Abstract
The Drosophila ovary is recognized as a powerful model to study stem cell self-renewal and differentiation. Decapentaplegic (Dpp) is secreted from the germline stem cell (GSC) niche to activate Bone Morphogenic Protein (BMP) signaling in GSCs for their self-renewal and is restricted in the differentiation niche for daughter cell differentiation. Here, we report that Switch/sucrose non-fermentable (SWI/SNF) component Osa depletion in escort cells (ECs) results in a blockage of GSC progeny differentiation. Further molecular and genetic analyses suggest that the defective germline differentiation is partially attributed to the elevated dpp transcription in ECs. Moreover, ectopic Engrailed (En) expression in osa-depleted ECs partially contributes to upregulated dpp transcription. Furthermore, we show that Osa regulates germline differentiation in a Brahma (Brm)-associated protein (BAP)-complex-dependent manner. Additionally, the loss of EC long cellular processes upon osa depletion may also partly contribute to the germline differentiation defect. Taken together, these data suggest that the epigenetic factor Osa plays an important role in controlling EC characteristics and germline lineage differentiation.
1. Introduction
The self-renewal of adult stem cells and the differentiation of their daughters are critical for homeostasis in many tissues, which are commonly regulated by microenvironments, also known as niches. The niche is composed of several different cell types of somatic lineage, which controls stem cell lineage extrinsically through intercellular signaling pathways [1,2]. It is important to understand how the niche functions to regulate tissue homeostasis.
Drosophila germline stem cells (GSCs) and their niches are an attractive system for studying the interactions between stem cells and the niche [3,4]. GSCs reside at the anterior tips of the ovaries in a structure called a germarium. The terminal filament (TF), cap cells (CpCs), and anterior-most escort cells (ECs) form a GSC niche [5,6]. Two to three GSCs are harbored in the niche via physical interactions with CpCs and anterior-most ECs [6,7]. The GSC daughters exit the GSC niche and are then enveloped by the cellular protrusions extended by the ECs, which transport the dividing germline cysts from the anterior portion of the germarium to the posterior [8,9] (Figure 1A). The GSCs are maintained by the Bone Morphogenic Protein (BMP) signaling activity, whose ligand, Decapentaplegic (Dpp), is principally emitted by the CpCs [7,10]. The differentiation factor bam is repressed by the phosphorylated Mothers against dpp (pMad), which is activated by BMP signaling activity [11]. When a GSC daughter cell exits the GSC niche, BMP signaling activity is diminished, and bam transcription is derepressed, which will promote the GSC daughter cell’s differentiation as a cystoblast (CB) [10,11,12,13,14]. The extrinsic cues of germline differentiation mainly come from the differentiation niche constituted by somatic ECs [8,9,15,16,17]. The expression of Thickveins (Tkv) [18] and suppression of Dally [19] in ECs are critical to restrict BMP signaling within a one-cell-diameter range to promote GSC daughter cell differentiation. EC protrusions are also essential for germ cell differentiation [8].
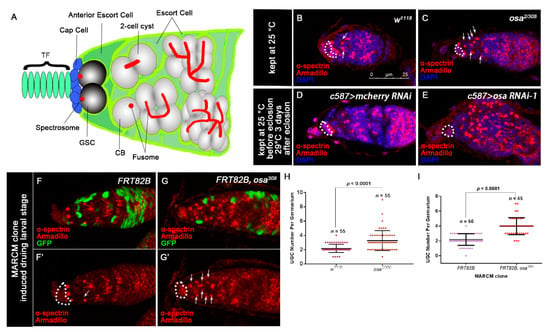
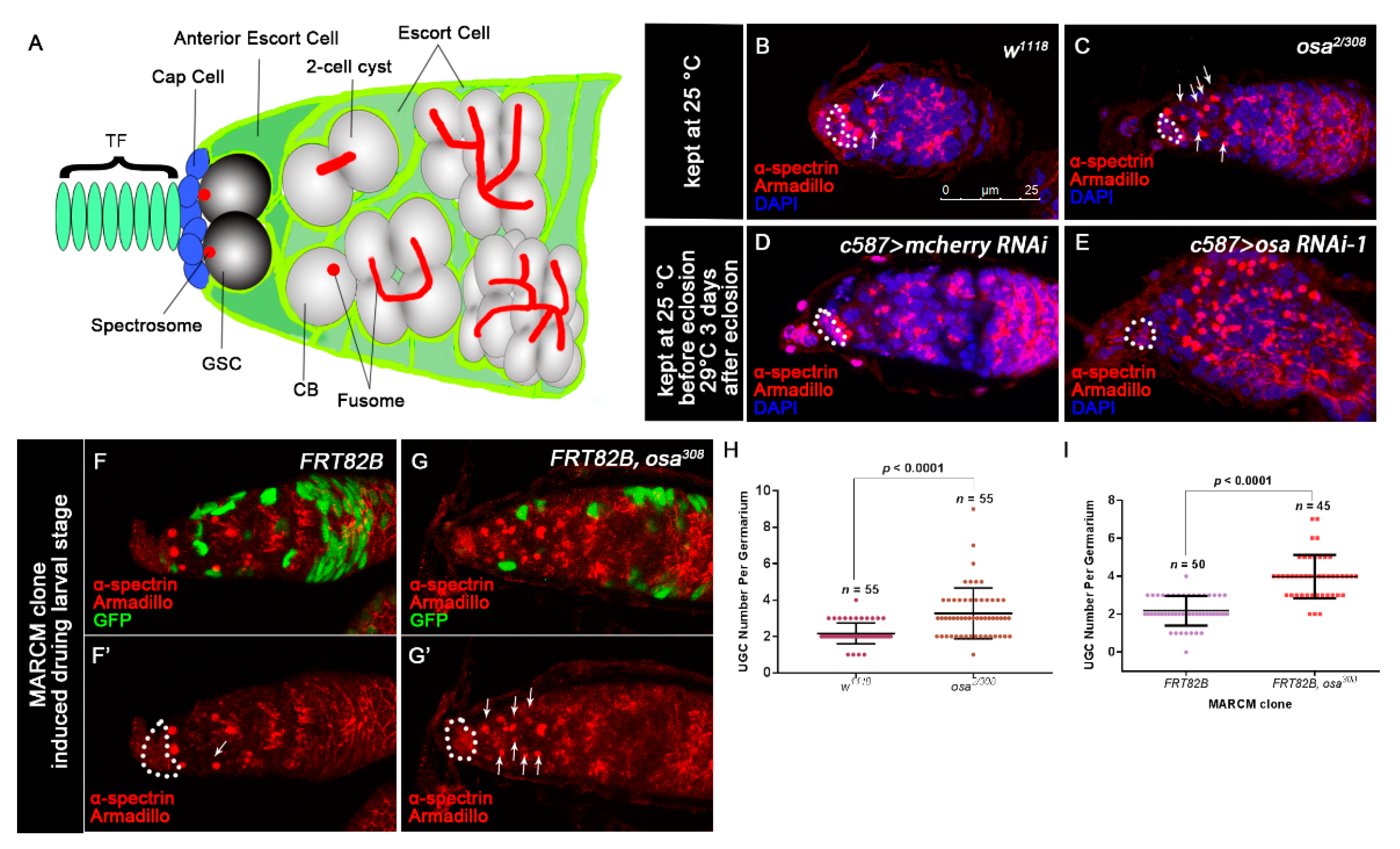
Figure 1.
Disruption of Osa results in the accumulation of round fusome-containing germ cells. (A) Schematic of a Drosophila germarium. (B–E) Germaria stained for α-spectrin (red), Armadillo (red) and DAPI (blue). CpCs are indicated by white dashed circles. (B) The wild-type (WT) (w1118) germarium contains three germline stem cells (GSCs) and two cystoblasts (CBs, indicated by white arrows). (C) The osa2/308 germarium displays an expanded number of UGCs outside the GSC niche (indicated by white arrows). The control germarium (D) contains a normal number of CBs. The osa KD germarium (E) displays an expanded number of UGCs. c587 > osa RNAi-2 gives a similar phenotype and is not shown in the figure. (F–G’) MARCM clones were stained for α-spectrin (red), Armadillo (red), and GFP (green). Clonal cells are marked by GFP. CpCs are indicated by white dashed circles. (F,F’) A germarium containing control clones displaying normal number of CBs (indicated by white arrows). (G,G’) A germarium containing the escort cell (EC) mutant for the osa308 exhibiting an expanded number of UGCs (indicated by white arrows). (H) A graph showing the quantification of UGCs for each germarium. (I) A graph showing the quantification of UGCs for each germarium. Error bars are presented as the Mean ± SD. Several compressed z-sections are shown in (B–G’). The scale bar is shown in (B).
Several epigenetic regulators have been reported to be active in ECs to restrict ectopic BMP signaling in the differentiation niche. Eggless (Egg), a H3K9 methyltransferase in Drosophila [20]; Lysine Specific Demethylase 1 (Lsd1) [21,22]; the piRNA pathway component, Piwi [23,24]; dSet-1 (Set1), a H3K4 trimethylase [25]; Polycomb and Trithorax group genes [26]; and Histone H1 [27] in ECs participate in dpp repression and thus secure germline differentiation. The Switch/sucrose non-fermentable (SWI/SNF) complex is a conserved chromatin remolding complex containing two subtypes in Drosophila, the Brahma (Brm)-associated protein (BAP) complex and the polybromo-containing BAP (PBAP) complex [28]. Brm, Moira (Mor), and Snf5-related 1 (Snr1) are the common components of both subtypes [29,30]. Brm is the catalytic subunit containing DNA-stimulated ATPase, while Moira and Snr1 are required for stable complex assembly [31,32,33,34]. Osa is specific to the BAP complex and is required for the recruitment of the complex to specific loci [35]. Brahma-associated protein 170kD (Bap170) and Polybromo are the signature subunits of the PBAP complex [35,36]. Osa has been reported to be an important epigenetic regulator for Drosophila development. In the Drosophila adult midgut, Osa activates the transcription of Delta (Dl) in intestinal stem cells (ISCs), which in turn stimulates the Notch signaling activity in enteroblast and promotes an enterocyte fate commitment [37]. In the Drosophila neural stem cell lineage, Osa limits intermediate neural progenitors (INPs), thus regaining neuroblast properties, and prevents brain tumor formation [38]. In eye imaginal discs, Osa interacts genetically and biochemically with Cyclin E to promote cell cycle progression [39]. In the wing disc, Osa is needed for vein patterning through regulating gene expression in response to Epidermal Growth Factor Receptor (EGFR) signaling and cooperation with Groucho (Gro) and Histone deacetylase 1 (Rpd3) to repress Wingless target genes [40,41]. In mammals, the mammalian homolog of Osa, ARID1A, contributes to liver regeneration in hepatocytes and has also been identified as a tumor suppressor in human cancers [42,43,44,45].
In this paper, we report evidence that Osa participates in GSC progeny differentiation in a BMP-signaling-dependent manner. Osa participates in the repression of dpp transcription in ECs, which facilitates BMP signaling activity outside the GSC niche. We also show that Osa represses Engrailed (En) expression in ECs. The ectopic En in osa-depleted ECs also partially contributes to activating the transcription of dpp. Furthermore, Osa regulates germline differentiation in a BAP-complex-dependent manner. Meanwhile, Osa controls EC protrusions. In addition, the adult EC knockdown of osa was observed to induce GSC loss. Taken together, our findings support that the epigenetic factor Osa plays an important role in controlling EC characteristics and germline lineage differentiation.
2. Materials and Methods
2.1. Fly Strains and Genetics
Flies were cultured at 25 °C on standard cornmeal media supplied with live yeast to the food surface unless otherwise stated.
To maximize the RNAi-mediated knockdown effect, newly enclosed flies were cultured at 29 °C for 3 days before dissection.
