Mitochondrial DNA Profiles of Individuals from a 12th Century Necropolis in Feldioara (Transylvania)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Background
2.2. Ancient DNA Analysis
2.3. Population Genetic Analyses
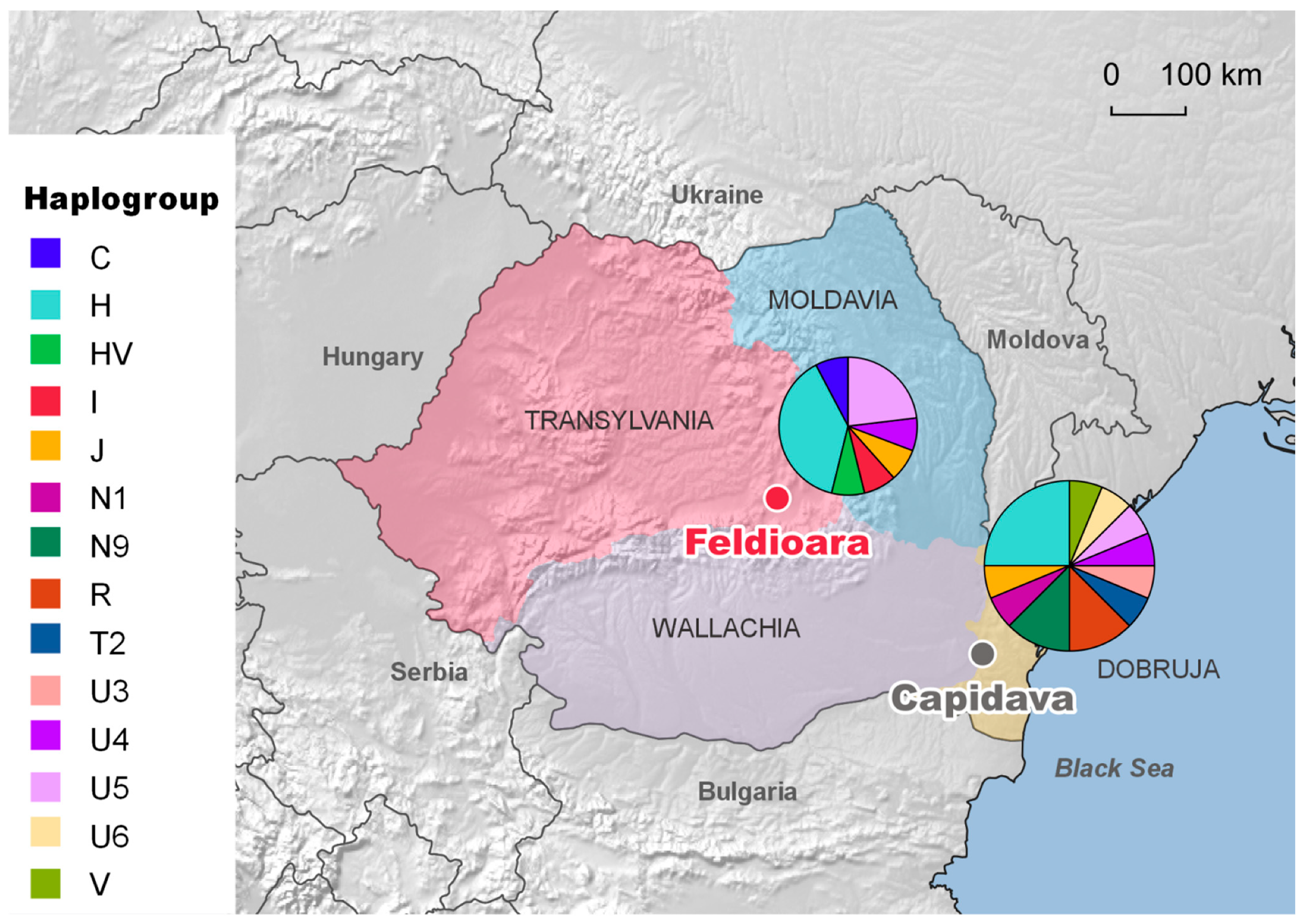
3. Results and Discussion
3.1. MtDNA Polymorphism
3.2. Population Genetic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Curta, F. Southeastern Europe in the Middle Ages, 500–1250; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]
- Cocos, R.; Schipor, S.; Hervella, M.; Cianga, P.; Popescu, R.; Banescu, C.; Constantinescu, M.; Martinescu, A.; Raicu, F. Genetic affinities among the historical provinces of Romania and Central Europe as revealed by an mtDNA analysis. BMC Genet. 2017, 18, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turchi, C.; Stanciu, F.; Paselli, G.; Buscemi, L.; Parson, W.; Tagliabracci, A. The mitochondrial DNA makeup of Romanians: A forensic mtDNA control region database and phylogenetic characterization. Forensic Sci. Int. Genet. 2016, 24, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Hervella, M.; Izagirre, N.; Alonso, S.; Ioana, M.; Netea, M.G.; de-la-Rua, C. The Carpathian range represents a weak genetic barrier in South-East Europe. BMC Genet. 2014, 15, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Göckenjan, H. Hilfsvölker und Grenzwächter im Mittelalterlichen Ungarn; Steiner: Wiesbaden, Germany, 1972; Volume 5. [Google Scholar]
- Nägler, T. Die Ansiedlung der Siebenbürger Sachsen; Kriterion-Verlag: Bucharest, Romania, 1979. [Google Scholar]
- Zimmermann, H. Der Deutsche Orden in Siebenbürgen: Eine Diplomatische Untersuchung; Böhlau Verlag Köln: Weimar, Fermany, 2011; Volume 26. [Google Scholar]
- Bosch, E.; Calafell, F.; Gonzalez-Neira, A.; Flaiz, C.; Mateu, E.; Scheil, H.G.; Huckenbeck, W.; Efremovska, L.; Mikerezi, I.; Xirotiris, N.; et al. Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann. Hum. Genet. 2006, 70, 459–487. [Google Scholar] [CrossRef] [PubMed]
- Brandstatter, A.; Egyed, B.; Zimmermann, B.; Duftner, N.; Padar, Z.; Parson, W. Migration rates and genetic structure of two Hungarian ethnic groups in Transylvania, Romania. Ann. Hum. Genet. 2007, 71, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Mendizabal, I.; Lao, O.; Marigorta, U.M.; Wollstein, A.; Gusmao, L.; Ferak, V.; Ioana, M.; Jordanova, A.; Kaneva, R.; Kouvatsi, A.; et al. Reconstructing the population history of European Romani from genome-wide data. Curr. Biol. CB 2012, 22, 2342–2349. [Google Scholar] [CrossRef] [Green Version]
- Csányi, B.; Bogácsi-Szabó, E.; Tömöry, G.; Czibula, Á.; Priskin, K.; Csõsz, A.; Mende, B.; Langó, P.; Csete, K.; Zsolnai, A.; et al. Y-Chromosome Analysis of Ancient Hungarian and Two Modern Hungarian-Speaking Populations from the Carpathian Basin. Ann. Hum. Genet. 2008, 72, 519–534. [Google Scholar] [CrossRef]
