Autosomal Microsatellite Investigation Reveals Multiple Genetic Components of the Highlanders from Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples, DNA Amplification and STR Typing
2.2. Statistical Analyses
3. Results
3.1. Genetic Diversity and Forensic Parameters
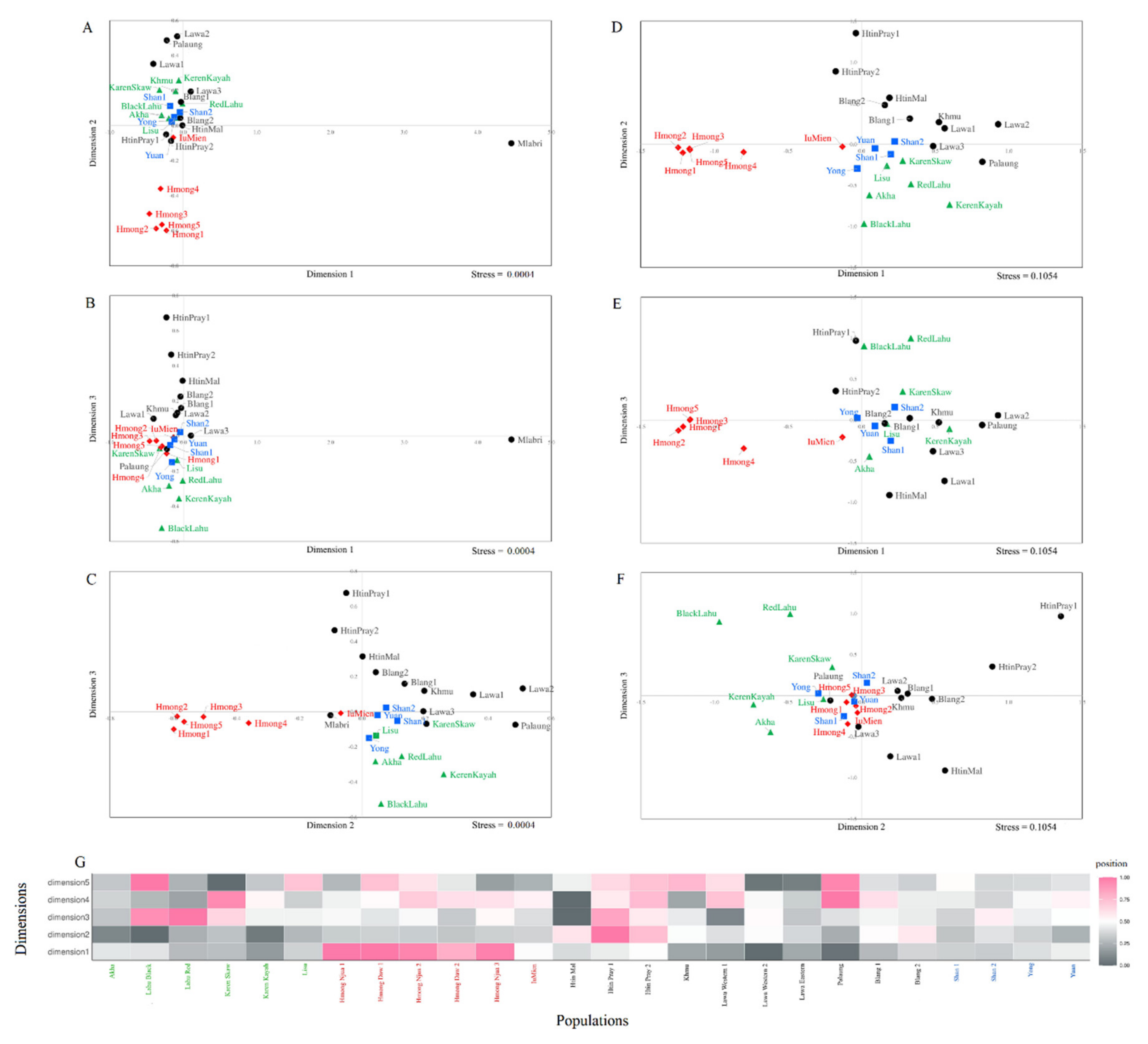
3.2. Genetic Relationship and Genetic Structure
3.3. AMOVA Results
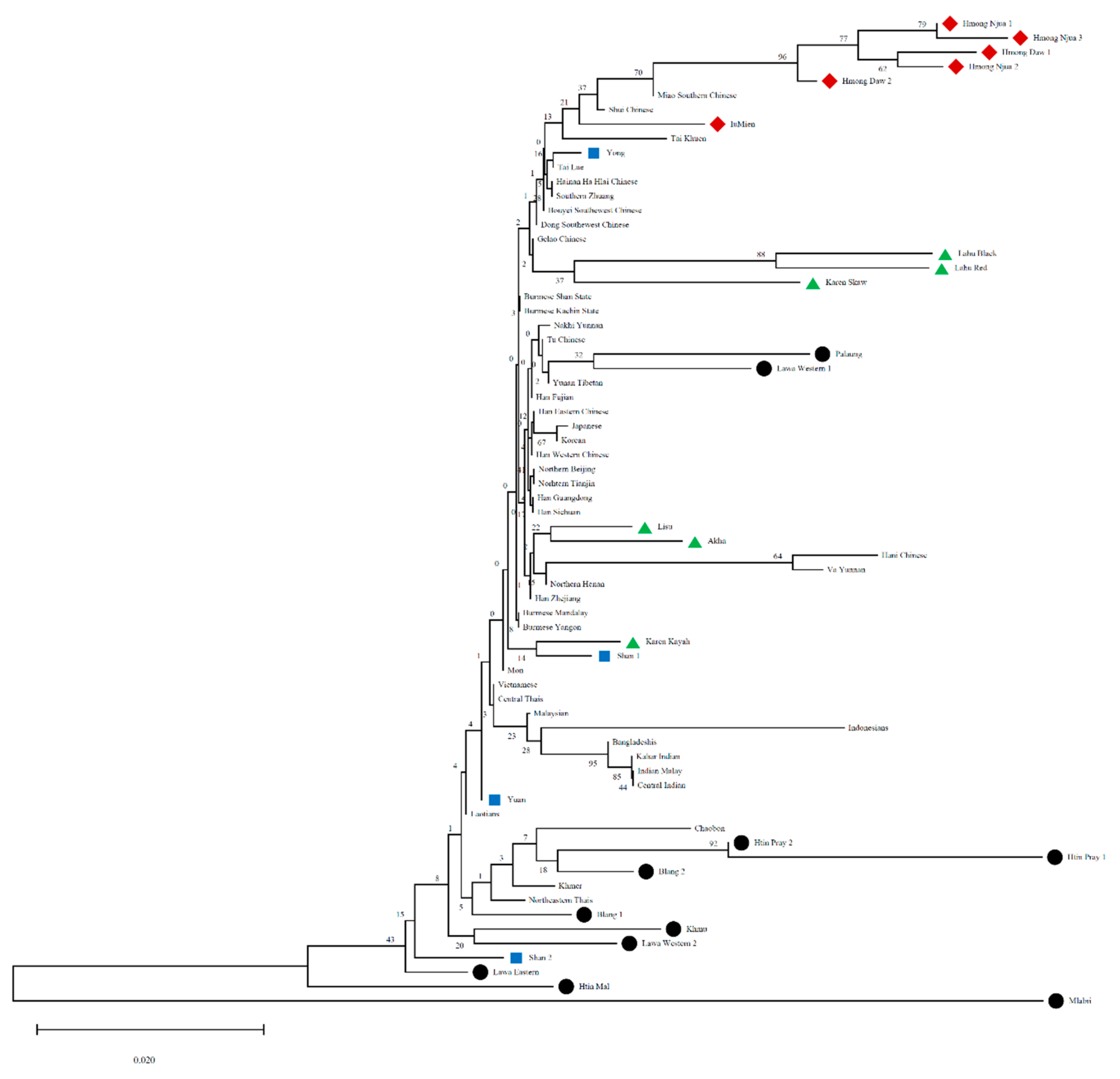
3.4. Asian Phylogenetic Tree
4. Discussion
4.1. The Genetic Structure of the Hmong and Their Linguistic Relative, IuMien
4.2. The Genetic Structure of the Lahu and Their Linguistic Relatives, Lisu and Akha
4.3. The Genetic Structure of the Htin and Their Linguistic Relatives, Khmu and Mlabri
4.4. The Genetic Structure of the Palaung and Their Linguistic Relatives, Blang and Lawa
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schliesinger, J. Ethnic Groups of Thailand: Non-Tai-Speaking Peoples; White Lotus Press: Bangkok, Thailand, 2000. [Google Scholar]
- Penth, H.; Forbes, A. The people of mountaintops. In A Brief History of Lan Na and the Peoples of Chiang Mai; Penth, H., Forbes, A., Eds.; Chiang Mai City Arts and Cultural Centre Chiang Mai Municipality: Chiang Mai, Thailand, 2004; pp. 247–254. [Google Scholar]
- Eberhard, D.M.; Simons, G.F.; Fennig, C.D. Ethnologue: Languages of the World, 23rd ed.; SIL International: Dallas, TX, USA, 2020. [Google Scholar]
- Suwanwela, C.; Kanchanahuta, S.; Onthuam, Y. Hill tribe opium addicts: A retrospective study of 1,382 patients. Bull. Narc. 1979, 31, 23–40. [Google Scholar]
- Saihoo, P. The Hill Tribes of Northern Thailand and the Opium Problem; Faculty of Political Science, Chulalongkorn University: Bangkok, Thailand, 1963; pp. 35–45. [Google Scholar]
- Vatahong, C. Drug Problems among Hill Tribe Thais; First National Academic Conference on Dependent Drug, Drug Suppression Unit; Prime Minister’s Office: Bangkok, Thailand, 2002; pp. 36–45. [Google Scholar]
- Marks, T.A. The Meo Hill Tribe Problem in North Thailand. Asian Surv. 1973, 13, 929–944. [Google Scholar] [CrossRef]
- Oota, H.; Settheetham-Ishida, W.; Tiwawech, D.; Ishida, T.; Stoneking, M. Human mtDNA and Y-chromosome variation is correlated with matrilocal versus patrilocal residence. Nat. Genet. 2001, 29, 20–21. [Google Scholar] [CrossRef] [PubMed]
- Apidechkul, T. A 20-year retrospective cohort study of TB infection among the Hill-tribe HIV/AIDS populations, Thailand. BMC Infect. Dis. 2016, 16, 72. [Google Scholar]
- Besaggio, D.; Fuselli, S.; Srikummool, M.; Kampuansai, J.; Castrì, L.; Tyler-Smith, C.; Seielstad, M.; Kangwanpong, D.; Bertorelle, G. Genetic variation in Northern Thailand Hill Tribes: Origins and relationships with social structure and linguistic differences. BMC Evol. Biol. 2007, 7 (Suppl. 2), S12. [Google Scholar] [CrossRef]