For pupal stage-specific expression of RNAi, c587 was used in combination with tubP-GAL80ts (c587ts for short), crosses were set up, and the progenies were kept at 18 °C until the early pupal stage before being transferred to 29 °C. The newly born females (<1 day old) or 3-day-old females were dissected for analysis.
For the adult-specific expression of RNAi, c587ts was used, crosses were set up, and the progenies were kept at 18 °C until eclosion, before being transferred to 29 °C for another 14 days prior to dissection.
Strains used in this study were as follows: w1118 was used as a control; osa308(BDSC 5949), osa2(BDSC 3616), nos-gal4(gift from Yu Cai), c587-gal4 (gift from Yu Cai), dad-lacZ (gift from Yu Cai), bamP-GFP (gift from Dennis M McKearin), dpphr4 (gift from Ting Xie), dppe90 (gift from Ting Xie), dpp2.0-lacZ (gift from Yu Cai), en4 (BDSC 1817), en7 (BDSC 1820), dpp RNAi-1 (BDSC 31531), dpp RNAi-2 (BDSC 31530), en RNAi-1 (VDRC 105678), en RNAi-2 (BDSC 33715), mcherry RNAi (BDSC 35785), hh-lacZ (BDSC 5330), UAS-mCD8-GFP (BDSC 5137), osa RNAi-1 (BDSC 31266), osa RNAi-2 (BDSC 35447), brm RNAi-1 (BDSC 34520), brm RNAi-2(BDSC 31712), mor RNAi-1 (BDSC 34919), mor RNAi-2 (VDRC 6969), snr1 RNAi (BDSC 32372), bap170 RNAi (BDSC 26308), bap180 RNAi-1 (BDSC 32840), FRT82B (BDSC 8216), and tubP- GAL80ts (BDSC 7017).
2.2. Clone Generation
MARCM clones of the osa mutant and control were generated by crossing FRT82B, osa308, or FRT82B with hs-Flp/FM7; UAS-GFPnls, tubP-GAL4/Cyo, y+; and tubP-GAL80, FRT82B.
The resulting larvae were heat shocked at 37 °C with three 60 min pulses at 5 h intervals each day from the mid-third larval instar (ML3, 96 h after egg laying (AEL)) to early pupal stage (EP, 144 h AEL). After heat shock, the flies were kept at 29 °C until dissection. Newly born (<1 day old) females were dissected and stained with the appropriate antibodies.
2.3. Pupae Staging
Staging of the pupae was performed as reported [46,47]. In short, synchronized eggs were collected in a fresh vial within 2 h. After the parents were removed, the offspring were cultured with optimal nutrition and uncrowded conditions. For flies carrying tub-Gal80ts, vials were kept at 18 °C until the temperature shift. Under these conditions, the EP was 12 days AEL.
2.4. Antibodies and Immunofluorescence
Staining of the ovaries was carried out as described previously [48]. In brief, females were dissected in PBS and fixed in 4% paraformaldehyde (Sigma, Darmstadt, Germany) in PBS for 30 min, rinsed with 0.3% PBST (PBS containing 0.3% Triton X-100 (Bio-Rad, Hercules, CA, USA)) three times, permeated with 1.0% PBST for 1 h, blocked with 10% goat serum (Life Technology, New York, NY, USA) in 0.3% PBST for 2 h, and stained overnight at 4 °C with primary antibodies. The ovaries were then washed three times in 0.3% PBST and incubated with secondary antibodies for 2 h at room temperature and stained with DAPI for 10 min. Finally, the ovaries were rinsed four times with 0.3% PBST and mounted.
The following primary antibodies were used in this work: Mouse anti-α-spectrin (1:20, DSHB 3A9), mouse anti-Armadillo (1:20, DSHB N2 7A1), rabbit anti-β-gal (1:1000, Cappel, Cochranville, PA, USA), rabbit anti-pMad (1:500, gift from ED. Laufer), rabbit anti-GFP (1:1000, Life Technology, New York, NY, USA), mouse anti-Engrailed (1:10, DSHB 4D9), rabbit anti-Engrailed (1:200, Santa Cruz, SCZ, USA), mouse anti-Osa (1:5, DSHB Osa). Secondary antibodies conjugated with Alexa Fluor 488 or 546 (Life Technology, New York, NY, USA) were used at 1:1000 dilutions. DAPI (Life Technology, New York, NY, USA,) was used to visualize the nuclei. Confocal images were captured on a Leica TCS SP8 WLL laser confocal microscope and obtained using the Leica AF Lite system (Leica, Weztlar, Germany). Images were processed in Adobe Photoshop CS6 (Version: 13.0 x 32).
2.5. Measurement of Fluorescence Intensity
To compare the fluorescence intensity between the gene knockout ECs and the control, all images were measured under the same parameters at the same time using the Leica SP8 quantification software (Version: LAS-AF-Lite_2.6.0). The selected regions within the ECs were randomly picked, and the Osa-staining mean fluorescence intensity was normalized to the fluorescence intensity of the adjacent germline cells, calculated, and compared. The hh-lacZ mean fluorescence intensity was normalized to the background, calculated, and compared.
2.6. EC Purification, RNA Isolation, and Quantitive PCR
GFP-positive ECs were isolated from the ovaries of c587-gal4/+; uas-mCD8-GFP/+; osa RNAi/+ and c587-gal4/+; uas-mCD8-GFP/+; and mcherry RNAi/+ virgins using FACS (BD FACSAria III, New Jersey, NJ, USA) according to the procedure published previously [19,49]. Total RNA was isolated with Trizol according to the manufacturer’s manual. cDNA was synthesized using a TOYOBO FSQ-301 kit (Osaka City, Japan), and qPCR was performed using SYBR Green (TOYOBO QPK-201, Osaka City, Japan) on a LightCycler 96 (Roche, Basel, Switzerland). The 2−ΔΔCt method was used to compare the gene expression levels. dpp RNA levels were normalized to rp49. Each reaction was performed in triplicate. Three biological replicates were performed.
2.7. Statistical Analysis
All statistical data were recorded in Excel (Microsoft, Redmond, WA, USA) and graphed in Prism 7.0 (GraphPad Software, Version: 7.00). Figure 1H,I: Student’s t test was used to compare the significant difference between two group. Figure 2H,I: chi-squared test was used when analyzing the categorical variable data. Figure 2G: one-way ANOVA test was applied when analyzing more than two groups. Figure 3A: paired-sample t-tests were used to compare the qRT-PCR results. Figure 3L,M: one-way ANOVA test was applied. Figure 4I–L: chi-squared test was used. Figure 5H,I,Q: one-way ANOVA test was applied. Figure 6K: one-way ANOVA test was applied. Figure 6L: chi-squared test was used. Figure 7D: one-way ANOVA test was applied. Figure S1D: one-way ANOVA test was applied. Figure S2E,I,J: one-way ANOVA test was applied. Figure S3C,D: Student’s t test was used. Figure S3E: one-way ANOVA test was applied. Figure S4C–E: one-way ANOVA test was applied. A value of p < 0.05 was considered statistically significant.
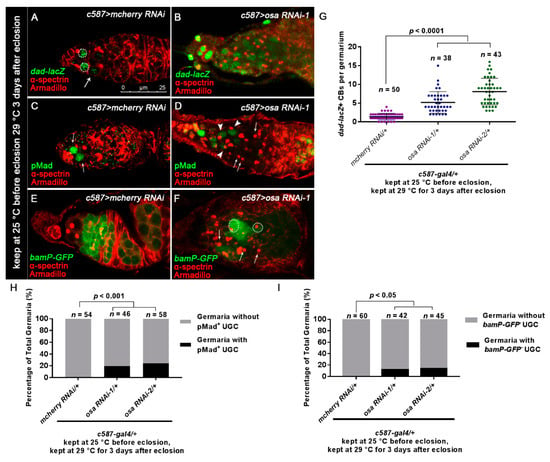
Figure 2.
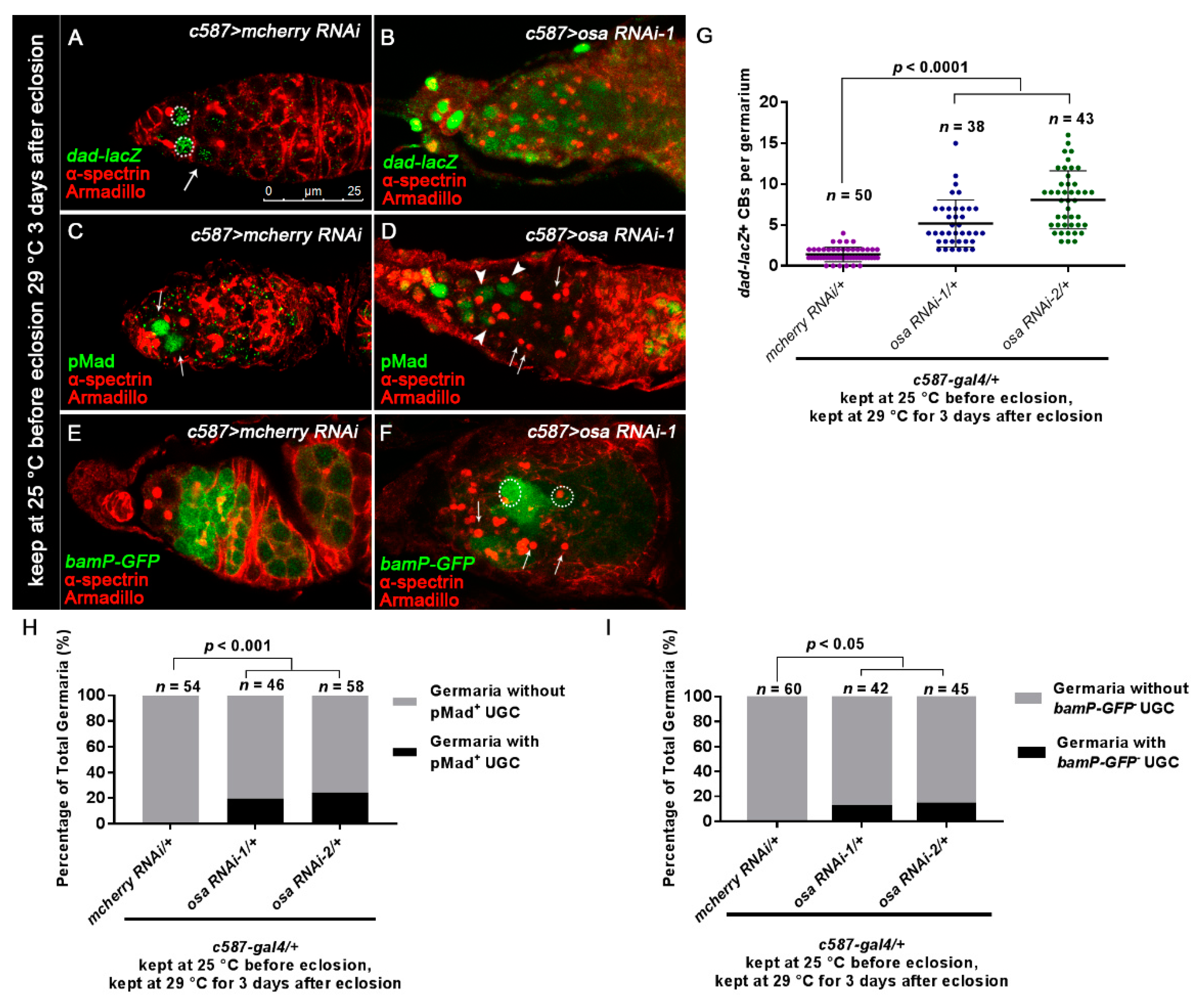
Loss of osa in ECs results in expanded BMP signaling. (A,B) Germaria stained for α-spectrin (red), Armadillo (red), and β-gal (green). BMP signal activity was monitored by dad-lacZ (detected by β-gal). The control germarium (A) exhibited three GSCs (indicated by white dashed circles) and one CB (indicated by white arrows) positive for dad-lacZ. The osa RNAi-1 germarium (B) exhibited dad-lacZ-positive UGCs. (C,D) Germaria were stained for α-spectrin (red), Armadillo (red), and pMad (green). The control germarium (C) exhibited strong pMad staining in GSCs (indicated by white arrows). In the osa RNAi-1 germarium (D), some of the accumulated UGCs exhibited pMad staining (indicated by white arrowheads), while others were negative for pMad (indicated by white arrows). (E,F) Germaria stained for α-spectrin (red), Armadillo (red), and GFP (green). bam transcription was monitored by bamP-GFP (detected by GFP). In the control germarium (E), bamP-GFP was negative in GSCs and upregulated in CB and the dividing cysts. In osa RNAi-1 germarium (F), there is a mixture of bamP-GFP-positive (indicated by white dashed circles) and bamP-GFP-negative (indicated by white arrows) UGCs. c587 > osa RNAi-2 has a similar phenotype (not shown). Several compressed z-sections are shown in (A–F). (G) Graph showing the quantification of dad-lacZ-positive UGCs for each germarium. (H) Graph showing the percentage quantification of germaria with pMad-positive UGCs. (I) Graph showing the percentage quantification of germaria with bamP-GFP-negative UGCs. The scale bar is shown in (A).
