Non-Target Effects of dsRNA Molecules in Hemipteran Insects
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects
2.2. RNA Extraction
2.3. cDNA Synthesis
2.4. Quantitative Real Time-PCR (qRT-PCR)
2.5. dsRNA Synthesis
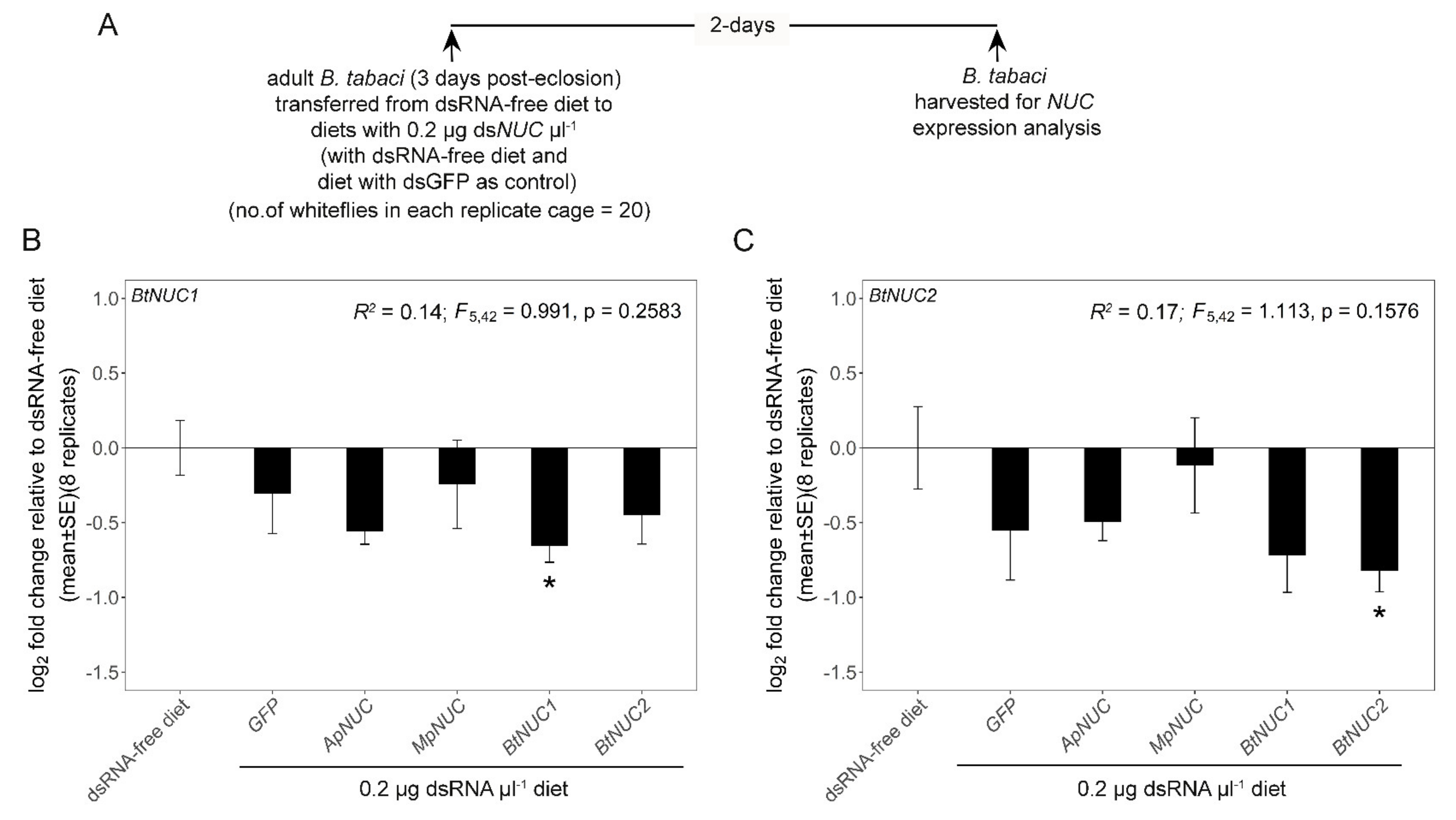
2.6. Experimental Designs
2.7. Statistical Analysis
3. Results
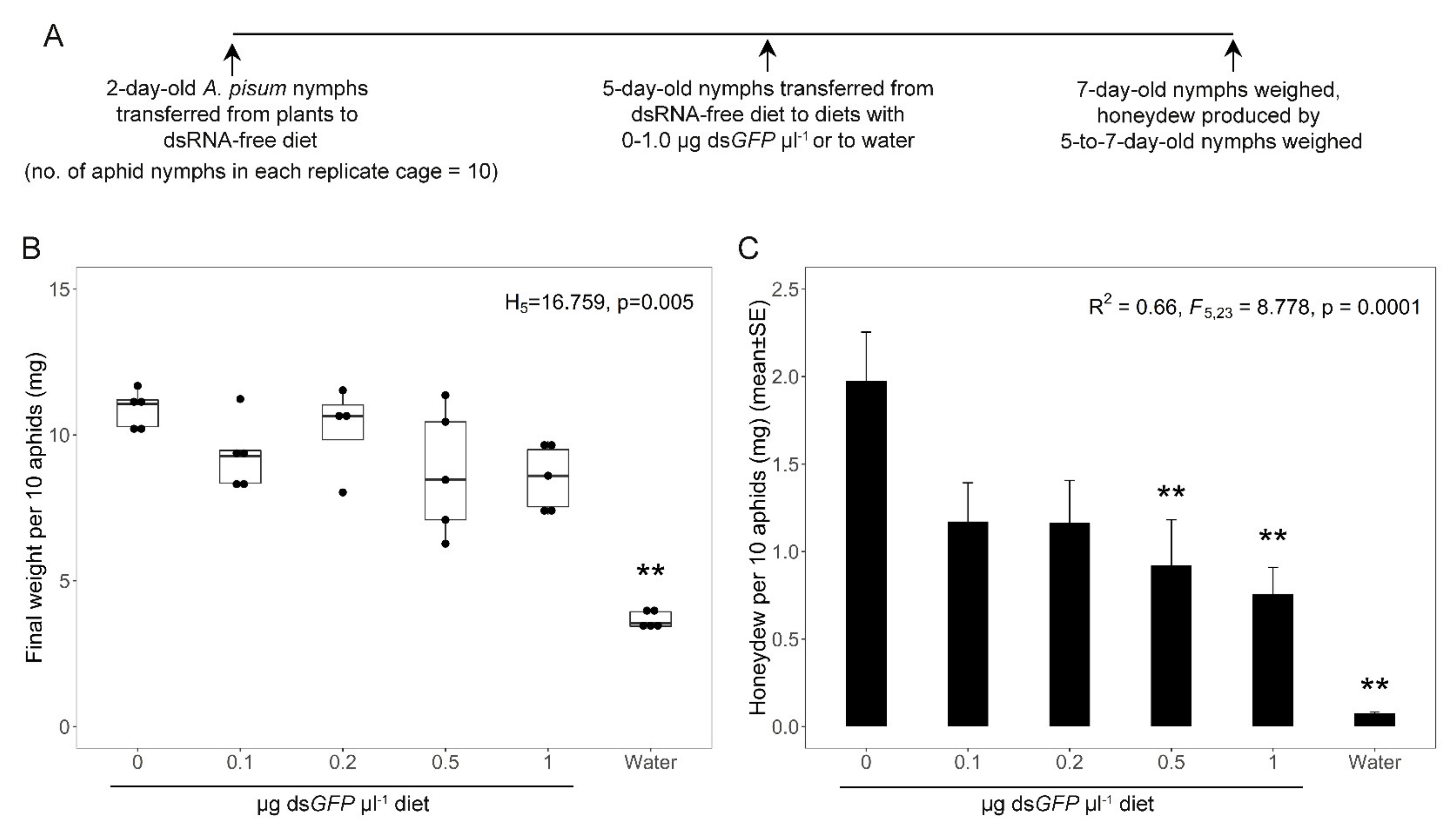
3.1. Optimization of the Dietary Concentration of dsRNA
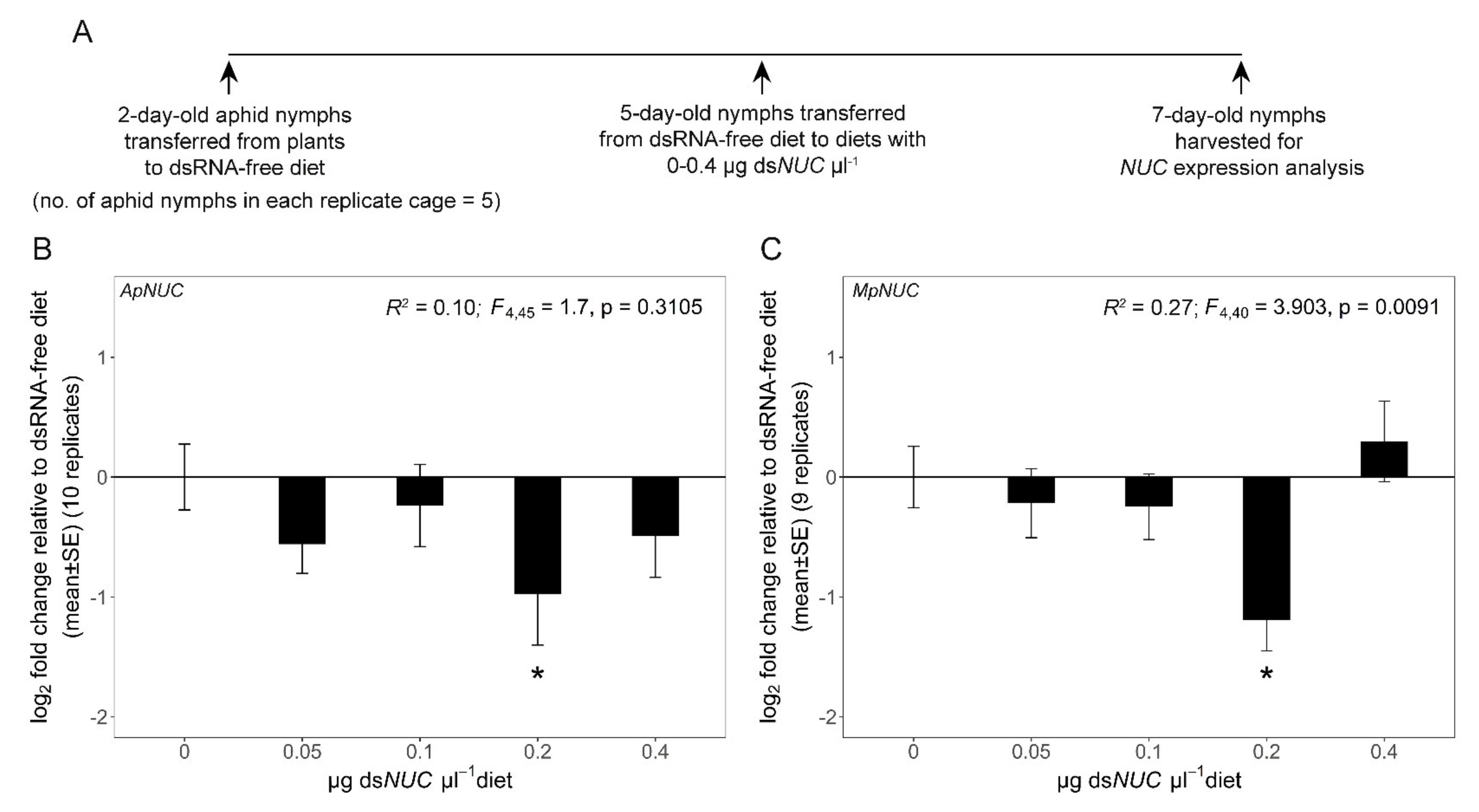
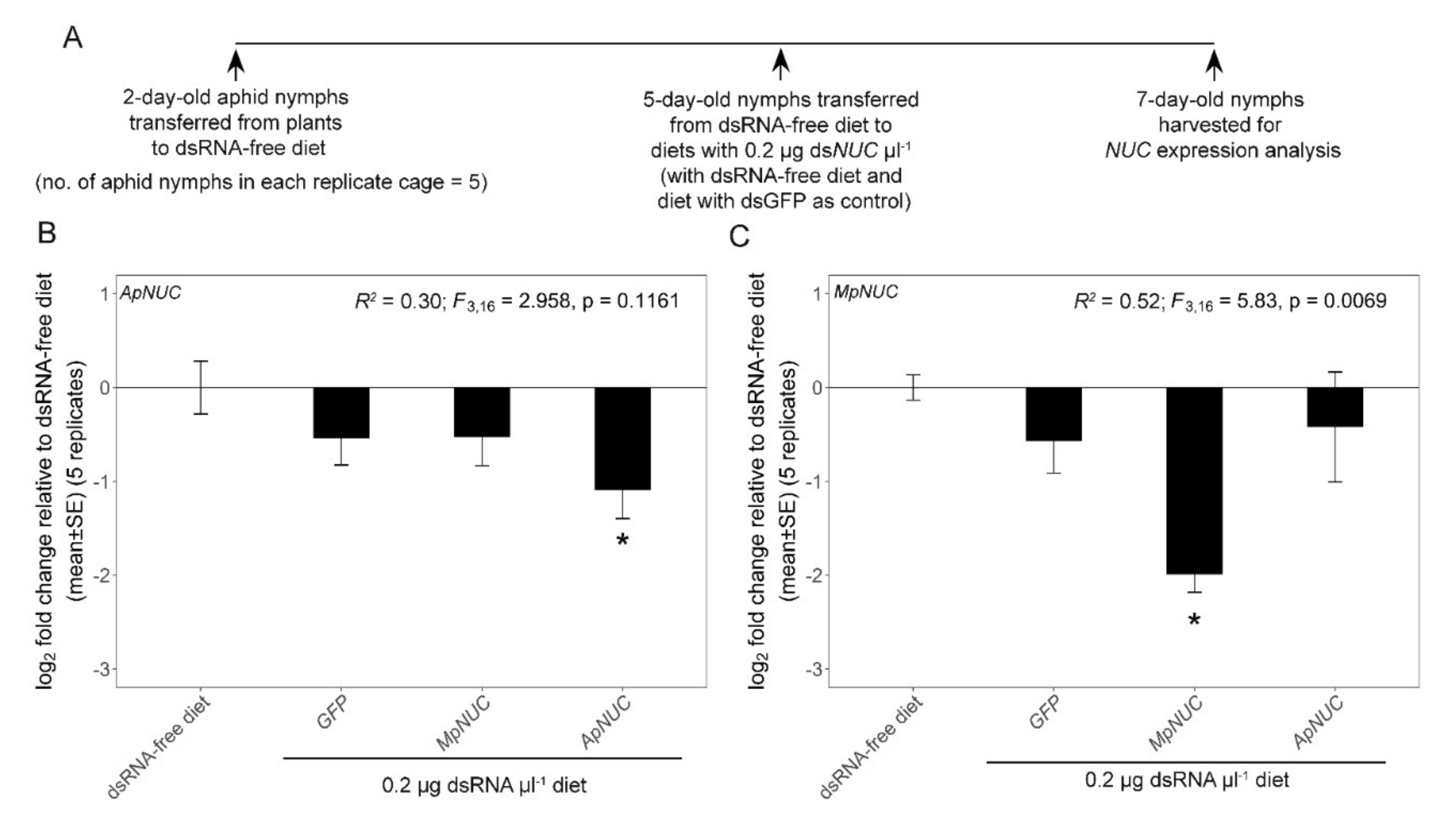
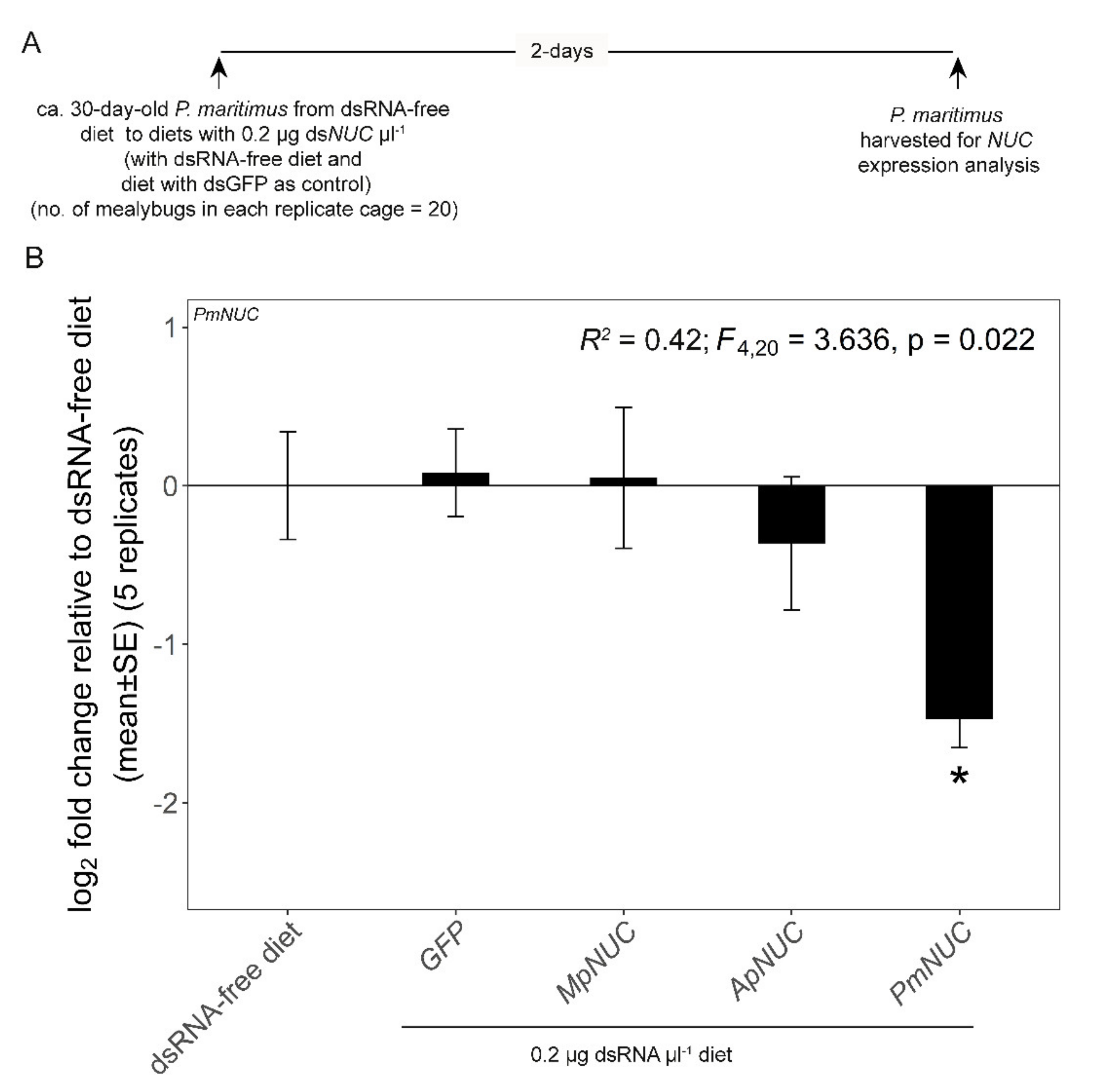
3.2. dsNUC Cross-Reactivity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Mao, Y.B.; Cai, W.J.; Wang, J.W.; Hong, G.J.; Tao, X.Y.; Wang, L.J.; Huang, Y.P.; Chen, X.Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef]
- Scott, J.G.; Michel, K.; Bartholomay, L.C.; Siegfried, B.D.; Hunter, W.B.; Smagghe, G.; Zhu, K.Y.; Douglas, A.E. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013, 59, 1212–1221. [Google Scholar] [CrossRef] [Green Version]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef] [Green Version]