- Ádám, V.; Bánfai, Z.; Maász, A.; Sümegi, K.; Miseta, A.; Melegh, B. Investigating the genetic characteristics of the Csangos, a traditionally Hungarian speaking ethnic group residing in Romania. J. Hum. Genet. 2020, 65, 1093–1103. [Google Scholar] [CrossRef]
- Barbarii, L.; Constantinescu, C.; Rolf, B. A study on Y-STR haplotypes in the Saxon population from Transylvania (Siebenburger Sachsen): Is there an evidence for a German origin? Rom. J. Leg. Med. 2004, 12, 247. [Google Scholar]
- Ioniță, A.; Căpățână, D.; Boroffka, N.G.O.; Boroffka, R.; Popescu, A. Feldioara-Marienburg: Contribuții Arheologice la istoria Țării Bârsei: Archaölogische Beiträge zur Geschichte des Burzenlandes; Editura Academiei Române: Bucharest, Romania, 2004. [Google Scholar]
- Muja, C.; Ioniță, A. Sexual dimorphism and general activity levels as revealed by the diaphyseal external shape and historical evidence: Case study on a medieval population from Transylvania. Dacia N.S. 2015, 59, 319–327. [Google Scholar]
- Ioniță, A. Die Besiedlung des Burzenlandes im 12.-13. Jahrhundert im Lichte der Archäologie. In Generalprobe Burzenland; Siebenbürgisches Archiv. Archiv des Vereins für Siebenbürgische Landeskunde; Böhlau Verlag: Vienna, Austria, 2013; Volume Band 42, pp. 107–124. [Google Scholar]
- REMPEL, H. Reihengräberfriedhöfe des 8. bis 11. Jahrhunderts aus Sachsen-Anhalt, Sachsen und Thüringen. Deutsche Akademie der Wissenschaften zu Berlin. Schr. Der Sekt. Für Vor-U. Frühgeschichte 1966, 20, 44–51. [Google Scholar]
- Gáll, E. The date of the appearance of the S-ended lock-rings in the Transylvanian Basin. Ephemer. Napoc. 2009, 19, 157–176. [Google Scholar]
- Giesler, J. Untersuchungen zur chronologie der Bijelo Brdo-Kultur. Ein beitrag zur archäologie des 10. und 11. jahrhunderts im Karpatenbecken. Prähistorische Z. 1981, 56, 3–221. [Google Scholar] [CrossRef]
- Rusu, I.; Modi, A.; Vai, S.; Pilli, E.; Mircea, C.; Radu, C.; Urduzia, C.; Pinter, Z.K.; Bodolica, V.; Dobrinescu, C.; et al. Maternal DNA lineages at the gate of Europe in the 10th century AD. PLoS ONE 2018, 13, e0193578. [Google Scholar] [CrossRef] [Green Version]
- Dabney, J.; Knapp, M.; Glocke, I.; Gansauge, M.T.; Weihmann, A.; Nickel, B.; Valdiosera, C.; Garcia, N.; Paabo, S.; Arsuaga, J.L.; et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. USA 2013, 110, 15758–15763. [Google Scholar] [CrossRef] [Green Version]
- Gabriel, M.N.; Huffine, E.F.; Ryan, J.H.; Holland, M.M.; Parsons, T.J. Improved MtDNA sequence analysis of forensic remains using a "mini-primer set" amplification strategy. J. Forensic Sci. 2001, 46, 247–253. [Google Scholar] [CrossRef]
- Tömöry, G.; Csányi, B.; Bogácsi-Szabó, E.; Kalmár, T.; Czibula, A.; Csősz, A.; Priskin, K.; Mende, B.; Langó, P.; Downes, C.S.; et al. Comparison of maternal lineage and biogeographic analyses of ancient and modern Hungarian populations. Am. J. Phys. Anthropol. 2007, 134, 354–368. [Google Scholar] [CrossRef] [Green Version]