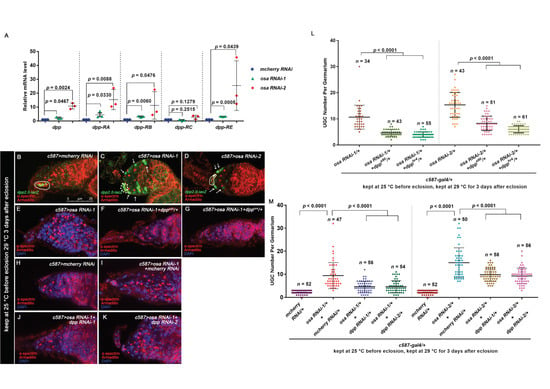
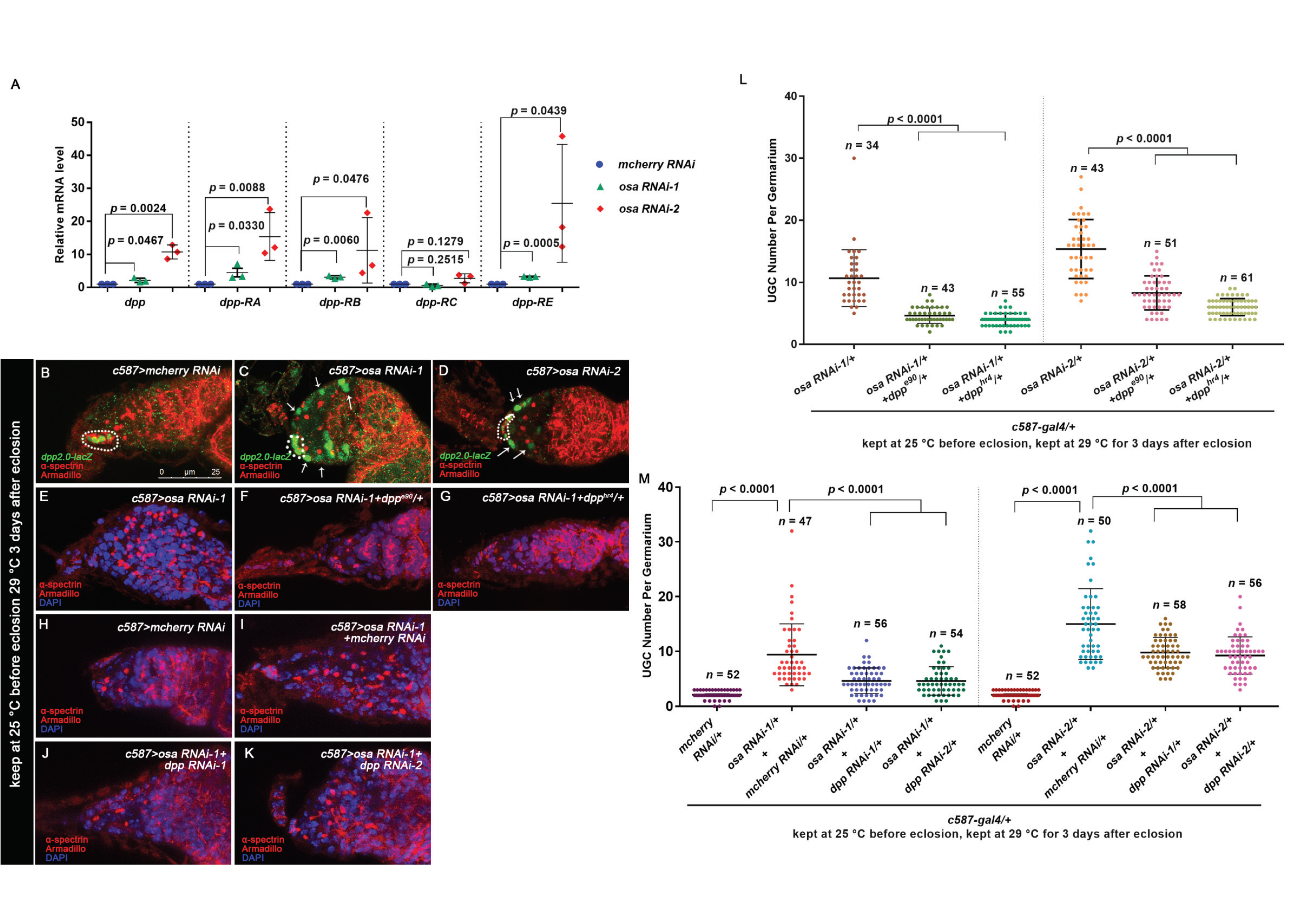
Figure 3.
Osa limits dpp transcription in ECs for germline differentiation control. (A) Quantitative PCR results for the purified ECs showing that mRNA levels for the annotated dpp transcripts were upregulated in osa KD ECs. (B,C,D) Germaria stained for α-spectrin (red), Armadillo (red), and β-gal (green). dpp transcription was monitored by dpp2.0-lacZ (detected by β-gal). CpCs are indicated by white dashed circles. In the control germarium (B), dpp2.0-lacZ was expressed in CpCs. In the osa RNAi-1 (C) and osa RNAi-2 (D) germaria, dpp2.0-lacZ was also expressed in ECs (indicated by white arrows). (E–K) Germaria stained for α-spectrin (red), Armadillo (red), and DAPI (blue). (H) The control germaria exhibiting a normal number of CBs. The osa RNAi-1 (E) and osa RNAi-1+ mcherry RNAi (I) germaria exhibiting UGC accumulation. The introduction of one copy of the dpp allele (F,G) into the background of c587 > osa RNAi-1 or dpp RNAi (J,K) into the background of c587>osa RNAi-1 relieved the UGC accumulation phenotype. c587>osa RNAi-2 provided similar phenotypes (not shown). (L,M) Graphs showing quantification of the UGC number per germarium. Error bars are presented as the Mean ± SD. Several compressed z-sections are shown (B–K). The scale bar is shown in (B).
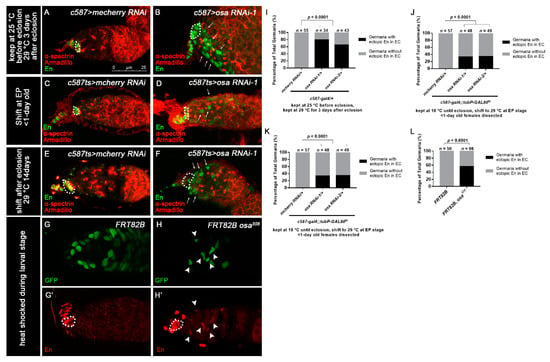
Figure 4.
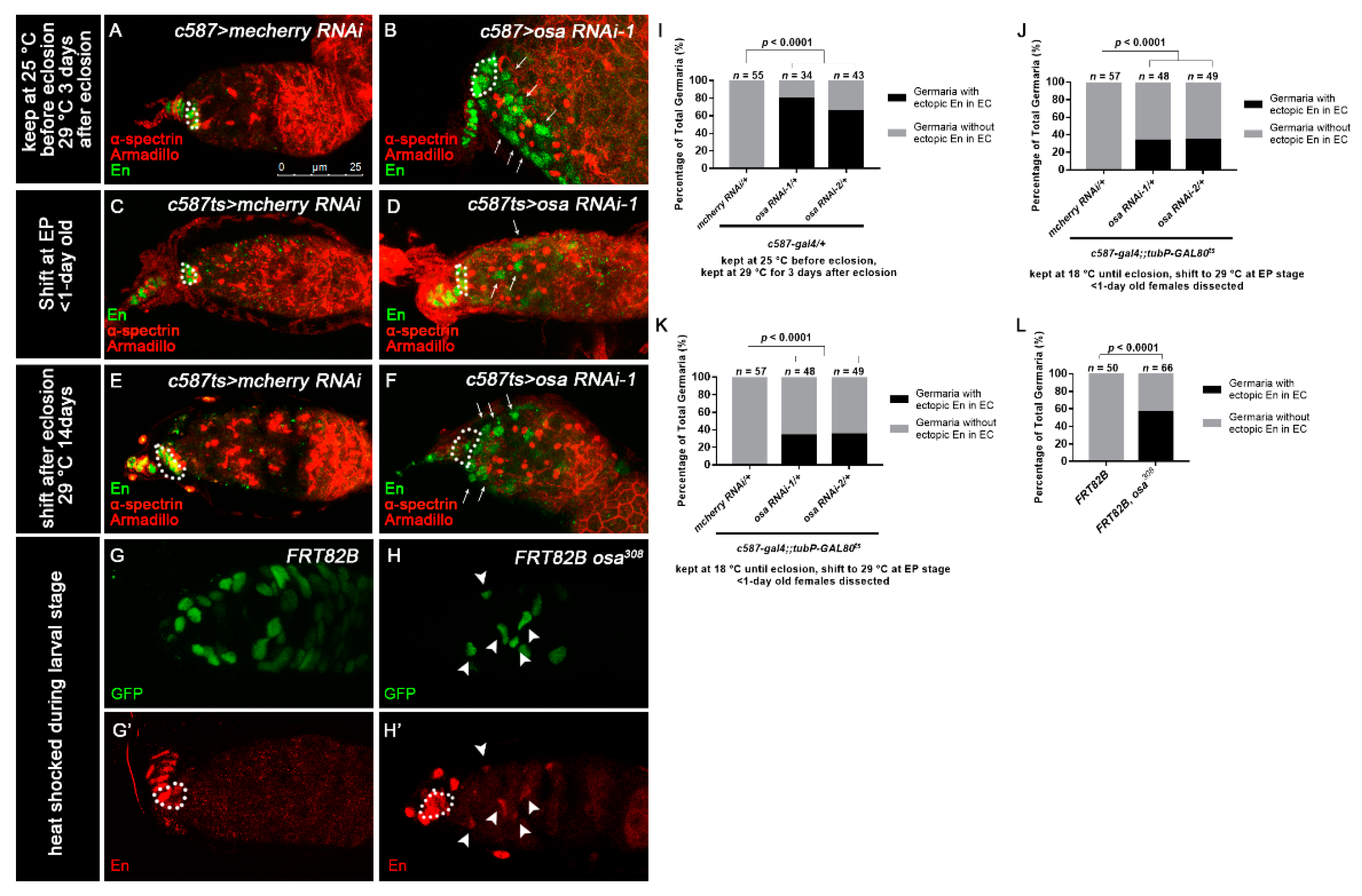
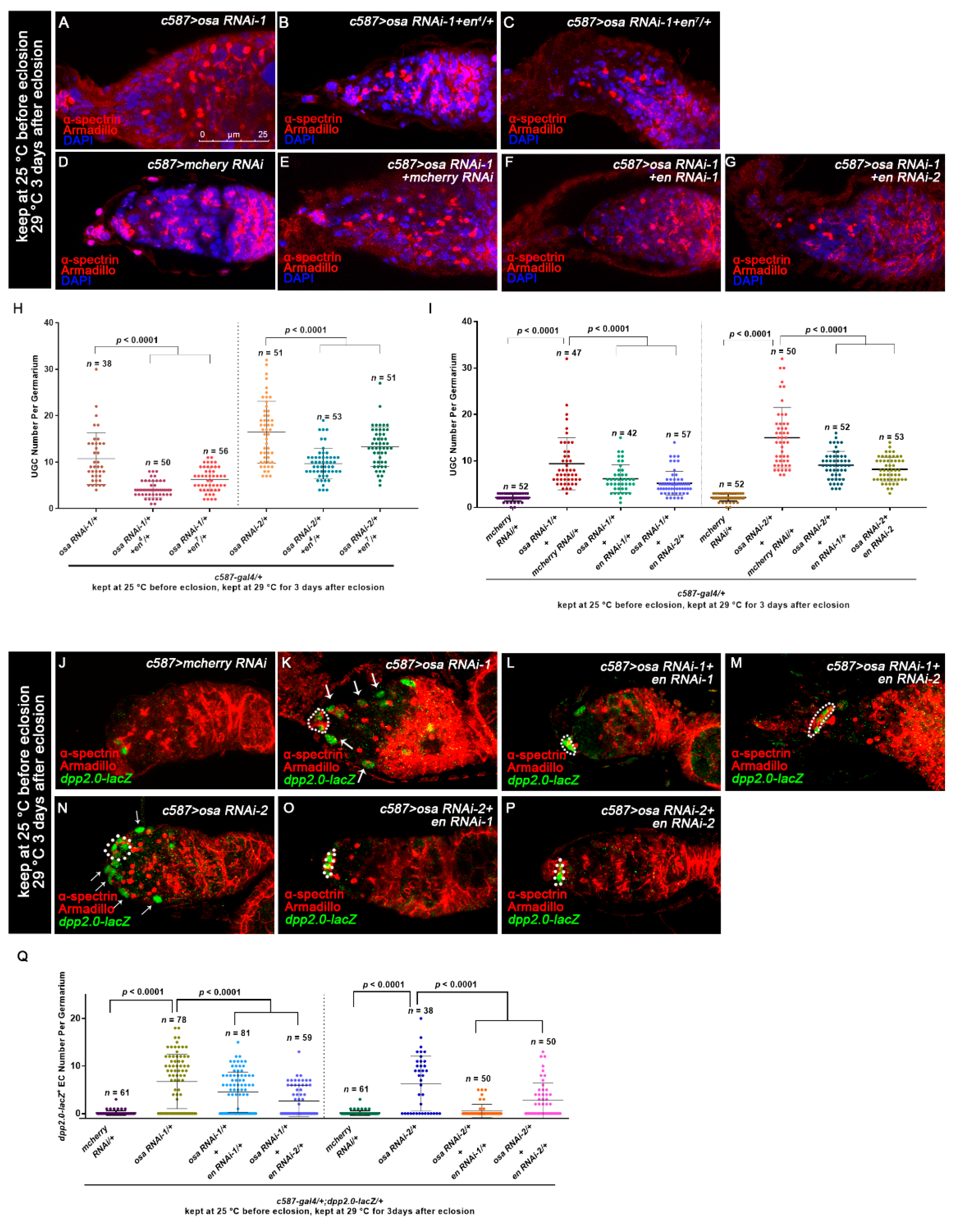
Osa represses engrailed expression in ECs. (A–F) Germaria stained for α-spectrin (red), Armadillo (red), and En (green). (A,B) Flies were initially kept at 25 °C and then shifted to 29 °C after eclosion. (C–F) c587-gal4;;tubP-GAL80ts was used for osa KD. (C,D) Flies were initially kept at 18 °C and then shifted to 29 °C at the EP stage. The newly born females were dissected. (E,F) Flies were initially kept at 18 °C until eclosion and then kept at 29 °C for another 14 days before dissection. The control germaria (A,C,E) exhibited specific En staining in TF and CpCs. The osa RNAi-1 germaria (B,D,F) exhibited ectopic En staining in the ECs (indicated by white arrows) in addition to CpCs. c587 > osa RNAi-2 gave a similar phenotype (not shown). (G–H’) MARCM clones stained for En (red). Clonal cells were marked by GFP (green). CpCs here are indicated by white dashed circles. The mosaic germarium with control clones (G,G’) exhibited specific En staining in TF and CpCs. The mosaic germarium with osa308 mutant ECs (H,H’) exhibited ectopic En staining in clonal ECs (indicated by white arrow heads) in addition to CpCs. Several compressed z-sections are shown in (A–H’). (I–L) Graph showing the percentage quantification of germaria with En ectopic expression. The scale bar is shown in (A).