- Kutanan, W.; Kampuansai, J.; Srikummool, M.; Brunelli, A.; Ghirotto, S.; Arias, L.; Macholdt, E.; Hübner, A.; Schröder, R.; Stoneking, M. Contrasting Paternal and Maternal Genetic Histories of Thai and Lao Populations. Mol. Biol. Evol. 2019, 3, 1490–1506. [Google Scholar] [CrossRef] [PubMed]
- Kutanan, W.; Shoocongdej, R.; Srikummool, M.; Hübner, A.; Suttipai, T.; Srithawong, S.; Kampuansai, J.; Stoneking, M. Cultural variation impacts paternal and maternal genetic lineages of the Hmong-Mien and Sino-Tibetan groups from Thailand. Eur. J. Hum. Genet. 2020, 28, 1563–1579. [Google Scholar] [CrossRef]
- Brunelli, A.; Kampuansai, J.; Seielstad, M.; Lomthaisong, K.; Kangwanpong, D.; Ghirotto, S.; Kutanan, W. Y chromosomal evidence on the origin of northern Thai people. PLoS ONE 2017, 12, e0181935. [Google Scholar] [CrossRef]
- Kampuansai, J.; Völgyi, A.; Kutanan, W.; Kangwanpong, D.; Pamjav, H. Autosomal STR variations reveal genetic heterogeneity in the Mon-Khmer speaking group of Northern Thailand. Forensic Sci. Int. Genet. 2017, 27, 92–99. [Google Scholar] [CrossRef]
- Kutanan, W.; Srikummool, M.; Pittayaporn, P.; Seielstad, M.; Kangwanpong, D.; Kumar, V.; Prombanchachai, T.; Chantawannakul, P. Admixed origin of the Kayah (Red Karen) in Northern Thailand Revealed by Biparental and Paternal Markers. Ann. Hum. Genet. 2015, 7, 108–122. [Google Scholar] [CrossRef]
- Listman, J.B.; Malison, R.T.; Sanichwankul, K.; Ittiwut, C.; Mutirangura, A.; Gelernter, J. Southeast Asian origins of five Hill Tribe populations and correlation of genetic to linguistic relationships inferred with genome-wide SNP data. Am. J. Phys. Anthropol. 2011, 144, 300–308. [Google Scholar] [CrossRef]
- Xu, S.; Kangwanpong, D.; Seielstad, M.; Srikummool, M.; Kampuansai, J.; Jin, L. Genetic evidence supports linguistic affinity of Mlabri-a hunter-gatherer group in Thailand. BMC Genet. 2010, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Kutanan, W.; Liu, D.; Kampuansai, J.; Srikummool, M.; Srithawong, S.; Shoocongdej, R.; Sangkhano, S.; Ruangchai, S.; Pittayaporn, P.; Arias, L.; et al. Reconstructing the human genetic history of mainland Southeast Asia: Insights from genome-wide data from Thailand and Laos. bioRxiv 2020. [Google Scholar] [CrossRef]
- Silva, N.M.; Pereira, L.; Poloni, E.S.; Currat, M. Human neutral genetic variation and forensic STR data. PLoS ONE 2012, 7, e49666. [Google Scholar] [CrossRef] [PubMed]
- Kutanan, W.; Kampuansai, J.; Changmai, P.; Flegontov, P.; Schröder, R.; Macholdt, E.; Hübner, A.; Kangwanpong, D.; Stoneking, M. Contrasting maternal and paternal genetic variation of hunter–gatherer groups in Thailand. Sci. Rep. 2018, 8, 1536. [Google Scholar] [CrossRef]