- Andrews, R.M.; Kubacka, I.; Chinnery, P.F.; Lightowlers, R.N.; Turnbull, D.M.; Howell, N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999, 23, 147. [Google Scholar] [CrossRef]
- van Oven, M. PhyloTree Build 17: Growing the human mitochondrial DNA tree. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e392–e394. [Google Scholar] [CrossRef] [Green Version]
- Weissensteiner, H.; Pacher, D.; Kloss-Brandstatter, A.; Forer, L.; Specht, G.; Bandelt, H.J.; Kronenberg, F.; Salas, A.; Schonherr, S. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016, 44, W58–W63. [Google Scholar] [CrossRef]
- Ottoni, C.; Ricaut, F.X.; Vanderheyden, N.; Brucato, N.; Waelkens, M.; Decorte, R. Mitochondrial analysis of a Byzantine population reveals the differential impact of multiple historical events in South Anatolia. Eur. J. Hum. Genet. Ejhg 2011, 19, 571–576. [Google Scholar] [CrossRef]
- Vai, S.; Ghirotto, S.; Pilli, E.; Tassi, F.; Lari, M.; Rizzi, E.; Matas-Lalueza, L.; Ramirez, O.; Lalueza-Fox, C.; Achilli, A.; et al. Genealogical relationships between early medieval and modern inhabitants of Piedmont. PLoS ONE 2015, 10, e0116801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vai, S.; Brunelli, A.; Modi, A.; Tassi, F.; Vergata, C.; Pilli, E.; Lari, M.; Susca, R.R.; Giostra, C.; Baricco, L.P.; et al. A genetic perspective on Longobard-Era migrations. Eur. J. Hum. Genet. 2019, 27, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Csősz, A.; Szécsényi-Nagy, A.; Csákyová, V.; Langó, P.; Bódis, V.; Köhler, K.; Tömöry, G.; Nagy, M.; Mende, B.G. Maternal Genetic Ancestry and Legacy of 10(th) Century AD Hungarians. Sci. Rep. 2016, 6, 33446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csáky, V.; Gerber, D.; Koncz, I.; Csiky, G.; Mende, B.G.; Szeifert, B.; Egyed, B.; Pamjav, H.; Marcsik, A.; Molnár, E.; et al. Genetic insights into the social organisation of the Avar period elite in the 7th century AD Carpathian Basin. Sci. Rep. 2020, 10, 948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šebest, L.; Baldovič, M.; Frtús, A.; Bognár, C.; Kyselicová, K.; Kádasi, Ľ.; Beňuš, R. Detection of mitochondrial haplogroups in a small avar-slavic population from the eigth–ninth century AD. Am. J. Phys. Anthropol. 2018, 165, 536–553. [Google Scholar] [CrossRef] [PubMed]
- Krzewińska, M.; Bjornstad, G.; Skoglund, P.; Olason, P.I.; Bill, J.; Götherström, A.; Hagelberg, E. Mitochondrial DNA variation in the Viking age population of Norway. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2015, 370, 20130384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melchior, L.; Lynnerup, N.; Siegismund, H.R.; Kivisild, T.; Dissing, J. Genetic diversity among ancient Nordic populations. PLoS ONE 2010, 5, e11898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alzualde, A.; Izagirre, N.; Alonso, S.; Alonso, A.; Albarrán, C.; Azkarate, A.; de la Rúa, C. Insights into the "isolation" of the Basques: mtDNA lineages from the historical site of Aldaieta (6th-7th centuries AD). Am. J. Phys. Anthropol. 2006, 130, 394–404. [Google Scholar] [CrossRef]