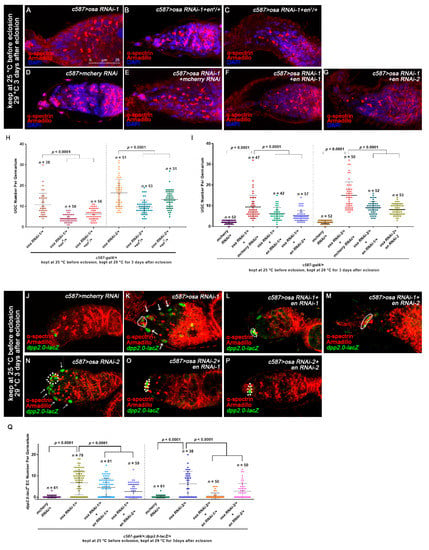
Figure 5.
Osa regulates dpp partially through Engrailed. (A–G) Germaria stained for α-spectrin (red), Armadillo (red), and DAPI (blue). The osa RNAi-1 (A) and osa RNAi-1+mcherry RNAi (E) germaria exhibited UGC accumulation. (D) The mcherry RNAi germaria exhibited a normal number of CBs. Introduction of one copy of an en allele (B,C) or en RNAi (F,G) significantly relieved the germ cell differentiation defect. (H,I) Graphs show the quantification of UGC number per germarium. Error bars are presented as the Mean ± SD. (J–P) Germaria stained for α-spectrin (red), Armadillo (red), and β-gal (green). dpp transcription was monitored by dpp2.0-lacZ (detected by β-gal). CpCs are indicated by white dashed circles. In the control germarium (J), dpp2.0-lacZ was expressed in CpCs. In osa RNAi-1 (K) and osa RNAi-2 (N) germaria, dpp2.0-lacZ was ectopically expressed in ECs (indicated by white arrows) in addition to CpCs. The introduction of en RNAi-1 (L,O) or en RNAi-2 (M,P) reduced the ectopic dpp2.0-lacZ in ECs. (Q) A graph showing the quantification of dpp2.0-lacZ-positive ECs for each germarium. Error bars are presented as the Mean ± SD. Several compressed z-sections are shown in (A–G,J–P). The scale bar is shown in (A).
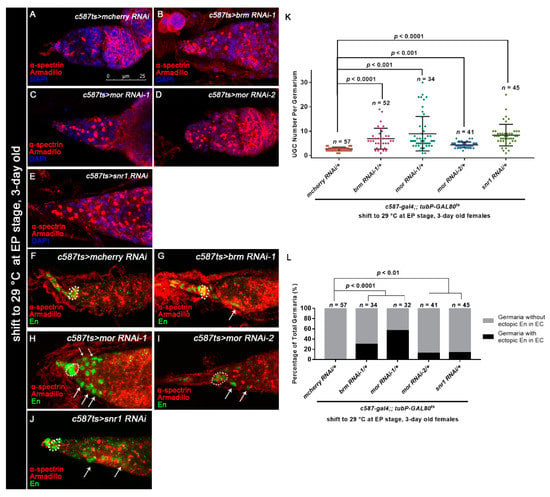
Figure 6.
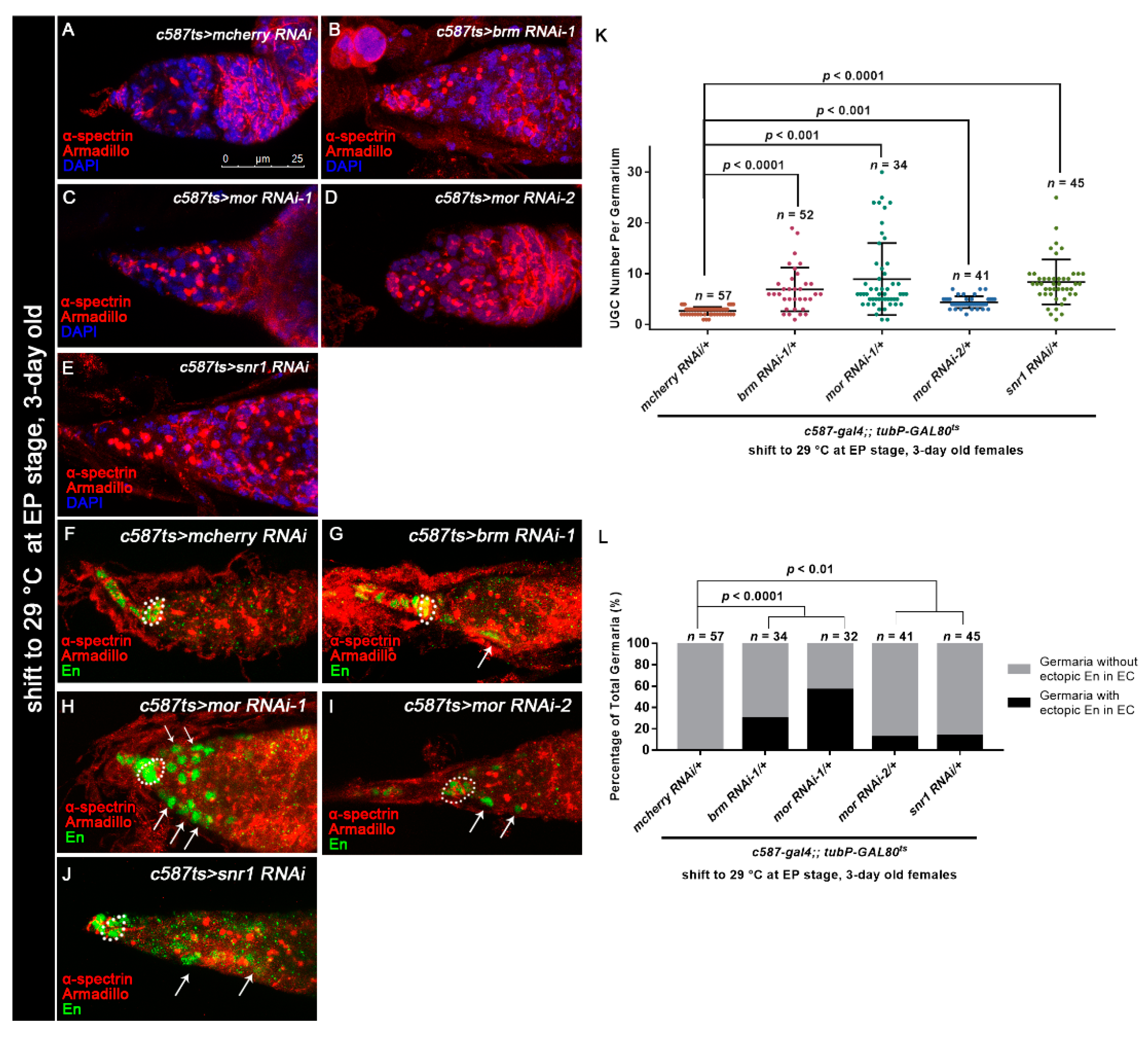
Osa functions in a BAP complex-dependent manner. (A–E) Germaria stained for α-spectrin (red), Armadillo (red), and DAPI (blue). c587-gal4;;tubP-GAL80ts was used for BAP complex component KD. The germarium from the control group (A) exhibited a normal number of CBs. The c587ts > brm RNAi-1 (B), c587ts > mor RNAi (C,D), and c587ts > snr1 RNAi (E) germaria exhibited UGC accumulation. (K) Graph showing the quantification of UGCs in each germarium. Error bars are presented as the Mean ± SD. (F–J) c587-gal4;;tubP-GAL80ts was used for BAP complex component KD. Germaria were stained for α-spectrin (red), Armadillo (red), and En (green). CpCs are indicated by white dashed circles. Germaria from the control group (F) exhibited exclusive expression of En in TF and CpCs. Germaria from c587ts > brm RNAi-1 (G), c587ts > mor RNAi (H,I), and c587ts > snr1 RNAi (J) exhibited ectopic En expression in ECs (indicated by white arrows) in addition to CpCs. (L) Graph showing the percentage quantification of germaria with En ectopic expression. Several compressed z-sections are shown in (A–J). The scale bar is shown in (A).