- Zhu, K.Y.; Palli, S.R. Mechanisms, Applications, and challenges of insect RNA interference. Annu. Rev. Entomol. 2020, 65, 293–311. [Google Scholar] [CrossRef] [Green Version]
- Cooper, A.M.W.; Silver, K.; Zhang, J.; Park, Y.; Zhu, K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 2019, 75, 18–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, A.E. Strategies for Enhanced crop resistance to insect pests. Annu. Rev. Plant Biol. 2018, 69, 637–660. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cagliari, D.; Dias, N.P.; Galdeano, D.M.; dos Santos, E.Á.; Smagghe, G.; Zotti, M.J. Management of pest insects and plant diseases by non-transformative RNAi. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Zotti, M.; dos Santos, E.A.; Cagliari, D.; Christiaens, O.; Taning, C.N.T.; Smagghe, G. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag. Sci. 2018, 74, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.; Widmer, F.; Siegfried, B.D.; Zhuo, X.; Romeis, J. Responses of two ladybird beetle species (Coleoptera: Coccinellidae) to dietary RNAi. Pest Manag. Sci. 2019, 75, 2652–2662. [Google Scholar] [CrossRef]
- Pan, H.; Yang, X.; Bidne, K.; Hellmich, R.L.; Siegfried, B.D.; Zhou, X. Dietary risk assessment of v-ATPase A dsRNAs on monarch butterfly larvae. Front. Plant Sci. 2017, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Good, R.T.; Varghese, T.; Golz, J.F.; Russell, D.A.; Papanicolaou, A.; Edwards, O.; Robin, C. OfftargetFinder: A web tool for species-specific RNAi design. Bioinformatics 2016, 32, 1232–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lück, S.; Kreszies, T.; Strickert, M.; Schweizer, P.; Kuhlmann, M.; Douchkov, D. siRNA-Finder (si-Fi) software for RNAi-target design and off-target prediction. Front. Plant Sci. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Bolognesi, R.; Ramaseshadri, P.; Anderson, J.; Bachman, P.; Clinton, W.; Flannagan, R.; Ilagan, O.; Lawrence, C.; Levine, S.; Moar, W.; et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 2012, 7, e47534. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Peng, Y.; Zhang, H.; Wang, K.; Zhao, C.; Zhu, G.; Reddy Palli, S.; Han, Z. Off-target effects of RNAi correlate with the mismatch rate between dsRNA and non-target mRNA. RNA Biol. 2021, 1–13. [Google Scholar] [CrossRef]
- Du, Q.; Thonberg, H.; Wang, J.; Wahlestedt, C.; Liang, Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005, 33, 1671–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, M.; Pyati, P.; Cao, M.; Bell, H.; Gatehouse, J.A.; Fitches, E. Insecticidal effects of dsRNA targeting the Diap1 gene in dipteran pests. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Virus and dsRNA-triggered transcriptional responses reveal key components of honey bee antiviral defense. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Flenniken, M.L.; Andino, R. Non-specific dsRNA-mediated antiviral response in the honey bee. PLoS ONE 2013, 8, e77263. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.; Terenius, O.; Li, W.; Faye, I. Baculovirus and dsRNA induce Hemolin, but no antibacterial activity, in Antheraea pernyi. Insect Mol. Biol. 2004, 13, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Smagghe, G.; Swevers, L. Transcriptional response of BmToll9-1 and RNAi machinery genes to exogenous dsRNA in the midgut of Bombyx mori. J. Insect Physiol. 