- Guimaraes, S.; Ghirotto, S.; Benazzo, A.; Milani, L.; Lari, M.; Pilli, E.; Pecchioli, E.; Mallegni, F.; Lippi, B.; Bertoldi, F.; et al. Genealogical discontinuities among Etruscan, Medieval, and contemporary Tuscans. Mol. Biol. Evol. 2009, 26, 2157–2166. [Google Scholar] [CrossRef] [Green Version]
- Nesheva, D.V.; Karachanak-Yankova, S.; Lari, M.; Yordanov, Y.; Galabov, A.; Caramelli, D.; Toncheva, D. Mitochondrial DNA Suggests a Western Eurasian Origin for Ancient (Proto-) Bulgarians. Hum. Biol. 2015, 87, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Neparaczki, E.; Maroti, Z.; Kalmar, T.; Kocsy, K.; Maar, K.; Bihari, P.; Nagy, I.; Fothi, E.; Pap, I.; Kustar, A.; et al. Mitogenomic data indicate admixture components of Central-Inner Asian and Srubnaya origin in the conquering Hungarians. PLoS ONE 2018, 13, e0205920. [Google Scholar] [CrossRef] [Green Version]
- Juras, A.; Dabert, M.; Kushniarevich, A.; Malmström, H.; Raghavan, M.; Kosicki, J.Z.; Metspalu, E.; Willerslev, E.; Piontek, J. Ancient DNA reveals matrilineal continuity in present-day Poland over the last two millennia. PLoS ONE 2014, 9, e110839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csákyová, V.; Szécsényi-Nagy, A.; Csősz, A.; Nagy, M.; Fusek, G.; Langó, P.; Bauer, M.; Mende, B.G.; Makovický, P.; Bauerová, M. Maternal Genetic Composition of a Medieval Population from a Hungarian-Slavic Contact Zone in Central Europe. PLoS ONE 2016, 11, e0151206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helgason, A.; Lalueza-Fox, C.; Ghosh, S.; Sigurethardóttir, S.; Sampietro, M.L.; Gigli, E.; Baker, A.; Bertranpetit, J.; Arnadóttir, L.; Thornorsteinsdottir, U.; et al. Sequences from first settlers reveal rapid evolution in Icelandic mtDNA pool. Plos Genet. 2009, 5, e1000343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusu, I.; Modi, A.; Radu, C.; Mircea, C.; Vulpoi, A.; Dobrinescu, C.; Bodolică, V.; Potârniche, T.; Popescu, O.; Caramelli, D.; et al. Mitochondrial ancestry of medieval individuals carelessly interred in a multiple burial from southeastern Romania. Sci. Rep. 2019, 9, 961. [Google Scholar] [CrossRef] [PubMed]
- Veeramah, K.R.; Rott, A.; Groß, M.; van Dorp, L.; López, S.; Kirsanow, K.; Sell, C.; Blöcher, J.; Wegmann, D.; Link, V.; et al. Population genomic analysis of elongated skulls reveals extensive female-biased immigration in Early Medieval Bavaria. Proc. Natl. Acad. Sci. USA 2018, 115, 3494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stolarek, I.; Handschuh, L.; Juras, A.; Nowaczewska, W.; Kóčka-Krenz, H.; Michalowski, A.; Piontek, J.; Kozlowski, P.; Figlerowicz, M. Goth migration induced changes in the matrilineal genetic structure of the central-east European population. Sci. Rep. 2019, 9, 6737. [Google Scholar] [CrossRef] [Green Version]