- Kutanan, W.; Kampuansai, J.; Colonna, V.; Nakbunlung, S.; Lertvicha, P.; Seielstad, M.; Bertorelle, G.; Kangwanpong, D. Genetic affinity and admixture of northern Thai people along their migration route in northern Thailand: Evidence from autosomal STR loci. J. Hum. Genet. 2011, 56, 130–137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Excoffier, L.; Lischer, H. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Promega. Powerstats Version 1.2, Tools for Analysis of Population Statistics. 1999. Available online: https://www.promega.com.cn/products/genetic-identity/ (accessed on 5 February 2021).
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Falush, D.; Stepheas, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 156–187. [Google Scholar]
- Hubisz, M.; Falush, D.; Stephens, M.; Pritchard, J. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population Structure. Mole Ecol. Notes 2003, 4, 137–138. [Google Scholar] [CrossRef]
- Batham, M.S.; Kushwaha, K.P.S.; Chauhan, T.; Kumawat, R.K.; Shrivastava, P. Autosomal STR allele frequencies in Kahars of Uttar Pradesh, India, drawn with PowerPlex® 21 multiplex system. Int. J. Legal Med. 2020, 134, 517–519. [Google Scholar] [CrossRef]
- Brinkmann, B.; Shimada, I.; Tuyen, N.; Hohoff, C. Allele frequency data for 16 STR loci in the Vietnamese population. Int. J. Legal Med. 2002, 116, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Duan, L.; Yuan, L.; Shen, Z.; He, W.; Zhai, D.; Huang, Y.; Xu, B. Genetic polymorphisms of 19 autosomal STR loci in the China Burmese immigrants. Forensic Sci. Int. Genet. 2017, 31, e46–e47. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhu, B.; Yu, X.; Li, Y.; Fang, J.; Xiong, X.; Mu, H.; Huang, Y.; Shi, X. Genetic polymorphisms of 15 STR loci of Chinese Dongxiang and Salar ethnic minority living in Qinghai Province of China. Leg Med. (Tokyo) 2007, 9, 38–42. [Google Scholar] [CrossRef]
- Dobashi, Y.; Kido, A.; Fujitani, N.; Susukida, R.; Oya, M. Population data of nine STR loci, D3S1358, vWA, FGA, TH01, TPOX, CSF1PO, D5S818, D13S317 and D7S820, in Bangladeshis and Indonesians. Forensic Sci. Int. 2003, 135, 72–74. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, Z.; Wang, Z.; Bian, S. Genetic data of 15 STR forensic loci in eastern Chinese population. Forensic Sci. Int. 2005, 154, 78–80. [Google Scholar] [CrossRef] [PubMed]
- Guo, F. Allele frequencies of 17 autosomal STR loci in the Va ethnic minority from Yunnan Province, Southwest China. Int. J. Legal Med. 2017, 131, 1251–1252. [Google Scholar] [CrossRef]