2013, 59, 646–654. [Google Scholar] [CrossRef]
- Nunes, F.M.F.; Aleixo, A.C.; Barchuk, A.R.; Bomtorin, A.D.; Grozinger, C.M.; Simões, Z.L.P. Non-target effects of green fluorescent protein (GFP)-derived double-stranded RNA (dsRNA-GFP) used in honey bee RNA interference (RNAi) assays. Insects 2013, 4, 90–103. [Google Scholar] [CrossRef] [Green Version]
- Piot, N.; Snoeck, S.; Vanlede, M.; Smagghe, G.; Meeus, I. The effect of oral administration of dsRNA on viral replication and mortality in Bombus terrestris. Viruses 2015, 7, 3172–3185. [Google Scholar] [CrossRef]
- Sigoillot, F.D.; King, R.W. Vigilance and validation: Keys to success in RNAi screening. ACS Chem. Biol. 2012, 6, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Perring, T.M.; Stansly, P.A.; Liu, T.X.; Smith, H.A.; Andreason, S.A. Whiteflies: Biology, Ecology, and Management; Elsevier Inc.: Amsterdam, The Netherlands, 2018; ISBN 9780128135082. [Google Scholar]
- Bass, C.; Puinean, A.M.; Zimmer, C.T.; Denholm, I.; Field, L.M.; Foster, S.P.; Gutbrod, O.; Nauen, R.; Slater, R.; Williamson, M.S. The evolution of insecticide resistance in the peach potato aphid, Myzus persicae. Insect Biochem. Mol. Biol. 2014, 51, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Sandhi, R.K.; Reddy, G.V.P. Biology, ecology, and management strategies for pea aphid (Hemiptera: Aphididae) in pulse crops. J. Integr. Pest Manag. 2020, 11. [Google Scholar] [CrossRef]
- Daane, K.M.; Almeida, R.P.P.; Bell, V.A.; Walker, J.T.S.; Botton, M.; Fallahzadeh, M.; Mani, M.; Miano, J.L.; Sforza, R.; Walton, V.M.; et al. Biology and management of mealybugs in vineyards. In Arthropod Management in Vineyards: Pests, Approaches, and Future Directions; Bostanian, N.J., Vincent, C., Isaacs, R., Eds.; Springer: Dordrecht, The Netherlands, 2012; Volume 9789400740, pp. 1–505. ISBN 9789400740327. [Google Scholar]
- Arthropod Pesticide Resistance Database. Available online: https://www.pesticideresistance.org/ (accessed on 15 February 2021).
