Kinetics and Topology of DNA Associated with Circulating Extracellular Vesicles Released during Exercise
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Subjects and Exercise Testing
2.3. Blood Sample Collection and Plasma Preparation
2.4. Extracellular Vesicle Isolation
2.5. Treatment of SEC- or CD9+/CD63+/CD81+EV Samples and DNA Isolation
2.6. Western Blotting (WB)
2.7. Cell-free DNA Measurement
2.8. Data Analysis
3. Results
3.1. Participants Characteristics and Exercise Performance
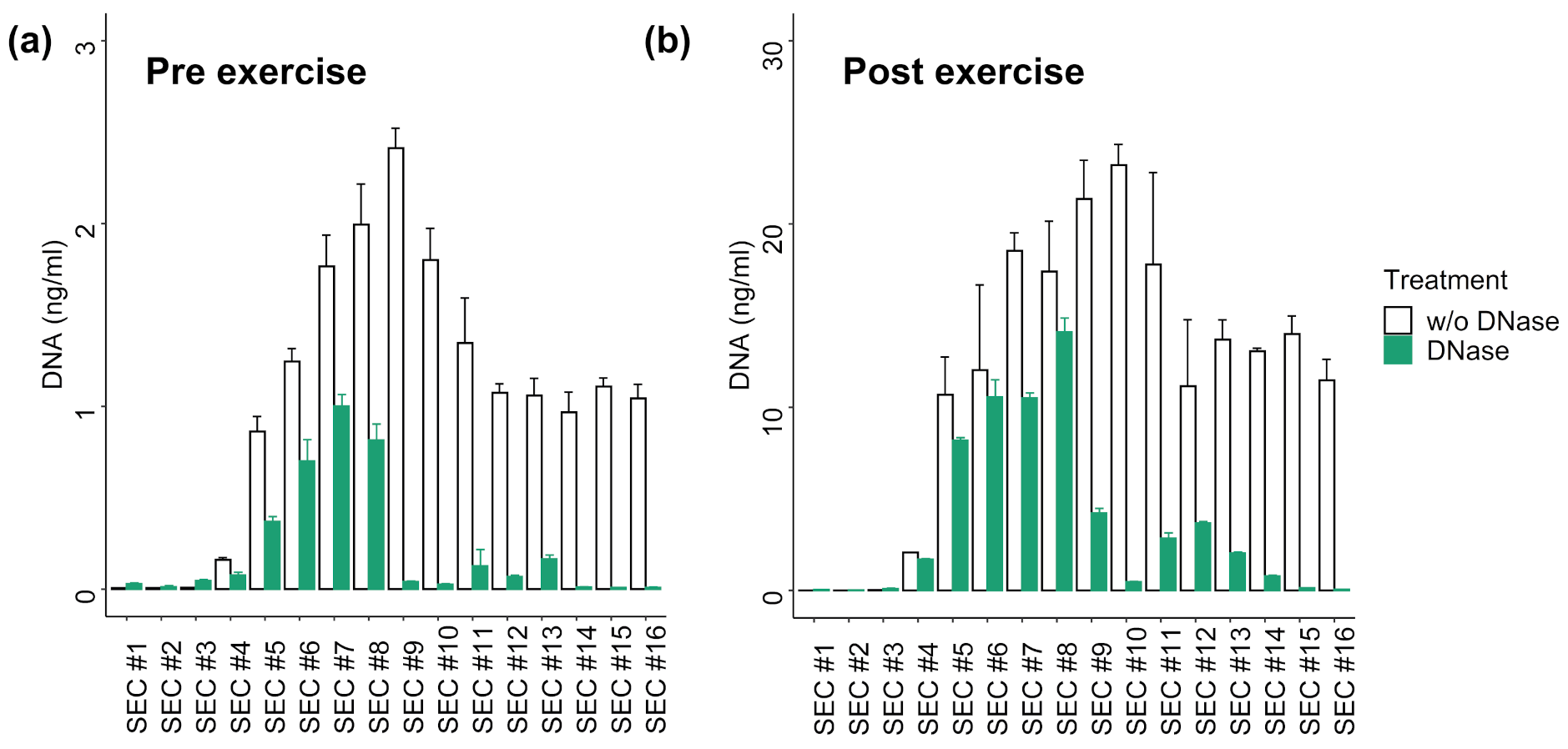
3.2. Distribution of EVs and cfDNA in SEC Fractions
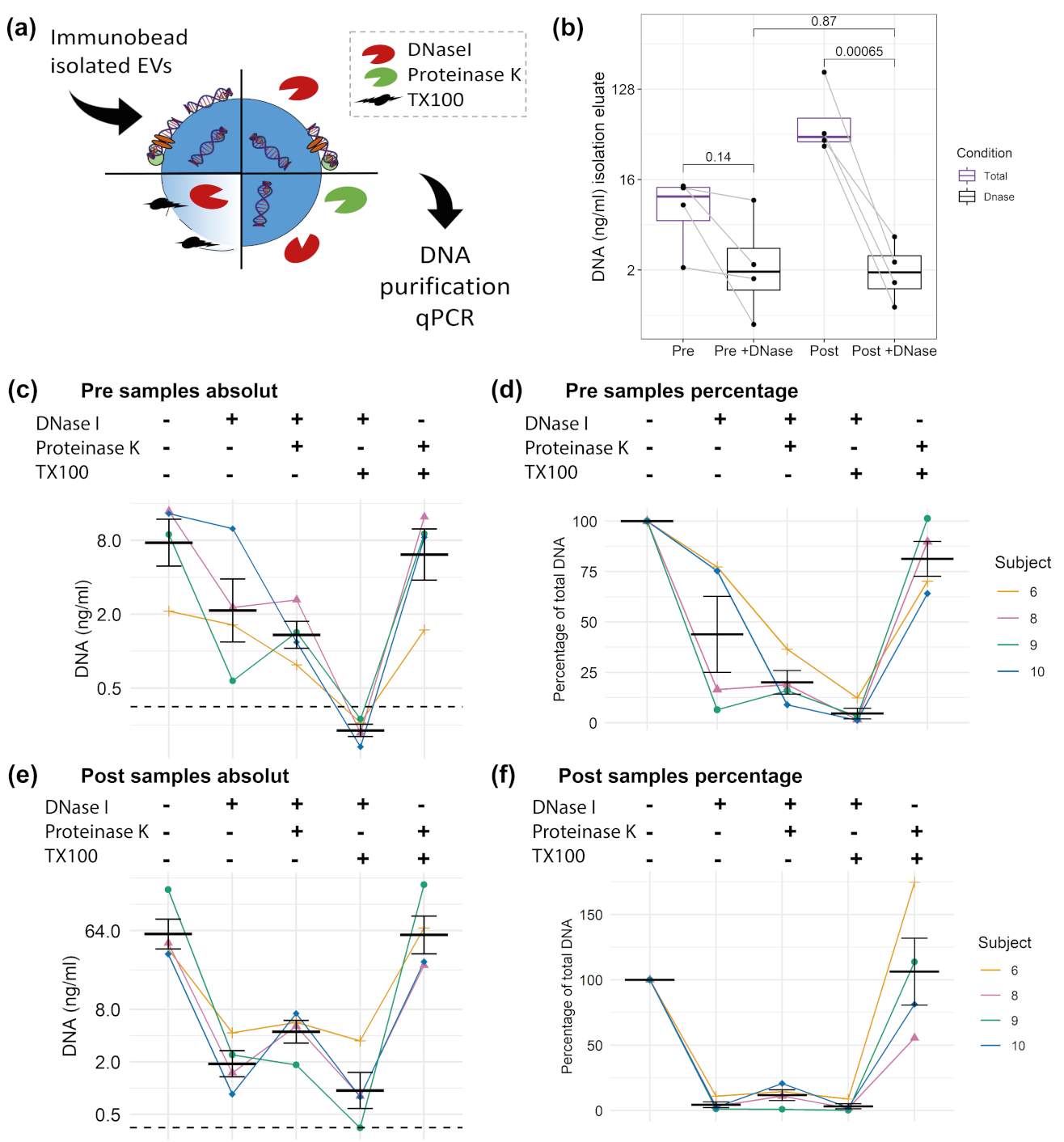
3.3. DNA Digestion in SEC Fractions
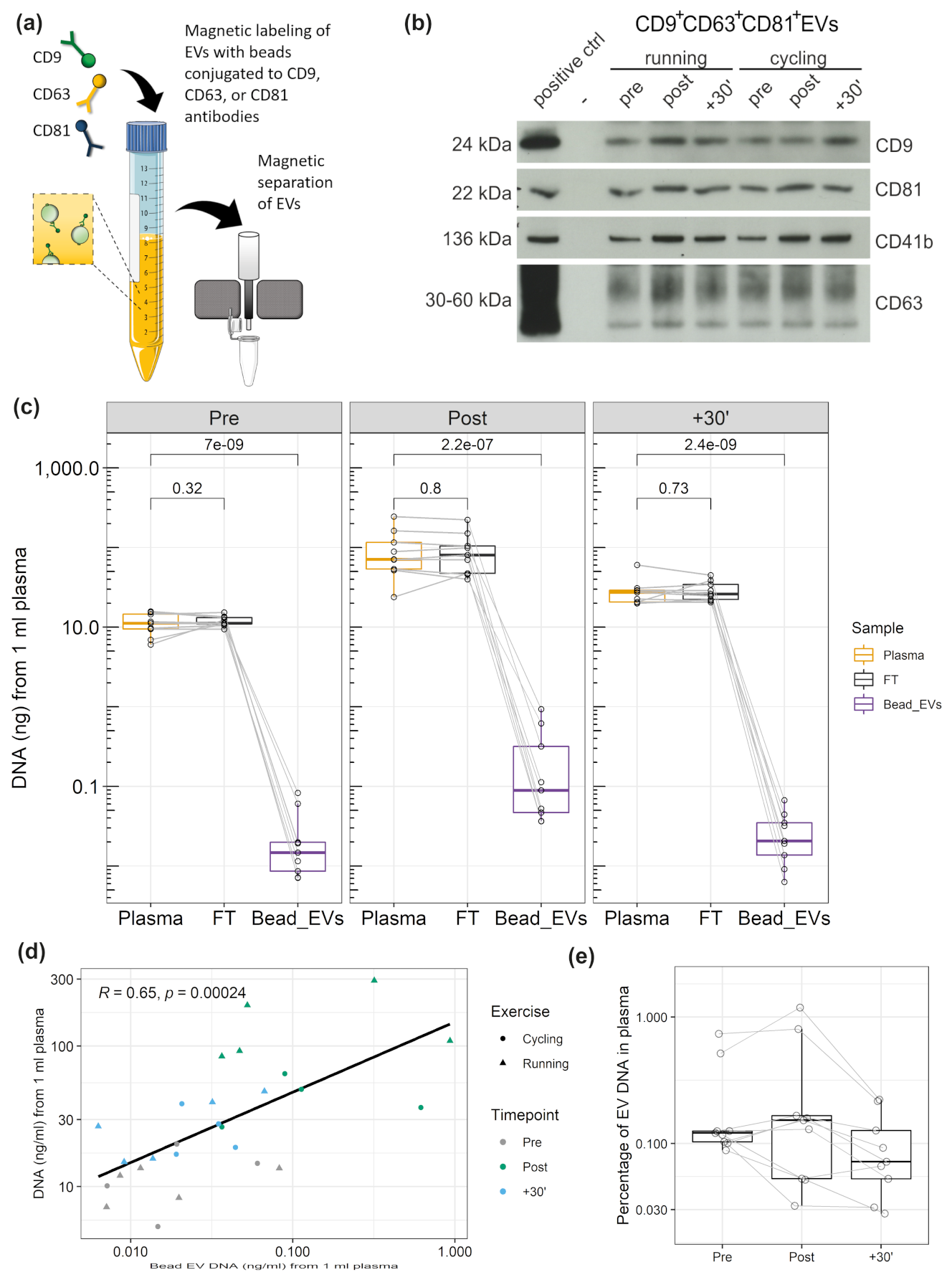
3.4. DNA Associated with CD9+/CD63+/CD81+EVs
3.5. DNA Cargo of EVs
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Malkin, E.Z.; Bratman, S.V. Bioactive DNA from extracellular vesicles and particles. Cell Death Dis. 2020, 11, 584. [Google Scholar] [CrossRef] [PubMed]
- Contrepois, K.; Wu, S.; Moneghetti, K.J.; Hornburg, D.; Ahadi, S.; Tsai, M.S.; Metwally, A.A.; Wei, E.; Lee-McMullen, B.; Quijada, J.V.; et al. Molecular Choreography of Acute Exercise. Cell 2020, 181, 1112–1130. [Google Scholar] [CrossRef]
- Uhlén, M.; Karlsson, M.J.; Hober, A.; Svensson, A.-S.; Scheffel, J.; Kotol, D.; Zhong, W.; Tebani, A.; Strandberg, L.; Edfors, F.; et al. The human secretome. Sci. Signal. 2019, 12, eaaz0274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stroun, M.; Anker, P. Nucleic acids spontaneously released by living frog auricles. Biochem. J. 1972, 128, 100P. [Google Scholar] [CrossRef]
- Anker, P.; Stroun, M.; Maurice, P.A. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res. 1975, 35, 2375–2382. [Google Scholar] [PubMed]