- Guo, F.; Li, J.; Wei, T.; Ye, Q.; Chen, Z. Genetic variation of 17 autosomal STR loci in the Zhuang ethnic minority from Guangxi Zhuang Autonomous Region in the south of China. Forensic Sci. Int. Genet. 2017, 28, e51–e52. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Chen, X.; Duan, Y.; Xia, Y.; He, W.; Wu, J.; Xu, B. Allele frequencies for fifteen autosomal STR loci in a Nakhi population from Yunnan Province, Southwest China. Forensic Sci. Int. Genet. 2016, 21, e13–e14. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yao, J.; Li, J.; Wen, J.; Yuan, X.; Xu, B. Population genetic data for 17 autosomal STR markers in the Hani population from China. Int. J. Legal Med. 2015, 129, 995–996. [Google Scholar] [CrossRef]
- Kim, Y.L.; Hwang, J.Y.; Kim, Y.J.; Lee, S.; Chung, N.G.; Goh, H.G.; Kim, C.C.; Kim, D.W. Allele frequencies of 15 STR loci using AmpF/STR Identifiler kit in a Korean population. Forensic Sci. Int. 2003, 136, 92–95. [Google Scholar] [CrossRef]
- Li, W.; Wang, X.; Wang, X.; Wang, F.; Du, Z.; Fu, F.; Wu, W.; Wang, S.; Mu, Z.; Chen, C.; et al. Forensic characteristics and phylogenetic analyses of one branch of Tai-Kadai language-speaking Hainan Hlai (Ha Hlai) via 23 autosomal STRs included in the Huaxia™ Platinum System. Mol. Genet. Genom. Med. 2020, 8, e1462. [Google Scholar] [CrossRef]
- Maruyama, S.; Minaguchi, K.; Takezaki, N.; Nambiar, P. Population data on 15 STR loci using AmpF/STR Identifiler kit in a Malay population living in and around Kuala Lumpur, Malaysia. Legal Med. 2008, 10, 160–162. [Google Scholar] [CrossRef][Green Version]
- Shrivastava, P.; Jain, T.; Trivedi, V.B. Genetic polymorphism study at 15 autosomal locus in central Indian population. SpringerPlus 2015, 4, 566. [Google Scholar] [CrossRef]
- Seah, L.H.; Jeevan, N.H.; Othman, M.I.; Jaya, P.; Ooi, Y.S.; Wong, P.C.; Kee, S.S. STR Data for the AmpFlSTR Identifiler loci in three ethnic groups (Malay, Chinese, Indian) of the Malaysian population. Forensic Sci. Int. 2003, 138, 134–137. [Google Scholar] [CrossRef]
- Song, X.B.; Zhou, Y.; Ying, B.W.; Wang, L.L.; Li, Y.S.; Liu, J.F.; Bai, X.G.; Zhang, L.; Lu, X.J.; Wang, J.; et al. Short-tandem repeat analysis in seven Chinese regional populations. Genet. Mol. Biol. 2010, 33, 605–609. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Srithawong, S.; Muisuk, K.; Srikummool, M.; Mahasirikul, N.; Triyarach, S.; Sriprasert, K.; Kutanan, W. Genetic structure of the ethnic Lao groups from mainland Southeast Asia revealed by forensic microsatellites. Ann. Hum. Genet. 2020, 84, 357–369. [Google Scholar] [CrossRef]