- Horowitz, A.R.; Ghanim, M.; Roditakis, E.; Nauen, R.; Ishaaya, I. Insecticide resistance and its management in Bemisia tabaci species. J. Pest Sci. 2020, 93, 893–910. [Google Scholar] [CrossRef]
- Jaouannet, M.; Rodriguez, P.A.; Thorpe, P.; Lenoir, C.J.G.; Macleod, R.; Escudero-Martinez, C.; Bos, J.I.B. Plant immunity in plant-aphid interactions. Front. Plant Sci. 2014, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wallingford, A.K.; Fuchs, M.F.; Martinson, T.; Hesler, S.; Loeb, G.M. Slowing the spread of grapevine leafroll-associated viruses in commercial vineyards with insecticide control of the vector, Pseudococcus maritimus (Hemiptera: Pseudococcidae). J. Insect Sci. 2015, 15. [Google Scholar] [CrossRef]
- Ibrahim, A.B.; Monteiro, T.R.; Cabral, G.B.; Aragão, F.J.L. RNAi-mediated resistance to whitefly (Bemisia tabaci) in genetically engineered lettuce (Lactuca sativa). Transgenic Res. 2017, 26, 613–624. [Google Scholar] [CrossRef]
- Killiny, N.; Kishk, A. Delivery of dsRNA through topical feeding for RNA interference in the citrus sap piercing-sucking hemipteran, Diaphorina citri. Arch. Insect Biochem. Physiol. 2017, 95, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kirfel, P.; Vilcinskas, A.; Skaljac, M. Lysine acetyltransferase p300/cbp plays an important role in reproduction, embryogenesis and longevity of the pea aphid Acyrthosiphon pisum. Insects 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Chen, Q.; Luan, J.; Chung, S.H.; Van Eck, J.; Turgeon, R.; Douglas, A.E. Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem. Mol. Biol. 2017, 88, 21–29. [Google Scholar] [CrossRef]
- Tzin, V.; Yang, X.; Jing, X.; Zhang, K.; Jander, G.; Douglas, A.E. RNA interference against gut osmoregulatory genes in phloem-feeding insects. J. Insect Physiol. 2015, 79, 105–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, A.K.; Clark, N.; Wentworth, K.S.; Hesler, S.; Fuchs, M.; Loeb, G.; Douglas, A.E. Evaluation of RNA interference for control of the grape mealybug Pseudococcus maritimus (Hemiptera: Pseudococcidae). Insects 2020, 11, 739. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.H.; Jing, X.; Luo, Y.; Douglas, A.E. Targeting symbiosis-related insect genes by RNAi in the pea aphid-Buchnera symbiosis. Insect Biochem. Mol. Biol. 2018, 95, 55–63. [Google Scholar] [CrossRef]
- Prentice, K.; Smagghe, G.; Gheysen, G.; Christiaens, O. Nuclease activity decreases the RNAi response in the sweetpotato weevil Cylas puncticollis. Insect Biochem. Mol. Biol. 2019, 110, 80–89. [Google Scholar] [CrossRef]
- Spit, J.; Philips, A.; Wynant, N.; Santos, D.; Plaetinck, G.; Vanden Broeck, J. Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2017, 81, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Bouvaine, S.; Behmer, S.T.; Lin, G.G.; Faure, M.L.; Grebenok, R.J.; Douglas, A.E. The physiology of sterol nutrition in the pea aphid Acyrthosiphon pisum. J. Insect Physiol. 2012, 58, 1383–1389. [Google Scholar] [CrossRef]
- Yoon, J.S.; Tian, H.G.; McMullen, J.G.; Chung, S.H.; Douglas, A.E. Candidate genetic determinants of intraspecific variation in pea aphid susceptibility to RNA interference. Insect Biochem. Mol. Biol. 2020, 123, 103408. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E.; Minto, L.B.; Wilkinson, T.L. Quantifying nutrient production by the microbial symbionts in an aphid. J. Exp. Biol. 2001, 204, 349–358. [Google Scholar]
- Pohlert, T. PMCMRplus: Calculate Pairwise Multiple Comparisons of Mean Rank Sums Extended 1.4.4. 2021. Available online: https://cran.r-project.org/web/packages/PMCMRplus/index.html (accessed on 20 January 2021).
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67. [Google Scholar] [CrossRef]
- Lenth, R.V. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. R Package Version 1.3.3. 2020. Available online: https://cran.r-project.org/web/packages/emmeans/index.html (accessed on 1 July 2020).