- Bronkhorst, A.J.; Wentzel, J.F.; Aucamp, J.; van Dyk, E.; du Plessis, L.; Pretorius, P.J. Characterization of the cell-free DNA released by cultured cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Galvanin, A.; Dostert, G.; Ayadi, L.; Marchand, V.; Velot, É.; Motorin, Y. Diversity and heterogeneity of extracellular RNA in human plasma. Biochimie 2019, 164, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Beiter, T.; Fragasso, A.; Hudemann, J.; Nieß, A.M.; Simon, P. Short-Term Treadmill Running as a Model for Studying Cell-Free DNA Kinetics In Vivo. Clin. Chem. 2011, 57, 633–636. [Google Scholar] [CrossRef] [Green Version]
- Breitbach, S.; Sterzing, B.; Magallanes, C.; Tug, S.; Simon, P. Direct measurement of cell-free DNA from serially collected capillary plasma during incremental exercise. J. Appl. Physiol. 2014, 117, 119–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, R.; Yeri, A.; Das, A.; Courtright-Lim, A.; Ziegler, O.; Gervino, E.; Ocel, J.; Quintero-Pinzon, P.; Wooster, L.; Bailey, C.S.; et al. Small RNA-seq during acute maximal exercise reveal RNAs involved in vascular inflammation and cardiometabolic health: Brief report. Am. J. Physiol. Hear. Circ. Physiol. 2017, 313, H1162–H1167. [Google Scholar] [CrossRef]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018, 29, 5068. [Google Scholar] [CrossRef] [Green Version]
- Lui, Y.Y.N.; Woo, K.-S.; Wang, A.Y.M.; Yeung, C.-K.; Li, P.K.T.; Chau, E.; Ruygrok, P.; Lo, Y.M.D. Origin of Plasma Cell-free DNA after Solid Organ Transplantation. Clin. Chem. 2003, 49, 495–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tug, S.; Helmig, S.; Deichmann, E.R.; Schmeier-Jürchott, A.; Wagner, E.; Zimmermann, T.; Radsak, M.; Giacca, M.; Simon, P. Exercise-induced increases in cell free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc. Immunol. Rev. 2015, 21, 164–173. [Google Scholar]