- Srithawong, S.; Muisuk, K.; Srikummool, M.; Kampuansai, J.; Pittayaporn, P.; Ruangchai, S.; Liu, D.; Kutanan, W. Close genetic relationship between central Thai and Mon people in Thailand revealed by autosomal microsatellites. Int. J. Legal Med. 2021, 135, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Wang, X.; Oxida, S. Genetic Polymorphisms of 15 STR Loci in a Japanese Population. Forensic Sci. Int. 2006, 51, 188–189. [Google Scholar] [CrossRef]
- Untoro, E.; Atmadja, D.S.; Pu, C.E.; Wu, F.C. Allele frequency of CODIS 13 in Indonesian population. Leg Med. (Tokyo) 2009, 11 (Suppl. 1), S203–S205. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, Y.; Liu, C.; Chan, D.W.; Chan, M.; He, M. Allele frequencies of 15 STRs in five ethnic groups (Han, Gelao, Jing, Shui and Zhuang) in South China. Forensic Sci. Int. Genet. 2013, 7, e9–e14. [Google Scholar] [CrossRef]
- Zhai, D.; Yang, J.; Huang, Y.; Chen, L.; Wu, D.; Wu, J.; Xu, B. The allele frequency of 15 STRs among three Tibeto-Burman-speaking populations from the southwest region of mainland China. Forensic Sci. Int. Genet. 2014, 13, e22–e24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L. Population data for 15 autosomal STR loci in the Bouyei ethnic minority from Guizhou Province, Southwest China. Forensic Sci. Int. Genet. 2015, 17, 108–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L. Population data for 15 autosomal STR loci in the Dong ethnic minority from Guizhou Province, Southwest China. Forensic Sci. Int. Genet. 2015, 16, 237–238. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Yan, J.; Shen, C.; Li, T.; Li, Y.; Yu, X.; Xiong, X.; Mu, H.; Huang, Y.; Deng, Y. Population genetic analysis of 15 STR loci of Chinese Tu ethnic minority group. Forensic Sci. Int. 2008, 174, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Takezaki, N.; Nei, M.; Tamura, K. POPTREEW: Webersion of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Mol. Biol. Evol. 2014, 31, 1622–1624. [Google Scholar] [CrossRef]
- Srithawong, S.; Srikummool, M.; Pittayaporn, P.; Ghirotto, S.; Chantawannakul, P.; Sun, J.; Eisenberg, A.; Chakraborty, R.; Kutanan, W. Genetic and linguistic correlation of the Kra-Dai-speaking groups in Thailand. J. Hum. Genet. 2015, 60, 371–380. [Google Scholar] [CrossRef]
- Ratliff, M.S. Hmong-Mien Language History; Pacific Linguistics: Canberra, Australia, 2010. [Google Scholar]
- Geddes, W.R. Migrants of the Mountains: The Cultural Ecology of the Blue Miao (Hmong Njua) of Thailand, 1st ed.; Clarendon Press: Oxford, UK, 1976. [Google Scholar]
- Jonsson, H. Thailand Mien Relations: Mountain People and State Control in Thailand; Cornell University Press: New York, NY, USA, 2005. [Google Scholar]
- Lewis, P.; Lewis, E. People of the Golden Triangle; Thames and Hudson Ltd.: London, UK, 1984. [Google Scholar]