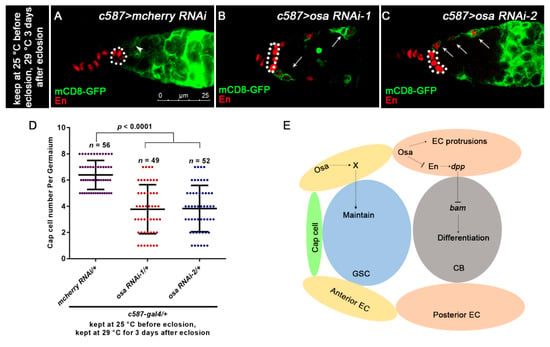
Figure 7.
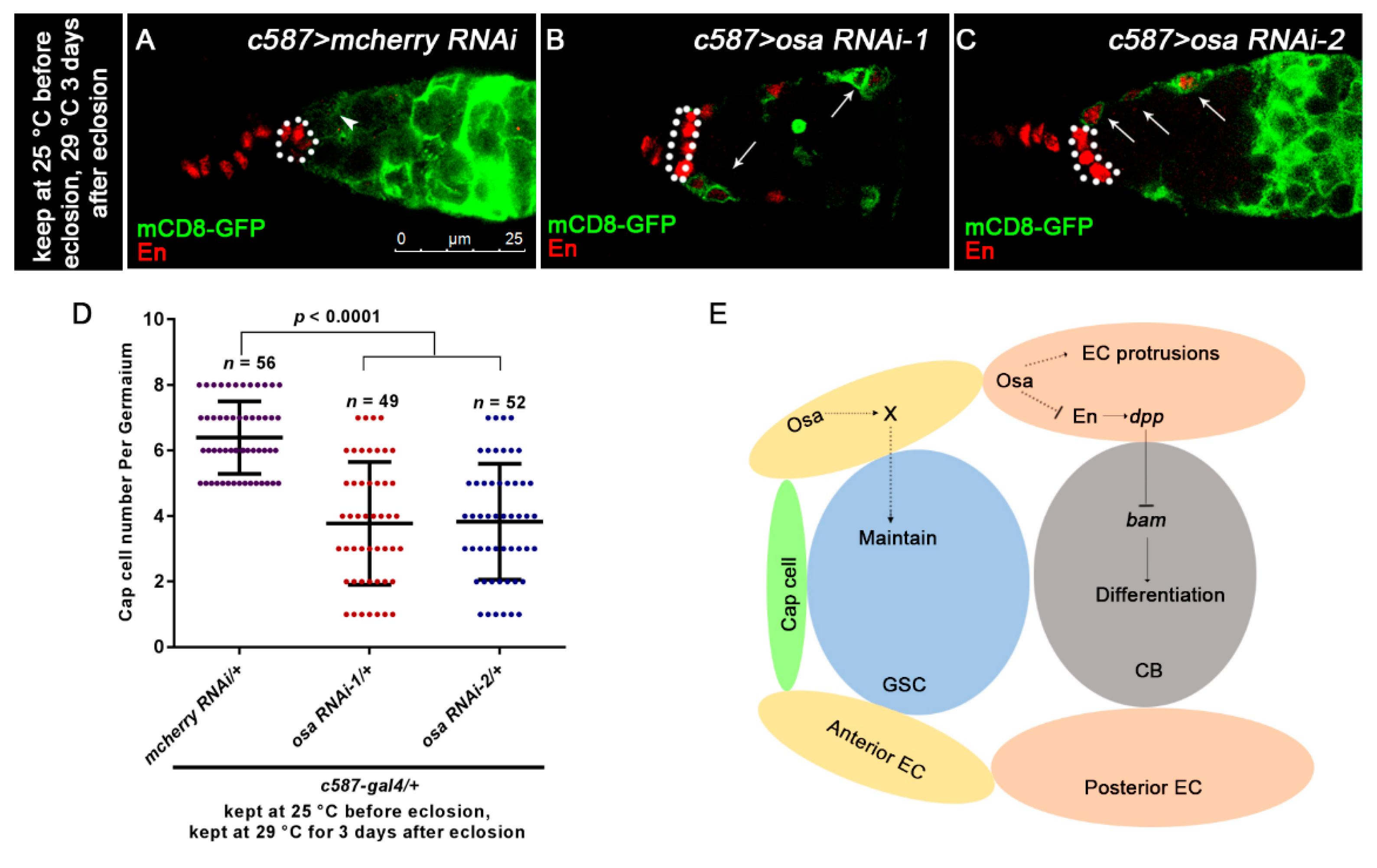
Osa regulates EC characteristics. (A,C) Germaria are stained for En (red) and GFP (green). EC membranes are marked by mCD8-GFP (detected by GFP). CpCs are indicated by white dashed circles. In control germarium (A), EC protrusions extend between cysts (indicated by white arrowhead). Additionally, en is exclusively expressed in TF and CpCs. In osa RNAi-1, osa RNAi-2 germarium (B,C), ECs lose their cellular processes (indicated by white arrows). Additionally, en is ectopically expressed in ECs, which is also marked by GFP (indicated by white arrows). (D) Graph shows the quantification of CpC per germarium. Error bars are presented as Mean ± SD. (E) Model illustrates Osa-mediated regulation of GSC maintenance and germ cell differentiation in ECs in germaria. Scale bar is shown in (A).
3. Results
3.1. Osa Is Required in Escort Cells for GSC Progeny Differentiation
Since Osa is an important epigenetic regulator for Drosophila development, we sought to identify whether Osa regulates the development of the germline. To evaluate the germline differentiation status in both wild-type (WT) and osa mutants (osa2/308), we stained the ovaries with α-spectrin and Armadillo to mark the spectrosome/fusome and the CpCs, respectively. The GSCs were identified by the spectrosomes, which directly contact the CpCs. Additionally, CBs were identified by round fusomes, which do not reside immediately adjacent to the CpCs [50] (Figure 1A). The control germaria typically contain 0–4 CBs outside the GSC niche (Figure 1B,H, white arrows). In contrast, the osa2/308 germaria contained significantly rounder fusome-containing germ cells than the control germaria (Figure 1C,H, white arrows). For quantification, the round fusome-containing germ cells outside the GSC niche were quantified as undifferentiated germ cells (UGCs). Altogether, these results indicate that decreased osa expression impairs differentiation of the germ cells.
GSC progeny differentiation is also controlled extrinsically by the differentiation niche formed by ECs. Thus, we sought to identify if Osa is required non-cell autonomously within ECs for GSC progeny differentiation. We first used c587-gal4, which is expressed in most somatic lineages in developing gonads but becomes largely restricted in ECs and early follicle cell progenitors in adulthood [51,52], to drive the expression of osa RNAi in ECs. Two individual RNAi lines (osa RNAi-1 and osa RNAi-2) were used to knockdown (KD) osa expression. To maximize KD efficiency moderately, the flies were initially kept at 25 °C until eclosion and then transferred to 29 °C for 3 days prior to dissection. First, the knockdown efficiency of two RNAi lines was confirmed by immunostaining. In the control germaria, Osa was ubiquitously expressed in both germline and somatic lineages, as detected by the Osa antibody (Figure S1A,A’, white arrows). In osa RNAi germaria, osa KD led to faint Osa staining in ECs (Figure S1B–D, white arrows). In addition, osa KD also induced UGC accumulation in the germaria (Figure 1D,E). These results indicate that knockdown osa in ECs may block germ cell differentiation.
It was previously reported that at two hours after puparium formation (APF), germaria were formed, and ECs could be identified by Tj and LamC antibody staining; at 48 h APF, germaria were completely formed [53]. Since c587-gal4 is known to be expressed in both somatic precursors and adult ECs [54], to avoid the influence of osa knockdown in somatic precursors before EC formation, we combined c587-gal4 with a temperature-sensitive mutant gal80 (tubP- GAL80ts) (referred to c587ts for short) to temporally control osa KD. GAL80ts is active at 18 °C and represses the activity of GAL4 but is inactive at 29 °C; thus, GAL4 could initiate the expression of RNAi [55]. First, we carried out the pupae-stage-specific KD of osa. To bypass the potential requirement of Osa before germarium formation, flies were initially kept at 18 °C and transferred to 29 °C during the early pupal stage after EC formation. The newly born females (<1 day old) were checked. Compared with the control germaria, the expression of osa RNAi led to a dramatic expansion of UGCs (Figure S2A,B,E). For the adult-specific KD, flies were initially kept at 18 °C until eclosion and then shifted to 29 °C for another 14 days before dissection. Compared with the control germaria, osa KD germaria were also filled with UGCs (Figure S2C–E). To rule out the leaky expression of osa RNAi before eclosion, the newly eclosed females were also checked, and their germaria appeared normal (Figure S2F–I). These findings suggest that Osa is required in ECs for germline differentiation control after EC formation. Interestingly, in addition to UGC accumulation upon the adult-specific KD of osa, these germaria also exhibited a significant GSC loss phenotype (Figure S2J). Since a recent study suggested that the anterior ECs are essential for GSC maintenance [6], we inferred that osa may be required in anterior ECs for GSC maintenance in adulthood.
To confirm the phenotypes of osa transheterozygotes and c587-gal4-driven osa RNAi, we performed a clonal analysis by inducing positively marked osa308 clones using the MARCM system [56] and then evaluated the UGC quantity in the mosaic germaria. Both the control and mutant clones were induced in parallel. The mosaic germaria with control clones (FRT82B) exhibited a normal number of CBs (Figure 1F,F’,I). However, the mosaic germaria with osa308 clonal ECs presented accumulated UGCs (Figure 1G,G’,I, white arrows). It was reported that brm’s loss of function in the germline does not disrupt germline differentiation [57]. Moreover, no UGC accumulation phenotype was found when we used nos-gal4 to knock down osa in the germline (unpublished data). These results indicate that Osa is required in ECs for germ cell differentiation control.
3.2. Osa Is Required for the Restriction of BMP Signaling Outside the GSC Niche
Previous studies have reported that the inactivation of BMP signaling is required for the differentiation of GSC daughters. For this purpose, we checked dad-lacZ, an enhancer trap line for the target gene of the BMP pathway [58]. Normally, dad-lacZ is confined to GSCs and CBs [59] (Figure 2A, white dashed circles and white arrows). By contrast, the number of dad-lacZ-positive UGCs was expanded in c587-gal4-driven osa KD germaria, although the intensity of β-gal staining was not as high as that in the control germaria (Figure 2B,G).