- Jiang, P.; Lo, Y.M.D. The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet. 2016, 32, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Grabuschnig, S.; Bronkhorst, A.J.; Holdenrieder, S.; Rosales Rodriguez, I.; Schliep, K.P.; Schwendenwein, D.; Ungerer, V.; Sensen, C.W. Putative origins of cell-free DNA in humans: A review of active and passive nucleic acid release mechanisms. Int. J. Mol. Sci. 2020, 21, 8062. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Brahmer, A.; Neuberger, E.; Esch-Heisser, L.; Haller, N.; Jorgensen, M.M.; Baek, R.; Möbius, W.; Simon, P.; Krämer-Albers, E.-M. Platelets, endothelial cells and leukocytes contribute to the exercise-triggered release of extracellular vesicles into the circulation. J. Extracell. Vesicles 2019, 8, 1615820. [Google Scholar] [CrossRef] [PubMed]
- Frühbeis, C.; Helmig, S.; Tug, S.; Simon, P.; Krämer-Albers, E.-M. Physical exercise induces rapid release of small extracellular vesicles into the circulation. J. Extracell. Vesicles 2015, 4, 28239. [Google Scholar] [CrossRef] [Green Version]
- Whitham, M.; Parker, B.L.; Friedrichsen, M.; Hingst, J.R.; Hjorth, M.; Hughes, W.E.; Egan, C.L.; Cron, L.; Watt, K.I.; Kuchel, R.P.; et al. Extracellular vesicles provide a means for tissue crosstalk during exercise. Cell Metab. 2018, 27, 237–251.e4. [Google Scholar] [CrossRef] [Green Version]
- Arraud, N.; Linares, R.; Tan, S.; Gounou, C.; Pasquet, J.M.; Mornet, S.; Brisson, A.R. Extracellular vesicles from blood plasma: Determination of their morphology, size, phenotype and concentration. J. Thromb. Haemost. 2014, 12, 614–627. [Google Scholar] [CrossRef]