- Blench, R. Stratification in the peopling of China: How far does the linguistic evidence match genetics and archaeology. In Past Human Migrations in Continental East Asia and Taiwan: Matching Archaeology, Linguistics and Genetics, 1st ed.; Alicia, S.M., Blench, R., Ross, M.D., Peiros, I., Marie, L., Eds.; Routledge: London, UK, 2008; pp. 105–132. [Google Scholar]
- Matisoff, J.A. Sino-Tibetan linguistics: Present state and future prospects. Annu. Rev. Anthropol. 1991, 20, 469–504. [Google Scholar] [CrossRef]
- Yang, X.; Wan, Z.; Perry, L.; Lu, H.; Wang, Q.; Zhao, C.; Li, J.; Xie, F.; Yu, J.; Cui, T.; et al. Early millet use in northern China. Proc. Natl. Acad. Sci. USA 2012, 109, 3726–3730. [Google Scholar] [CrossRef] [PubMed]
- Sagart, L.; Jacques, G.; Lai, Y.; Ryder, R.J.; Thouzeau, V.; Greenhill, S.J.; List, J.M. Dated language phylogenies shed light on the ancestry of Sino-Tibetan. Proc. Natl. Acad. Sci. USA 2019, 116, 10317–10322. [Google Scholar] [CrossRef]
- Zhang, M.; Yan, S.; Pan, W.; Jin, L. Phylogenetic evidence for Sino-Tibetan origin in northern China in the Late Neolithic. Nature 2019, 569, 112–115. [Google Scholar] [CrossRef]
- Li, Y.; Hong, Y.; Li, X.; Yang, J.; Li, L.; Huang, Y.; Wang, C.; Li, H.; Xu, B. Allele frequency of 19 autosomal STR loci in the Bai population from the southwestern region of mainland China. Electrophoresis 2015, 36, 2498–2503. [Google Scholar] [CrossRef]
- Yao, H.B.; Wang, C.C.; Wang, J.; Tao, X.; Shang, L.; Wen, S.Q.; Du, Q.; Deng, Q.; Xu, B.; Huang, Y.; et al. Genetic structure of Tibetan populations in Gansu revealed by forensic STR loci. Sci. Rep. 2017, 3, 41195. [Google Scholar] [CrossRef]
- Oota, H.; Pakendorf, B.; Weiss, G.; von Haeseler, A.; Pookajorn, S.; Settheetham-Ishida, W.; Tiwawech, D.; Ishida, T.; Stoneking, M. Recent origin and cultural reversion of a hunter-gatherer group. PLoS Biol. 2005, 3, 0536–0542. [Google Scholar] [CrossRef] [PubMed]
- Schrock, L.J. Minority Groups in Thailand; Washington Headquarters Department of the Army, Pamphlet: Washington, DC, USA, 1970; No. 550-107. [Google Scholar]
- Penth, H. A Brief History of Lanna: Civilizations of North. Thailand, 2nd ed.; Silkworm Books: Chiang Mai, Thailand, 2000. [Google Scholar]



| Ethnicity | Populations | Sample Size | Language | References | Gene Diversity (SD) | Average HE | Total Allele | CMP | CPD | CPE | Loci Departed from HWE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Akha | Akha | 38 | Sino-Tibetan | Present study | 0.766 (0.390) | 0.773 | 110 | 1.28 × 10−15 | 0.9999888 | 0.999999999999999 | |
| Lahu | Lahu Black | 25 | Sino-Tibetan | Present study | 0.729 (0.373) | 0.735 | 94 | 1.02 × 10−13 | 0.9999955 | 0.999999999999898 | |
| Lahu Red | 24 | Sino-Tibetan | Present study | 0.707 (0.363) | 0.715 | 91 | 5.88 × 10−13 | 0.9999930 | 0.999999999999412 | ||