Activation of BMP signaling in GSCs resulted in the phosphorylation of Mad (pMad). Then, pMad translocated into the GSC nucleus and repressed the differentiation factor bam [11,14,60]. pMad staining is normally restricted in GSCs (Figure 2C, white arrows). In the germaria of osa KD, pMad staining was strong in the GSCs. A part of UGCs in a subset of germaria expressed pMad at a low level (Figure 2D, white arrowheads; Figure 2H, osa RNAi-1 germaria(9/46), osa RNAi-2 germaria(14/58)).
Next, we monitored the bam transcription activity with bamP-GFP (a GFP reporter of bam transcription) [60]. bamP-GFP expression is normally absent in GSCs; then, it becomes detectable in CB and reaches its highest levels in dividing cysts (Figure 2E). However, in a subset of germaria with osa KD, several UGCs were bamP-GFP-negative outside the GSC niche (Figure 2F, white arrows), while others exhibited upregulated bamP-GFP expression (Figure 2F, white dashed circles). 11.9%(5/42) of osa RNAi-1 germaria and 13.8% (9/65) of osa RNAi-2 germaria contained bamP-GFP negative UGCs (Figure I). These results indicate that the accumulated UGCs represent a mixture of GSC-like and/or CB-like stages. Collectively, osa-depletion in ECs induces ectopic BMP signaling activity outside the GSC niche.
3.3. Osa Restricts Dpp Transcription in Escort Cells
There is growing evidence that Dpp is regulated precisely in ECs to facilitate homeostasis of the germline [18,22,26,61]. Moreover, the upregulation of dpp transcription in ECs is responsible for the enlargement of BMP activity outside the GSC niche. To determine if Osa is required in ECs to repress dpp transcription, we performed qRT-PCR to examine the transcription levels of dpp in the FACS-sorted control and osa KD ECs. The primer sequences are provided in Table 1. Since there are four annotated dpp isoforms in Drosophila, first, we detected the dpp mRNA level using primer pairs recognizing all four isoforms. Then, dpp was increased in the EC samples with osa KD than that of control ones. To determine if the elevation is isoform-specific, we next examined dpp expression using isoform-specific primer pairs. We found significant upregulation of dpp-RA, dpp-RB, and dpp-RE in osa KD ECs (Figure 3A). These results indicate that Osa is required in ECs to repress dpp transcription.

Table 1.
Primer sequences used for quantitative real-time polymerase chain reaction (qRT-PCR).
To visualize dpp expression in the germaria, we introduced dpp2.0-lacZ [62], which is used to monitor dpp transcription, into the context of the c587-gal4-driven osa KD. As previously reported, dpp2.0-lacZ is highly expressed in CpCs (Figure 3B, white dashed circle) and sporadically expressed in ECs in control germaria [62]. However, osa KD induced the ectopic expression of dpp2.0-lacZ in not only the anterior-most ECs but also in the posterior ones (Figure 3C,D, white arrows). Collectively, these results indicate that dpp transcription is upregulated in osa-deficient ECs.
To further demonstrate the link between elevated dpp in ECs and UGC formation, we performed a genetic analysis by reducing dpp in the background of c587-gal4-driven osa KD and assessed the UGC number. We found that either of the two dpp alleles, dppe90 or dpphr4, could partially rescue the UGC accumulation phenotype induced by osa KD (Figure 3E–G,L). Consistently, the double knockdown of osa dpp also partially reduced the UGC number in osa KD germaria (Figure 3H–K,M). In summary, these results show that Osa regulates GSC progeny differentiation partly by modulating dpp in ECs.
3.4. Osa Limits Engrailed Expression in Escort Cells
Next, we sought to investigate how Osa limits dpp expression outside the GSC niche. It was reported that Osa may contribute to restricting engrailed (en) expression in embryo [63], indicating that Osa is a repressor of en. Meanwhile, ectopic expression of en in ECs leads to ectopic BMP signaling, resulting in an expanded GSC-like cell tumor phenotype [22], and En regulates dpp2.0-lacZ reporter activity in CpCs [62]. Thus, we tested whether en is upregulated in ECs upon c587-gal4-driven osa KD. For this purpose, we carried out immunostaining of En in the germaria of the control and osa KD females. Flies were initially kept at 25 °C until eclosion and then transferred to 29 °C for another 3 days prior to dissection. The control germaria exhibited the specific expression of En in TF and CpCs (Figure 4A, white dashed circle). Interestingly, some c587-gal4-driven osa KD germaria exhibited ectopic En expression in ECs (osa RNAi-1: 27/34 of total germaria, osa RNAi-2: 28/43 of total germaria, Figure 4B, white arrows; Figure 4I).
We subsequently investigated temporally controlled manipulations of osa. For the pupal-stage-specific expression of RNAi, flies were initially kept at 18 °C until the early pupal stage and then transferred to 29 °C for RNAi expression. The newly born females (<1 day old) were checked. For the adult-specific expression of RNAi, flies were raised at 18 °C and then shifted to 29 °C after eclosion. The flies were aged at 29 °C for 14 days prior to dissection. In the control germaria, En was restricted to the TF and CpC (Figure 4C,E, white dashed circles). Surprisingly, the germaria of both pupal-specific and adult-specific osa depletion exhibited ectopic en expression in the ECs (pupae-specific osa KD: osa RNAi-1: 16/48 of total germaria, osa RNAi-2: 17/49 of total germaria; adult-specific osa KD: osa RNAi-1: 51/51 of total germaria, osa RNAi-2: 58/59 of total germaria; Figure 4D,F, white arrows; Figure 4J,K). Then, we further examined the en expression patterns in the osa mosaic germaria. en was also ectopically expressed in osa mutant clonal ECs (37/66 of total clonal germaria, Figure 4G–H’, white arrowheads; Figure 4L), which confirmed the results of the RNAi KD. These results suggest that Osa may repress en expression in ECs after EC formation in both the pupal stage and adulthood.
To test if the upregulated en in ECs is essential for the UGC formation induced by osa KD, we reduced en expression by introducing one copy of the en allele into the background of the osa RNAi. Indeed, both en4 and en7 could partially suppress the UGC accumulation induced by osa KD (Figure 5A–C,H). A similar effect was also found when en RNAi was co-expressed with osa RNAi (Figure 5D–G,I). In summary, the ectopic expression of en in ECs is partially attributed to the UGC accumulation in c587-driven osa KD germaria.
To further determine if the ectopic expression of en in ECs is responsible for the ectopic dpp2.0-lacZ activity caused by osa KD in ECs, we co-expressed en RNAi and osa RNAi with c587-gal4 and then quantified dpp2.0-lacZ activity. Interestingly, two individual en RNAi lines could partially suppress the ectopic β-gal staining in ECs in comparison with osa KD alone (Figure 5J–Q). These results indicate that ectopic expression of en is at least partially responsible for the increased dpp transcription caused by osa KD in ECs.
3.5. Osa Regulates Germline Differentiation through the BAP Complex
Osa is a component of the BAP complex, which also includes Brm, Snr1, and Mor. Therefore, we wondered if Osa regulates GSC progeny differentiation in a BAP-complex-dependent manner. To assess this possibility, we tested c587-gal4-driven RNAi against brm, snr1, or mor. Since overexpression of these RNAi lines (brm RNAi-1, snr1 RNAi-1, mor RNAi-1) with c587-gal4 caused pupal lethality or the emergence of abnormal ovaries at 25 °C, we used c587ts to bypass the potential developmental effects. Flies were initially kept at 18 °C and then transferred to 29 °C at the early pupal stage until dissection. Germaria from 3-day-old females exhibited significant expansion of UGCs in the germaria, which was similar to the effects in osa KD (Figure 6A–E, Figure S2A,B,E). The c587-gal4 > brm RNAi-2 background germaria cultured at 25 °C didn’t exhibited significant expansion of the UGCs, but when transferred to 29 °C the significant expansion of the UGCs phenotype was also exhibited (Figure S3A–D). Meanwhile, the knockdown of bap170 or bap180 with c587-gal4 did not induce any obvious germline differentiation defects (Figure S3E). These results suggest that the Osa-containing BAP complex participates in germline differentiation control non-cell autonomously.
Furthermore, germaria with BAP complex component KD exhibited ectopic En in ECs (brm RNAi-1: 10/34 of total germaria, snr1 RNAi: 6/45 of total germaira, mor RNAi-1: 18/32 of total germaria, mor RNAi-2: 5/41 of total germaria Figure 6F–J, white arrows; Figure 6L), leading to an osa KD phenocopy. The brm KD germaria also exhibited ectopic dpp2.0-lacZ in ECs (Figure S3F,G), suggesting that Brm may repress dpp transcription in ECs. Taken together, these results show that Osa regulates germ cell differentiation in a BAP-complex-dependent manner.
3.6. Osa Maintains Escort Cell Characteristics
Since EC protrusions are required to promote germ cell differentiation [8], we sought to determine if Osa regulates EC morphology. Thus, we introduced the UAS-mCD8-GFP/+ into the background of c587-gal4 to outline the morphology of ECs. In the control germaria, ECs extended protrusions to encapsulate the germ cells underneath (Figure 7A, white arrowhead). However, osa KD ECs failed to extend protrusions (Figure 7B,C white arrows). These results indicate that Osa may help to maintain EC protrusions. Moreover, we observed ectopic En staining and dpp transcription in osa-depleted ECs, which suggests a transition from EC to CpC. To determine whether osa mutant ECs completely switched their identities, we examined c587-gal4 and en expression patterns in the control and osa RNAi ovaries. Normally, there is a mutually exclusive expression pattern of c587-gal4 and en in control germaria (Figure 7A). Interestingly, osa KD induced En-positive c587-gal4-positive ECs (Figure 7B,C white arrows). Secondly, we checked the EC identity with that of another CpC marker, hh-lacZ. In the control germaria, hh-lacZ was expressed in TF and CpC at high levels and in ECs at low or undetectable levels. The average hh-lacZ intensity in ECs in the c587-gal4-driven osa depletion germaria was comparable to that in the control ones (Figure S4A–C). This result suggests that the expression level of hh-lacZ in the ECs was not altered upon osa depletion. Altogether, osa-depleted ECs displayed characteristics of both cell types in adult germaria. The osa-depleted EC obtained the ability of dpp production, which in turn activated BMP signaling activity outside the GSC niche. It should be noted that an enlargement of the GSC niche also induced UGC accumulation, as previously reported [64]. As c587-gal4 was active in the precursor cells of both CpC and EC during the developmental stage, we could not rule out the possibility that c587-gal4-deriven osa-depletion induced an enlargement of the GSC niche. To rule out this possibility, we counted the CpC number in osa-depleted germaria and in the control ones. Interestingly, the CpC number was significantly reduced in the osa-depleted germaria, as highlighted by En staining (Figure 7D) and hh-lacZ (Figure S4D). Accordingly, the GSC number was also decreased by c587-gal4-driven osa RNAi (Figure S4E), supporting the decrease in CpC number. In summary, UGC accumulation in c587-gal4-driven osa RNAi germaria was elicited by the cell characteristics transition of ECs.
4. Discussion
Multiple studies have demonstrated that GSC progeny differentiation is a highly regulated process. Limiting dpp expression in the germline differentiation niche is crucial for proper germline development. In this study, we provided evidence that SWI/SNF component Osa depletion in escort cells (ECs) results in the blockage of GSC progeny differentiation. UGC accumulation occurs upon the loss of Osa in ECs containing a mixture of GSCs, pre-CB cells, and CB-like cells. Molecular and genetic studies suggest a link between ectopic dpp transcription in ECs and UGC accumulation elicited by c587-gal4-driven osa KD. Moreover, ectopic en expression in osa-depleted ECs partially contributes to upregulated dpp transcription. Osa in ECs also contributes to EC property regulation, such as regulation of EC protrusions. We also provided evidence that Osa in the anterior-most ECs may participate in GSC maintenance (Figure 7E).