- Berckmans, R.J.; Lacroix, R.; Hau, C.M.; Sturk, A.; Nieuwland, R. Extracellular vesicles and coagulation in blood from healthy humans revisited. J. Extracell. Vesicles 2019, 8, 1688936. [Google Scholar] [CrossRef] [PubMed]
- Palviainen, M.; Saraswat, M.; Varga, Z.; Kitka, D.; Neuvonen, M.; Puhka, M.; Joenväärä, S.; Renkonen, R.; Nieuwland, R.; Takatalo, M.; et al. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo—Implications for biomarker discovery. PLoS ONE 2020, 15, e0236439. [Google Scholar] [CrossRef] [PubMed]
- Buzás, E.I.; Tóth, E.Á.; Sódar, B.W.; Szabó-Taylor, K.É. Molecular interactions at the surface of extracellular vesicles. Semin. Immunopathol. 2018, 40, 453–464. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; LeBleu, V.S. Discovery of double-stranded genomic DNA in circulating exosomes. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of exosome composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.H.; Chennakrishnaiah, S.; Audemard, E.; Montermini, L.; Meehan, B.; Rak, J. Oncogenic ras-driven cancer cell vesiculation leads to emission of double-stranded DNA capable of interacting with target cells. Biochem. Biophys. Res. Commun. 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, S.; Cornils, K.; Speiseder, T.; Badbaran, A.; Reimer, R.; Indenbirken, D.; Grundhoff, A.; Brunswig-Spickenheier, B.; Alawi, M.; Lange, C. Indication of horizontal DNA gene transfer by extracellular vesicles. PLoS ONE 2016, 11, e0163665. [Google Scholar] [CrossRef] [Green Version]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Montermini, L.; Jeong, H.; Sharma, S.; Meehan, B.; Rak, J. Mapping subpopulations of cancer cell-derived extracellular vesicles and particles by nano-flow cytometry. ACS Nano 2019, 13, 10499–10511. [Google Scholar] [CrossRef] [PubMed]