| Karen | Karen Skaw | 23 | Sino-Tibetan | Present study | 0.762 (0.390) | 0.767 | 97 | 5.67 × 10−14 | 0.9999942 | 0.999999999999943 | |
| Keren Kayah | 46 | Sino-Tibetan | Kutanan et al. (2015) | 0.752 (0.381046) | 0.752 | 103 | 4.58 × 10−15 | 0.9999984 | 0.999999999999995 | ||
| Lisu | Lisu | 26 | Sino-Tibetan | Present study | 0.759 (0.388) | 0.761 | 100 | 1.58 × 10−14 | 0.9999884 | 0.999999999999984 | |
| Hmong | Hmong Njua 1 | 58 | Hmong–Mien | Present study | 0.719 (0.365) | 0.721 | 111 | 1.76 × 10−14 | 0.9999462 | 0.999999999999982 | |
| Hmong Daw 1 | 21 | Hmong–Mien | Present study | 0.717 (0.371) | 0.726 | 91 | 3.29 × 10−13 | 0.9999813 | 0.999999999999671 | ||
| Hmong Njua 2 | 29 | Hmong–Mien | Present study | 0.736 (0.376) | 0.740 | 102 | 5.39 × 10−14 | 0.9999968 | 0.999999999999946 | ||
| Hmong Daw 2 | 32 | Hmong–Mien | Present study | 0.746 (0.388) | 0.764 | 117 | 4.83 × 10−15 | 0.9999886 | 0.999999999999995 | vWA | |
| Hmong Njua 3 | 17 | Hmong–Mien | Present study | 0.720 (0.372) | 0.720 | 81 | 7.16 × 10−12 | 0.9999871 | 0.999999999992835 | FGA | |
| IuMien | IuMien | 35 | Hmong–Mien | Present study | 0.761 (0.392) | 0.767 | 111 | 5.42 × 10−15 | 0.9999113 | 0.999999999999995 | D18S51 |
| Mlabri | Mlabri | 19 | Austro-Asiatic | Present study | 0.547 (0.288) | 0.5470 | 51 | 8.83 × 10−9 | 0.9986837 | 0.999999991173593 | |
| Htin | Htin Mal | 37 | Austro-Asiatic | Present study | 0.719 (0.366) | 0.733 | 108 | 6.47 × 10−14 | 0.9999270 | 0.999999999999935 | D19S433 |
| Htin Pray 1 | 26 | Austro-Asiatic | Present study | 0.723 (0.370) | 0.739 | 92 | 1.52 × 10−13 | 0.9999562 | 0.999999999999848 | TH01, D18S51 | |
| Htin Pray 2 | 41 | Austro-Asiatic | Present study | 0.765 (0.388) | 0.765 | 106 | 1.8 × 10−15 | 0.9999912 | 0.999999999999998 | ||
| Khmu | Khmu | 26 | Austro-Asiatic | Present study | 0.737 (0.379) | 0.749 | 95 | 3.94 × 10−14 | 0.9999682 | 0.999999999999961 | FGA |
| Lawa | Lawa Western 1 | 39 | Austro-Asiatic | Present study | 0.752 (0.385) | 0.768 | 108 | 5.31 × 10−15 | 0.9999816 | 0.999999999999995 | |
| Lawa Western 2 | 47 | Austro-Asiatic | Kutanan et al. 2011) | 0.751 (0.381) | 0.753 | 103 | 3.82 × 10−15 | 0.9999987 | 0.999999999999996 | ||
| Lawa Eastern | 50 | Austro-Asiatic | Kutanan et al. (2011) | 0.767(0.388) | 0.767 | 114 | 8.99 × 10−16 | 0.9999996 | 0.999999999999999 | ||
| Palaung | Palaung | 54 | Austro-Asiatic | Present study | 0.747 (0.378) | 0.754 | 119 | 2.71 × 10−15 | 0.9999782 | 0.999999999999997 | |
| Blang | Blang 1 | 35 | Austro-Asiatic | Present study | 0.788 (0.400) | 0.797 | 125 | 5.72 × 10−16 | 0.9999717 | 0.999999999999999 | |
| Blang 2 | 27 | Austro-Asiatic | Present study | 0.774 (0.395) | 0.776 | 118 | 2.00 × 10−15 | 0.9999930 | 0.999999999999998 | ||