- Emery, M.V.; Duggan, A.T.; Murchie, T.J.; Stark, R.J.; Klunk, J.; Hider, J.; Eaton, K.; Karpinski, E.; Schwarcz, H.P.; Poinar, H.N.; et al. Ancient Roman mitochondrial genomes and isotopes reveal relationships and geographic origins at the local and pan-Mediterranean scales. J. Archaeol. Sci. Rep. 2018, 20, 200–209. [Google Scholar] [CrossRef]
- R Core, T. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Ward, J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Suzuki, R.; Terada, Y.; Shimodaira, H. pvclust: Hierarchical Clustering with P-Values via Multiscale Bootstrap Resampling, R package version 2.2-0; 2019. Available online: https://CRAN.R-project.org/package=pvclust (accessed on 25 January 2021).
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2019, 37, 291–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R package version 2.5-6; 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 25 January 2021).
- Nikitin, A.G.; Newton, J.R.; Potekhina, I.D. Mitochondrial haplogroup C in ancient mitochondrial DNA from Ukraine extends the presence of East Eurasian genetic lineages in Neolithic Central and Eastern Europe. J. Hum. Genet. 2012, 57, 610–612. [Google Scholar] [CrossRef] [Green Version]
- Duggan, A.T.; Whitten, M.; Wiebe, V.; Crawford, M.; Butthof, A.; Spitsyn, V.; Makarov, S.; Novgorodov, I.; Osakovsky, V.; Pakendorf, B. Investigating the Prehistory of Tungusic Peoples of Siberia and the Amur-Ussuri Region with Complete mtDNA Genome Sequences and Y-chromosomal Markers. PLoS ONE 2013, 8, e83570. [Google Scholar] [CrossRef] [PubMed]
- Pilipenko, A.S.; Trapezov, R.O.; Cherdantsev, S.V.; Babenko, V.N.; Nesterova, M.S.; Pozdnyakov, D.V.; Molodin, V.I.; Polosmak, N.V. Maternal genetic features of the Iron Age Tagar population from Southern Siberia (1st millennium BC). PLoS ONE 2018, 13, e0204062. [Google Scholar] [CrossRef]
- de Barros Damgaard, P.; Martiniano, R.; Kamm, J.; Moreno-Mayar, J.V.; Kroonen, G.; Peyrot, M.; Barjamovic, G.; Rasmussen, S.; Zacho, C.; Baimukhanov, N.; et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science 2018, 360, eaar7711. [Google Scholar] [CrossRef] [Green Version]
- Nikitin, A.G.; Ivanova, S.; Kiosak, D.; Badgerow, J.; Pashnick, J. Subdivisions of haplogroups U and C encompass mitochondrial DNA lineages of Eneolithic–Early Bronze Age Kurgan populations of western North Pontic steppe. J. Hum. Genet. 2017, 62, 605–613. [Google Scholar] [CrossRef]
- Csáky, V.; Gerber, D.; Szeifert, B.; Egyed, B.; Stégmár, B.; Botalov, S.G.e.; Grudochko, I.V.e.; Matveeva, N.P.; Zelenkov, A.S.; Sleptsova, A.V.; et al. Early medieval genetic data from Ural region evaluated in the light of archaeological evidence of ancient Hungarians. Sci. Rep. 2020, 10, 19137. [Google Scholar] [CrossRef]
- Damgaard, P.d.B.; Marchi, N.; Rasmussen, S.; Peyrot, M.; Renaud, G.; Korneliussen, T.; Moreno-Mayar, J.V.; Pedersen, M.W.; Goldberg, A.; Usmanova, E.; et al. 137 ancient human genomes from across the Eurasian steppes. Nature 2018, 557, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Jeong, C.; Wang, K.; Wilkin, S.; Taylor, W.T.T.; Miller, B.K.; Bemmann, J.H.; Stahl, R.; Chiovelli, C.; Knolle, F.; Ulziibayar, S.; et al. A Dynamic 6,000-Year Genetic History of Eurasia’s Eastern Steppe. Cell 2020, 183, 890–904.e829. [Google Scholar] [CrossRef]