In FACS-purified ECs, we observed significant upregulation in three of the four isoforms of dpp transcripts. This result was further confirmed by the dpp transcription reporter dpp2.0-lacZ. However, what motivates dpp transcription outside the GSC niche? Engrailed is known to be essential for dpp transcription in CpCs [62], and ectopic en expression in ECs leads to ectopic BMP signaling, resulting in the UGC accumulation phenotype [22]. These reports suggest that maintaining en expression in cap cells and suppressing en expression in ECs are critical steps for germline cell homeostasis. How en is regulated in cap cells and ECs remains a fundamental question in stem cell biology. Here, we identified that the Osa-containing BAP complex is required in ECs to limit en expression, and ectopic En in osa-depleted ECs also induces dpp transcription. Lsd1 was reported to limit dpp expression in ECs by repressing en ectopic expression in ECs [22], and there is a possibility that Osa cooperates with Lsd1 to regulate en expression in ECs. More investigation is needed to explore the molecular mechanism that determines how Osa regulates en expression in ECs.
A prior study reported that inhibiting the brm function in ECs alone did not produce any visible phenotype [57] and that c587-gal4-deriven depletion of brm can inhibit dpp derepression in ECs, which is caused by the c587-gal4-deriven depletion of PRC1 [26]. In this study, however, we obtained a contradictory result showing that brm-depletion in ECs induces UGC accumulation and that ectopic dpp2.0-lacZ activity outside the GSC niche uses the same brm RNAi line (brm RNAi-2) (Figure S3A–C,E,F). We previously studied the c587-gal4-driven brm RNAi line at 25 °C and failed to find obvious germline differentiation defects (Figure S3D). To increase KD efficiency, flies were initially kept at 25 °C until eclosion and then transferred to 29 °C for 3 days prior dissection, at which point the UGC accumulation phenotype could be detected. The phenotype was also confirmed with two individual brm RNAi lines (Figure 6A,B,K). This result is consistent with the finding that Osa regulates germline differentiation through the BAP complex, as the RNAi-mediated inactivation of either BAP component leads to UGC accumulation.
A recent study suggested that the GSC-contacting anterior ECs are essential for GSC maintenance [6]. We carried out an adult-specific KD of osa with c587-gal4 to avoid the defects that arise during GSC niche formation. Further, c587-gal4 was found to be restrictively expressed in ECs and early follicle cells in adult ovaries. Indeed, we observed significant UGC accumulation accompanied by a GSC loss phenotype. Thus, it is conceivable that Osa may be required in anterior ECs for GSC maintenance. Besides Dpp, some other factors, such as DE-cadherin and Wnt6, are also required in anterior ECs for GSC maintenance [6], but further investigation is needed to explore the molecular mechanisms by which Osa acts in adult ECs to modulate GSC maintenance. We also found that the CpC number was significantly reduced in osa-depleted germaria. It was reported that the activation of Notch signaling is essential for cap cell formation and adult cap cell maintenance [64,65]. During the late larva 3 (LL3) stage, newly formed TF secretes the ligand of Notch, Dl, thereby activating Notch signaling activity in the adjacent ICs and inducing cap cell formation. The overexpression of Dl in basal TFs increases the cap cell number [66], and Dl mutant basal TFCs induce fewer cap cells [65]. In the Drosophila adult midgut, Osa activated Dl expression in ISCs by binding to the Dl promoter region and then expanded Notch signaling activity in enteroblast, leading enteroblast to differentiate into enterocyte [37]. Osa might regulate the cap cell number by regulating the expression of Dl and other components of the Notch signaling pathway. However, further investigation is needed to explore this possibility.
5. Conclusions
In this work, SWI/SNF complex protein Osa was identified as a regulator of germline differentiation in the Drosophila female germline differentiation niche (posterior escort cells). Osa functions in escort cells to limit dpp transcription to prevent ectopic BMP signaling in the differentiating germline in part via suppressing EC-specific ectopic activation of Engrailed. Depletion of brm or other components of the BAP complex phenocopied the UGC accumulation phenotype, concluding a role that BAP chromatin remodeling complex in escort cells suppresses BMP signaling and facilitates cystoblast differentiation. In addition, Osa maintains the cellular extension of ECs which wraps and plays a role in promoting germline differentiation. Taken together, the epigenetic factor Osa plays an important role in controlling EC characteristics and germline lineage differentiation. We also provide evidence that Osa influences cap cell and GSC number, and Osa functions in adult ECs for GSC maintenance. The new findings would contribute to our understanding on how the Drosophila ovarian germline niche is established and in a broader view, the role of epigenetic machinery in defining the niche activity.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/12/3/363/s1. Figure S1: c587-gal4-driven osa RNAi knock down Osa in ECs efficiently. (A–C’) Germaria are stained for Osa (red) and GFP (green). mCD8-GFP is used to mark the EC cell membranes (detected by GFP). c587-gal4 is used for osa KD. The control germarium (A,A’) exhibit Osa staining in both germline and somatic cells, including ECs (indicated by white arrows). The osa KD (B–C’) germaria exhibited faint Osa staining in ECs (indicated by white arrows). (D) Graph shows the quantification of Osa staining mean fluorescence intensity ratio compared with the adjacent germline cells. Scale bar is shown in (A). Figure S2: Osa functions in both pupal stage and adulthood after EC formation for germline differentiation control. (A–H) Germaria are stained for α-spectrin (red), Armadillo (red) and DAPI (blue). c587ts is used for osa KD. (A, B) Flies are raised at 18 °C up to early pupal stage and then shifted to 29°C. The newly born females are dissected. (C,D) Flies are raised at 18 °C up to eclosion, and then shifted to 29°C for 14 days prior to dissection. (F–H) Flies are kept at 18 °C and dissected immediately after eclosion. The control germaria (A,C,F) exhibit normal number of CBs. The osa RNAi-1 germaria (B,D) exhibit UGCs accumulation. c587ts >osa RNAi-2 gives the similar phenotype and figures are not shown. (G,H) osa RNAi-1 (G) and osa RNAi-2 (H) groups exhibit normal procedure of germline differentiation. (E,I) Graph shows the quantification of UGCs per germarium. Error bars are presented as Mean ± SD. Several compressed z-sections are shown in (A–D,F–G). Scale bar is shown in (A). (J) Graph shows the quantification of GSC number per germarium. Figure S3: c587-gal4-driven Brm KD germarium exhibits UGCs accumulation and ectopic dpp2.0-lacZ activity in ECs. (A,B) Germaria are stained for α-spectrin (red), Armadillo (red) and DAPI (blue). CpCs are indicated by dashed circles. c587-gal4 is used for osa KD. The control germarium (A) exhibits normal number of CB. The brm KD germarium transferred to 29 °C (B) exhibits UGCs accumulation. (C,D) Graph shows the quantification of UGC per germarium. Error bars are presented as Mean ± SD. (E) Graph shows the quantification of UGC per germarium. Error bars are presented as Mean ± SD. (F,G) Germaria are stained for α-spectrin (red), Armadillo (red) and β-gal (green). dpp transcription is monitored by dpp2.0-lacZ (detected by β-gal). CpCs are indicated by dashed circles. The control germarium (F) exhibits specific dpp2.0-lacZ activity in CpCs. The brm RNAi-2 germarium (G) exhibits ectopic dpp2.0-lacZ activity in ECs. Several compressed z-sections are shown in (A,B,F,G). Scale bar is shown in (A). Figure S4: c587-gal4-driven osa RNAi didn’t change hh-lacZ expression level in ECs and result in reduced cap cell and GSC number. (A,B) Germaria are stained for α-spectrin (red) and β-gal (green). TF and CpCs are marked by hh-lacZ (detected by β-gal). CpCs are indicated by white dashed circles. c587-gal4 is used for osa KD. In control germarium (A) and osa KD germarium (B), hh-lacZ is highly expressed in TF and CpCs. C587>osa RNAi-2 gives the similar phenotype and figure is not shown. (C) Graph shows the quantification of hh-lacZ mean fluorescence intensity. (D) Graph shows the quantification of CpC per germarium. Error bars are presented as Mean ± SD. (E) Graph shows the quantification of GSC per germarium. Error bars are presented as Mean ± SD. (E) Graph shows the quantification of GSC per germarium.
Author Contributions
X.H. (Xiaolong Hu) and M.L. conceived and designed the study; X.H. (Xiaolong Hu) performed the experiments and analyzed the data; X.H. (Xiaolong Hu) and M.L. wrote the manuscript; X.H. (Xue Hao) and Y.L. provided technical support; L.Z. provided critical suggestions; L.Z. and G.W. provided the funding. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2019YFA0802001 and 2017YFA0103601 for Lei Zhang.), National Natural Science foundation of China (No. 31530043 and 31625017 for Lei Zhang.), “Strategic Priority Research Program” of Chinese Academy of Sciences (XDB19000000 for Lei Zhang.), and Shanghai Leading Talents Program for Lei Zhang. National Key R&D Program of China (YS2020YFA090044), National Natural Science Foundation of China (grant numbers 31670106, 31872627, and 31470223), and Shanghai Jiao Tong University Scientific and Technological Innovation Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.