| Shan | Shan 1 | 44 | Tai-Kadai | Kutanan et al. (2011) | 0.783 (0.396) | 0.783 | 117 | 5.83 × 10−16 | 0.9999939 | 0.999999999999999 | |
| Shan 2 | 22 | Tai-Kadai | Present study | 0.762 (0.390) | 0.768 | 110 | 2.68 × 10−14 | 0.9999996 | 0.999999999999973 | ||
| Yuan | Yuan | 87 | Tai-Kadai | Kutanan et al. (2011) | 0.781 (0.393) | 0.781 | 126 | 7.16 × 10−17 | 0.9999973 | 0.999999999999999 | |
| Yong | Yong | 55 | Tai-Kadai | Kutanan et al. (2011) | 0.776 (0.392) | 0.734 | 125 | 5.23 × 10−16 | 0.9999974 | 0.999999999999999 |
| No. of Groups | No. of Populations | % of Variance | |||
|---|---|---|---|---|---|
| Within Populations | Among Populations within Groups | Among Groups | |||
| All studied sample | 1 | 27 | 96.09 | 3.91 * | |
| Sino-Tibetan (ST) | 1 | 6 | 96.69 | 3.31 * | |
| Hmong–Mien (HM) | 1 | 6 | 98.61 | 1.39 * | |
| Austroasiatic (AA) | 1 | 11 | 95.07 | 4.93 * | |
| Austroasiatic (excluding the Mlabri) | 1 | 10 | 96.27 | 3.73 * | |
| Tai-Kadai (TK) | 1 | 4 | 99.46 | 0.54 * | |
| ST/HM/AA/TK | 4 | 27 | 95.83 | 3.11 * | 1.06 * |
| AA vs. ST | 2 | 17 | 94.95 | 4.46 * | 0.59 * |
| AA vs. TK | 2 | 12 | 96.35 | 3.79 | −0.14 |
| AA vs. HM | 2 | 14 | 93.67 | 3.87 | 2.45 * |
| HM vs. TK | 2 | 10 | 97.28 | 0.83 * | 1.90 * |
| HM vs. ST | 2 | 12 | 95.20 | 2.37 * | 2.42 * |
| ST vs. TK | 2 | 10 | 98.02 | 1.95 * | 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mawan, A.; Prakhun, N.; Muisuk, K.; Srithawong, S.; Srikummool, M.; Kampuansai, J.; Shoocongdej, R.; Inta, A.; Ruangchai, S.; Kutanan, W. Autosomal Microsatellite Investigation Reveals Multiple Genetic Components of the Highlanders from Thailand. Genes 2021, 12, 383. https://doi.org/10.3390/genes12030383
Mawan A, Prakhun N, Muisuk K, Srithawong S, Srikummool M, Kampuansai J, Shoocongdej R, Inta A, Ruangchai S, Kutanan W. Autosomal Microsatellite Investigation Reveals Multiple Genetic Components of the Highlanders from Thailand. Genes. 2021; 12(3):383. https://doi.org/10.3390/genes12030383
Chicago/Turabian StyleMawan, Aornpriya, Nonglak Prakhun, Kanha Muisuk, Suparat Srithawong, Metawee Srikummool, Jatupol Kampuansai, Rasmi Shoocongdej, Angkhana Inta, Sukhum Ruangchai, and Wibhu Kutanan. 2021. "Autosomal Microsatellite Investigation Reveals Multiple Genetic Components of the Highlanders from Thailand" Genes 12, no. 3: 383. https://doi.org/10.3390/genes12030383
APA StyleMawan, A., Prakhun, N., Muisuk, K., Srithawong, S., Srikummool, M., Kampuansai, J., Shoocongdej, R., Inta, A., Ruangchai, S., & Kutanan, W. (2021). Autosomal Microsatellite Investigation Reveals Multiple Genetic Components of the Highlanders from Thailand. Genes, 12(3), 383. https://doi.org/10.3390/genes12030383






