | |||||
| 100136064 | TNFa2 2 | Forward | GGAGGCTGTGTGGCGTTCT | 60 | 73 | NM_001124374.1 |
| Reverse | TGCTGACACCAGGCAAAGAG | |||||
| 100136017 | MMP13 2 | Forward | GCACCTTCTCTCTGCCCCGC | 60 | 235 | XM_021618131.2 |
| Reverse | AGGCTCTGTTGTGGTTTGCTGC | |||||
| 110499402 | IRF11-1 (a1) | Forward | TTGATGAGACAGCTCAAGTTTTC | 55 | 146 | XM_021576528.1 |
| Reverse | CTTAGGATCAGGTTCGTCTTTC | |||||
| 110502724 | IRF11-2 (a2) | Forward | CCAGGGGTCACCTGGCG | 62 | 169 | XM_021580977.1 |
| Reverse | TCCATGTCTTCGGATCAGGC | |||||
| 110533376 | IRF1-1 | Forward | TTACAAAATGCTGAGCGTCAG | 55 | 237 | XM_021617512.1 |
| Reverse | GTCTCCCCTACGTTGTCTGA | |||||
| 100135950 | IRF1-2 | Forward | GATGAAGAACGTCCACTCC | 55 | 231 | NM_001124293.1 |
| Reverse | AAATCATCTAGGCTGTCTGT | |||||
| 110519953 | IRF2-1 | Forward | ATGCGAATGCGACCATGGC | 55 | 195 | XM_021596860.1 |
| Reverse | GTATGAATGGCCCAGTTCTTG | |||||
| 100136151 | IRF2-2 | Forward | TGGAACAGATAAACTCTTC | 62 | 130 | NM_001124438.1 |
| Reverse | ATAAATAAAGGAGCGTCTTTC | |||||
| 100750229 | IRF3 | Forward | AGCAATGGTAGGGTTCAAGG | 60 | 179 | NM_001257262.1 |
| Reverse | CATCTGGCCACTGGAACAG | |||||
| 100499175 | IRF4-a1 | Forward | CCCACATGAGCTCAGTCAATAG | 60 | 139 | XM_021613502.1 |
| Reverse | GGGTCGGCTGAGTGGCTG | |||||
| 100499174 | IRF4-a2 | Forward | CGATCAGATTAACAGCAGTAG | 60 | 130 | NM_001310139.1 |
| Reverse | CATCCTCCTCTCGATTGTAG | |||||
| 110521762 | IRF4-b1 | Forward | GCTCGTGCAGCGAAGTCAG | 60 | 181 | XM_021599601.1 |
| Reverse | AGGCATCTGTGTCTGCAGG | |||||
| 110524663 | IRF4-b2 | Forward | TCCGGATTCGGACTACGGC | 60 | 110 | XM_021604510.1 |
| Reverse | TCTCCCACACGAGGCCTGC | |||||
| 110500261 | IRF5-1 | Forward | AGCATTACCATGGCAGCGC | 60 | 130 | XM_021577521.1 |
| Reverse | TGTTGGAGGGTCCTACCG | |||||
| 110500261 | IRF5-2 | Forward | AGCATTACCATGGCAGCGC | 60 | 130 | XM_021577521.1 |
| Reverse | TGTTGGAGGGTCCTACCG | |||||
| 110528098 | IRF6-1 | Forward | GGATGAAGATGAATCAGATGGC | 60 | 209 | XM_021609915.1 |
| Reverse | GGGACGAAGGCTGCATCTC | |||||
| 110494340 | IRF6-2 | Forward | AGACAACAAGCGCTTCAGGG | 60 | 111 | XM_021569306.1 |
| Reverse | TGGAACTTTCCTGTCTCCAC | |||||
| 100750228 | IRF7-1 (a) | Forward | AGCAATACACTGGTTTGTTC | 60 | 145 | XM_021600499 |
| Reverse | GTGGGATGCTCATTGATTTTC | |||||
| 110497044 | IRF7-2 (b) | Forward | GCCGGGTTGTGTTTTGTG | 60 | 144 | XM_021573049.1 |
| Reverse | CTTGTCATTGGGATGCGTG | |||||
| 110526480 | IRF8-1 | Forward | TGGGAGGACGACAGTCGCAC | 60 | 95 | XM_021607480.1 |
| Reverse | GCCTTGAAGATAGAGGCGTCG | |||||
| 110506608 | IRF8-2 | Forward | GTCTGGGAGGACGACAGC | 60 | 98 | XM_021586344.1 |
| Reverse | GCCTTGAAGATAGAAGCGTCT | |||||
| 110535315 | IRF9-1 | Forward | TCCGATGGGGGTCGTGTG | 60 | 160 | XM_021620234.1 |
| Reverse | CCAACACTTGTTCATTCATC | |||||
| 110489699 | IRF9-2 | Forward | TGTCTGAGGGGTGTCATGC | 60 | 146 | XM_021562471 |
| Reverse | GATGGGTACGAGGCGGTAG | |||||
| 110492403 | IRF10-1 (a) | Forward | CTTACCTGGGAGAACGAAG | 60 | 151 | XM_021566691.1 |
| Reverse | GACGAGTCTTCCAGGTG | |||||
| 110532004 | IRF10-2 (b) | Forward | ATCTGAATGAAGATGCAGCC | 60 | 172 | XM_021615574.1 |
| Reverse | CGCTCTGGGACCTCCTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clark, T.C.; Boudinot, P.; Collet, B. Evolution of the IRF Family in Salmonids. Genes 2021, 12, 238. https://doi.org/10.3390/genes12020238
Clark TC, Boudinot P, Collet B. Evolution of the IRF Family in Salmonids. Genes. 2021; 12(2):238. https://doi.org/10.3390/genes12020238
Chicago/Turabian StyleClark, Thomas C., Pierre Boudinot, and Bertrand Collet. 2021. "Evolution of the IRF Family in Salmonids" Genes 12, no. 2: 238. https://doi.org/10.3390/genes12020238
APA StyleClark, T. C., Boudinot, P., & Collet, B. (2021). Evolution of the IRF Family in Salmonids. Genes, 12(2), 238. https://doi.org/10.3390/genes12020238