- Antonio, M.L.; Gao, Z.; Moots, H.M.; Lucci, M.; Candilio, F.; Sawyer, S.; Oberreiter, V.; Calderon, D.; Devitofranceschi, K.; Aikens, R.C.; et al. Ancient Rome: A genetic crossroads of Europe and the Mediterranean. Science 2019, 366, 708. [Google Scholar] [CrossRef] [PubMed]
- Klunk, J.; Duggan, A.T.; Redfern, R.; Gamble, J.; Boldsen, J.L.; Golding, G.B.; Walter, B.S.; Eaton, K.; Stangroom, J.; Rouillard, J.-M.; et al. Genetic resiliency and the Black Death: No apparent loss of mitogenomic diversity due to the Black Death in medieval London and Denmark. Am. J. Phys. Anthropol. 2019, 169, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Olalde, I.; Brace, S.; Allentoft, M.E.; Armit, I.; Kristiansen, K.; Booth, T.; Rohland, N.; Mallick, S.; Szecsenyi-Nagy, A.; Mittnik, A.; et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 2018, 555, 190–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margaryan, A.; Lawson, D.J.; Sikora, M.; Racimo, F.; Rasmussen, S.; Moltke, I.; Cassidy, L.M.; Jørsboe, E.; Ingason, A.; Pedersen, M.W.; et al. Population genomics of the Viking world. Nature 2020, 585, 390–396. [Google Scholar] [CrossRef]
- Mathieson, I.; Lazaridis, I.; Rohland, N.; Mallick, S.; Patterson, N.; Roodenberg, S.A.; Harney, E.; Stewardson, K.; Fernandes, D.; Novak, M.; et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 2015, 528, 499–503. [Google Scholar] [CrossRef] [Green Version]
- Malyarchuk, B.; Derenko, M.; Grzybowski, T.; Perkova, M.; Rogalla, U.; Vanecek, T.; Tsybovsky, I. The peopling of Europe from the mitochondrial haplogroup U5 perspective. PLoS ONE 2010, 5, e10285. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gînguță, A.; Rusu, I.; Mircea, C.; Ioniță, A.; Banciu, H.L.; Kelemen, B. Mitochondrial DNA Profiles of Individuals from a 12th Century Necropolis in Feldioara (Transylvania). Genes 2021, 12, 436. https://doi.org/10.3390/genes12030436
Gînguță A, Rusu I, Mircea C, Ioniță A, Banciu HL, Kelemen B. Mitochondrial DNA Profiles of Individuals from a 12th Century Necropolis in Feldioara (Transylvania). Genes. 2021; 12(3):436. https://doi.org/10.3390/genes12030436
Chicago/Turabian StyleGînguță, Alexandra, Ioana Rusu, Cristina Mircea, Adrian Ioniță, Horia L. Banciu, and Beatrice Kelemen. 2021. "Mitochondrial DNA Profiles of Individuals from a 12th Century Necropolis in Feldioara (Transylvania)" Genes 12, no. 3: 436. https://doi.org/10.3390/genes12030436
APA StyleGînguță, A., Rusu, I., Mircea, C., Ioniță, A., Banciu, H. L., & Kelemen, B. (2021). Mitochondrial DNA Profiles of Individuals from a 12th Century Necropolis in Feldioara (Transylvania). Genes, 12(3), 436. https://doi.org/10.3390/genes12030436