Acknowledgments
We sincerely thank ED. Laufer, Ting Xie, Dennis M McKearin, Yu Cai, the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, and the Developmental Studies Hybridoma Bank for providing fly stocks and antibodies.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, L.; Xie, T. STEM CELL NICHE: Structure and Function. Annu. Rev. Cell Dev. Biol. 2005, 21, 605–631. [Google Scholar] [CrossRef]
- Morrison, S.J.; Spradling, A.C. Stem Cells and Niches: Mechanisms That Promote Stem Cell Maintenance throughout Life. Cell 2008, 132, 598–611. [Google Scholar] [CrossRef]
- Lin, H. The stem-cell niche theory: Lessons from flies. Nat. Rev. Genet 2002, 3, 931–940. [Google Scholar] [CrossRef]
- Chen, D.; McKearin, D. Gene Circuitry Controlling a Stem Cell Niche. Curr. Biol. 2005, 15, 179–184. [Google Scholar] [CrossRef]
- Liu, Z.; Zhong, G.; Chai, P.C.; Luo, L.; Liu, S.; Yang, Y.; Baeg, G.-H.; Cai, Y. Coordinated niche-associated signals promote germline homeostasis in the Drosophila ovary. J. Cell Biol. 2015, 211, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Page-McCaw, A. Wnt6 maintains anterior escort cells as an integral component of the germline stem cell niche. Development 2018, 145, dev158527. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Spradling, A.C. A niche maintaining germ line stem cells in the Drosophila ovary. Science 2000, 290, 328–330. [Google Scholar] [CrossRef]
- Kirilly, D.; Wang, S.; Xie, T. Self-maintained escort cells form a germline stem cell differentiation niche. Developement 2011, 138, 5087–5097. [Google Scholar] [CrossRef]
- Morris, L.X.; Spradling, A.C. Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development 2011, 138, 2207–2215. [Google Scholar] [CrossRef]
- Xie, T.; Spradling, A.C. Decapentaplegic is Essential for the Maintenance and Division of Germline Stem Cells in the Drosophila Ovary. Cell 1998, 94, 251–260. [Google Scholar] [CrossRef]
- Chen, D.; McKearin, D. Dpp Signaling Silences bam Transcription Directly to Establish Asymmetric Divisions of Germline Stem Cells. Curr. Biol. 2003, 13, 1786–1791. [Google Scholar] [CrossRef] [PubMed]
- McKearin, D.M.; Spradling, A.C. bag-of-marbles: A Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990, 4, 2242–2251. [Google Scholar] [CrossRef] [PubMed]
- Ohlstein, B.; McKearin, D. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development 1997, 124, 3651–3662. [Google Scholar]
- Song, X. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development 2004, 131, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Konig, A. Ecdysteroids affect Drosophila ovarian stem cell niche formation and early germline differentiation. EMBO J. 2011, 30, 1549–1562. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, M.; Cortez, Y.M.; Wong-Deyrup, S.; Tavares, L.; Schowalter, S.; Flora, P.; Hill, C.; Nasrallah, M.A.; Chittur, S.; Rangan, P. Transposon Dysregulation Modulates dWnt4 Signaling to Control Germline Stem Cell Differentiation in Drosophila. PLoS Genet. 2016, 12, e1005918. [Google Scholar] [CrossRef]
- Banisch, T.U.; Maimon, I.; Dadosh, T.; Gilboa, L. Escort cells generate a dynamic compartment for germline stem cell differentiation via combined Stat and Erk signalling. Development 2017, 144, 1937–1947. [Google Scholar] [CrossRef]
- Luo, L.; Wang, H.; Fan, C.; Liu, S.; Cai, Y. Wnt ligands regulate Tkv expression to constrain Dpp activity in the Drosophila ovarian stem cell niche. J. Cell Biol. 2015, 209, 595–608. [Google Scholar] [CrossRef]
- Liu, M.; Lim, T.M.; Cai, Y. The Drosophila Female Germline Stem Cell Lineage Acts to Spatially Restrict DPP Function Within the Niche. Sci. Signal. 2010, 3, ra57. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pan, L.; Wang, S.; Zhou, J.; McDowell, W.; Park, J.; Haug, J.; Staehling, K.; Tang, H.; Xie, T. Histone H3K9 Trimethylase Eggless Controls Germline Stem Cell Maintenance and Differentiation. PLoS Genet. 2011, 7, e1002426. [Google Scholar] [CrossRef]
- Eliazer, S.; Shalaby, N.A.; Buszczak, M. Loss of lysine-specific demethylase 1 nonautonomously causes stem cell tumors in the Drosophila ovary. Proc. Natl. Acad. Sci. USA 2011, 108, 7064–7069. [Google Scholar] [CrossRef] [PubMed]
- Eliazer, S.; Palacios, V.; Wang, Z.; Kollipara, R.K.; Kittler, R.; Buszczak, M. Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells. PLoS Genet. 2014, 10, e1004200. [Google Scholar] [CrossRef]
- Jin, Z.; Flynt, A.S.; Lai, E.C. Drosophila piwi mutants exhibit germline stem cell tumors that are sustained by elevated Dpp signaling. Curr. Biol. 2013, 23, 1442–1448. [Google Scholar] [CrossRef]
- Ma, X.; Wang, S.; Do, T.; Song, X.; Inaba, M.; Nishimoto, Y.; Liu, L.-P.; Gao, Y.; Mao, Y.; Li, H.; et al. Piwi Is Required in Multiple Cell Types to Control Germline Stem Cell Lineage Development in the Drosophila Ovary. PLoS ONE 2014, 9, e90267. [Google Scholar] [CrossRef] [PubMed]
- Xuan, T.; Xin, T.; He, J.; Tan, J.; Gao, Y.; Feng, S.; He, L.; Zhao, G.; Li, M. dBre1/dSet1-dependent pathway for histone H3K4 trimethylation has essential roles in controlling germline stem cell maintenance and germ cell differentiation in the Drosophila ovary. Dev. Biol. 2013, 379, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Li, X. Control of germline stem cell differentiation by Polycomb and Trithorax group genes in the niche microenvironment. Development 2016, 143, 3449–3458. [Google Scholar] [CrossRef]
- Yang, Z.; Sun, J.; Hu, Y.; Wang, F.; Wang, X.; Qiao, H.-H.; Xu, J.; Mao, D.; Ren, X.; Pan, L.-X.; et al. Histone H1 defect in escort cells triggers germline tumor in Drosophila ovary. Dev. Biol. 2017, 424, 40–49. [Google Scholar] [CrossRef]
- Mohrmann, L.; Langenberg, K.; Krijgsveld, J.; Kal, A.J.; Heck, A.J.R.; Verrijzer, C.P. Differential Targeting of Two Distinct SWI/SNF-Related Drosophila Chromatin-Remodeling Complexes. Mol. Cell. Biol. 2004, 24, 3077–3088. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.T.; Furukawa, T.; Tanese, N.; Treisman, J.E. Osa associates with the Brahma chromatin remodeling complex and romotes the activation of some target genes. EMBO J. 1999, 18, 7029–7040. [Google Scholar] [CrossRef]
- Mohrmann, L.; Verrijzer, C.P. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 2005, 1681, 59–73. [Google Scholar] [CrossRef]
- Reisman, D.; Glaros, S.; Thompson, E. The SWI/SNF complex and cancer. Oncogene 2009, 28, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Tamkun, J.W.; Deuring, R.; Scott, M.P.; Kissinger, M.; Pattatucci, A.M.; Kaufman, T.C.; Kennison, J.A. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2SWI2. Cell 1992, 68, 561–572. [Google Scholar] [CrossRef]
- Crosby, M.A. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 1999, 19, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Dingwall, A.K.; Beek, S.J.; McCallum, C.M.; Tamkun, J.W.; Kalpana, G.V.; Goff, S.P.; Scott, M.P. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell 1995, 6, 777–791. [Google Scholar] [CrossRef]
- Moshkin, Y.M.; Mohrmann, L.; van Ijcken, W.F.; Verrijzer, C.P. Functional Differentiation of SWI/SNF Remodelers in Transcription and Cell Cycle Control. Mol. Cell. Biol. 2006, 27, 651–661. [Google Scholar] [CrossRef]
- Vázquez, M.; Moore, L.; Kennison, J.A. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development 1999, 126, 733–742. [Google Scholar] [PubMed]
- Zeng, X.; Lin, X.; Hou, S.X. The Osa-containing SWI/SNF chromatin-remodeling complex regulates stem cell commitment in the adult Drosophila intestine. Development 2013, 140, 3532–3540. [Google Scholar] [CrossRef]
- Eroglu, E.; Burkard, T.R.; Jiang, Y.; Saini, N.; Homem, C.C.F.; Reichert, H.; Knoblich, J.A. SWI/SNF Complex Prevents Lineage Reversion and Induces Temporal Patterning in Neural Stem Cells. Cell 2014, 156, 1259–1273. [Google Scholar] [CrossRef] [PubMed]
- Baig, J.; Chanut, F.; Kornberg, T.B.; Klebes, A. The Chromatin-Remodeling Protein Osa Interacts With CyclinE in Drosophila Eye Imaginal Discs. Genetics 2010, 184, 731–744. [Google Scholar] [CrossRef]
- Terriente-Felix, A.; de Celis, J.F. A subunit of the BAP chromatin-remodelling complex, participates in the regulation of gene expression in response to EGFR signalling in the Drosophila wing. Dev. Biol. 2009, 329, 350–361. [Google Scholar] [CrossRef]
- Collins, R.T.; Treisman, J.E. Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev. 2000, 14, 3140–3152. [Google Scholar] [CrossRef]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.A.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Shain, A.H.; Pollack, J.R. The Spectrum of SWI/SNF Mutations, Ubiquitous in Human Cancers. PLoS ONE 2013, 8, e55119. [Google Scholar] [CrossRef] [PubMed]
- Ronan, J.L.; Wu, W.; Crabtree, G.R. From neural development to cognition: Unexpected roles for chromatin. Nat. Rev. Genet. 2013, 14, 347–359. [Google Scholar] [CrossRef]
- Mathur, R. ARID1A loss in cancer: Towards a mechanistic understanding. Pharmacol. Ther. 2018, 190, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Gancz, D.; Gilboa, L. Insulin and Target of rapamycin signaling orchestrate the development of ovarian niche-stem cell units in Drosophila. Development 2013, 140, 4145–4154. [Google Scholar] [CrossRef]
- Gancz, D.; Lengil, T.; Gilboa, L. Coordinated Regulation of Niche and Stem Cell Precursors by Hormonal Signaling. PLoS Biol. 2011, 9, e1001202. [Google Scholar] [CrossRef]
- Li, Q.; Xin, T.; Chen, W.; Zhu, M.; Li, M. Lethal(2)giant larvae is required in the follicle cells for formation of the initial AP asymmetry and the oocyte polarity during Drosophila oogenesis. Cell Res. 2008, 18, 372–384. [Google Scholar] [CrossRef]
- Kai, T.; Williams, D.; Spradling, A.C. The expression profile of purified Drosophila germline stem cells. Dev. Biol. 2005, 283, 486–502. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Yue, L.; Spradling, A.C. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 1994, 120, 947–956. [Google Scholar] [PubMed]
- Kai, T.; Spradling, A. An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc. Natl. Acad. Sci. USA 2003, 100, 4633–4638. [Google Scholar] [CrossRef]
- Decotto, E.; Spradling, A.C. The Drosophila Ovarian and Testis Stem Cell Niches: Similar Somatic Stem Cells and Signals. Dev. Cell 2005, 9, 501–510. [Google Scholar] [CrossRef]
- Lai, C.-M.; Lin, K.-Y.; Kao, S.-H.; Chen, Y.-N.; Huang, F.; Hsu, H.-J. Hedgehog signaling establishes precursors for germline stem cell niches by regulating cell adhesion. J. Cell Biol. 2017, 216, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.-H.; Xie, T. Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development 2003, 130, 2579–2588. [Google Scholar] [CrossRef]
- McGuire, S.E.; Mao, Z.; Davis, R.L. Spatiotemporal Gene Expression Targeting with the TARGET and Gene-Switch Systems in Drosophila. Sci. Signal. 2004, 2004, pl6. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Winter, C.; Marticke, S.S.; Lee, A.; Luo, L. Essential Roles of Drosophila RhoA in the Regulation of Neuroblast Proliferation and Dendritic but Not Axonal Morphogenesis. Neuron 2000, 25, 307–316. [Google Scholar] [CrossRef]
- He, J.; Xuan, T.; Xin, T.; An, H.; Wang, J.; Zhao, G.; Li, M. Evidence for Chromatin-Remodeling Complex PBAP-Controlled Maintenance of the Drosophila Ovarian Germline Stem Cells. PLoS ONE 2014, 9, e103473. [Google Scholar] [CrossRef] [PubMed]
- Tsuneizumi, K.; Nakayama, T.; Kamoshida, Y.; Kornberg, T.B.; Christian, J.L.; Tabata, T. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nat. Cell Biol. 1997, 389, 627–631. [Google Scholar] [CrossRef]
- Zhao, R.; Xuan, Y.; Li, X.; Xi, R. Age-related changes of germline stem cell activity, niche signaling activity and egg production in Drosophila. Aging Cell 2008, 7, 344–354. [Google Scholar] [CrossRef]
- Chen, D.; McKearin, D.M. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 2003, 130, 1159–1170. [Google Scholar] [CrossRef]
- Wang, S.; Gao, Y.; Song, X.; Ma, X.; Zhu, X.; Mao, Y.; Yang, Z.; Ni, J.; Li, H.; Malanowski, K.E.; et al. Wnt signaling-mediated redox regulation maintains the germ line stem cell differentiation niche. eLife 2015, 4, e08174. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Siah, C.K.; Cai, Y. Engrailed acts with Nejire to control decapentaplegic expression in the Drosophila ovarian stem cell niche. Development 2017, 144, 3224–3231. [Google Scholar] [CrossRef]
- Treisman, J.E.; Luk, A.; Rubin, G.M.; Heberlein, U. eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev. 1997, 11, 1949–1962. [Google Scholar] [CrossRef]
- Song, X.; Call, G.B.; Kirilly, D.; Xie, T. Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development 2007, 134, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.-J.; Drummond-Barbosa, D. Insulin signals control the competence of the Drosophila female germline stem cell niche to respond to Notch ligands. Dev. Biol. 2011, 350, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Yatsenko, A.S.; Shcherbata, H.R. Stereotypical architecture of the stem cell niche is spatiotemporally established by miR-125-dependent coordination of Notch and steroid signaling. Development 2018, 145, dev159178. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).