- Douglas, A.E. Nutritional physiology of aphids. Adv. Insect Physiol. 2003, 31, 73–140. [Google Scholar]
- Wang, K.; Peng, Y.; Pu, J.; Fu, W.; Wang, J.; Han, Z. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 2016, 77, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.C.; Sun, W.; Spencer, J.L.; Pittendrigh, B.R.; Seufferheld, M.J. Differential effects of RNAi treatments on field populations of the western corn rootworm. Pestic. Biochem. Physiol. 2014, 110, 1–6. [Google Scholar] [CrossRef]
- Sugahara, R.; Tanaka, S.; Jouraku, A.; Shiotsuki, T. Geographic variation in RNAi sensitivity in the migratory locust. Gene 2017, 605, 5–11. [Google Scholar] [CrossRef]
- Kingsolver, J.G.; Daniel, T.L. Mechanics of food handling by fluid-feeding insects. In Regulatory Mechanisms in Insect Feeding; Chapman, R.F., de Boer, G., Eds.; Springer: Boston, MA, USA, 1995; pp. 32–73. ISBN 9788578110796. [Google Scholar]
- Dadd, R.H. Insect nutrition: Current developments and metabolic implications. Annu. Rev. Entomol. 1973, 18, 381–420. [Google Scholar] [CrossRef]
- Bachman, P.; Fischer, J.; Song, Z.; Urbanczyk-Wochniak, E.; Watson, G. Environmental fate and dissipation of applied dsRNA in soil, aquatic systems, and plants. Front. Plant Sci. 2020, 11, 1–6. [Google Scholar] [CrossRef]
- Fletcher, S.J.; Reeves, P.T.; Hoang, B.T.; Mitter, N. A perspective on RNAi-based biopesticides. Front. Plant Sci. 2020, 11, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Jaouannet, M.; Dempsey, D.A.; Imani, J.; Coustau, C.; Kogel, K.H. RNA-based technologies for insect control in plant production. Biotechnol. Adv. 2020, 39, 107463. [Google Scholar] [CrossRef]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef] [Green Version]
- Shang, F.; Ding, B.Y.; Ye, C.; Yang, L.; Chang, T.Y.; Xie, J.; Tang, L.D.; Niu, J.; Wang, J.J. Evaluation of a cuticle protein gene as a potential RNAi target in aphids. Pest Manag. Sci. 2020, 76, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Bachman, P.; Fridley, J.; Mueller, G.; Moar, W.; Levine, S.L. Sequence–activity relationships for the Snf7 insecticidal dsRNA in Chrysomelidae. Front. Plant Sci. 2020, 11, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, N.; Devos, Y.; Álvarez-Alfageme, F.; Lanzoni, A.; Waigmann, E. Risk assessment considerations for genetically modified RNAi plants: EFSA’s Activities and Perspective. Front. Plant Sci. 2020, 11, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Insect Species | % Sequence Identity of NUC Gene Sequences/dsNUC Sequences | dsNUC Length (nt) | Predicted Cross Reactivity of dsNUC Number of Perfect Matching 21 nt siRNA Molecules (% of Total siRNA Molecules) 1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Ap | Mp | Pm | Bt (NUC1) | Ap | Mp | Pm | Bt (NUC1) | ||
| Ap | 328 | ||||||||
| Mp | 88/92 | 328 | 77 (25%) | ||||||
| Pm | 39/35 | 39/38 | 250 | 0 | 0 | ||||
| Bt (NUC1) | 43/47 | 43/47 | 31/41 | 402 | 0 | 0 | 0 | ||
| Bt (NUC2) | 46/40 | 45/40 | 43/34 | 38/40 | 400 | 0 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arora, A.K.; Chung, S.H.; Douglas, A.E. Non-Target Effects of dsRNA Molecules in Hemipteran Insects. Genes 2021, 12, 407. https://doi.org/10.3390/genes12030407
Arora AK, Chung SH, Douglas AE. Non-Target Effects of dsRNA Molecules in Hemipteran Insects. Genes. 2021; 12(3):407. https://doi.org/10.3390/genes12030407
Chicago/Turabian StyleArora, Arinder K., Seung Ho Chung, and Angela E. Douglas. 2021. "Non-Target Effects of dsRNA Molecules in Hemipteran Insects" Genes 12, no. 3: 407. https://doi.org/10.3390/genes12030407
APA StyleArora, A. K., Chung, S. H., & Douglas, A. E. (2021). Non-Target Effects of dsRNA Molecules in Hemipteran Insects. Genes, 12(3), 407. https://doi.org/10.3390/genes12030407