- Németh, A.; Orgovan, N.; Sódar, B.W.; Osteikoetxea, X.; Pálóczi, K.; Szabó-Taylor, K.É.; Vukman, K.V.; Kittel, Á.; Turiák, L.; Wiener, Z.; et al. Antibiotic-induced release of small extracellular vesicles (exosomes) with surface-associated DNA. Sci. Rep. 2017, 7, 8202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Higginbotham, J.N.; Jeppesen, D.K.; Yang, Y.-P.; Li, W.; McKinley, E.T.; Graves-Deal, R.; Ping, J.; Britain, C.M.; Dorsett, K.A.; et al. Transfer of functional cargo in exomeres. Cell Rep. 2019, 27, 940–954. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Erratum to: Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2018, 9, 4109. [Google Scholar] [CrossRef]
- Allenson, K.; Castillo, J.; San Lucas, F.A.; Scelo, G.; Kim, D.U.; Bernard, V.; Davis, G.; Kumar, T.; Katz, M.; Overman, M.J.; et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017, 28, 741–747. [Google Scholar] [CrossRef]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lázaro-Ibáñez, E.; Lässer, C.; Shelke, G.V.; Crescitelli, R.; Jang, S.C.; Cvjetkovic, A.; García-Rodríguez, A.; Lötvall, J. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J. Extracell. Vesicles 2019, 8, 1656993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Che, S.P.Y.; Kurywchak, P.; Tavormina, J.L.; Gansmo, L.B.; Correa de Sampaio, P.; Tachezy, M.; Bockhorn, M.; Gebauer, F.; Haltom, A.R.; et al. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol. Ther. 2017, 18, 158–165. [Google Scholar] [CrossRef] [Green Version]
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De, S.; et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef] [Green Version]
- Klump, J.; Phillipp, U.; Follo, M.; Eremin, A.; Lehmann, H.; Nestel, S.; von Bubnoff, N.; Nazarenko, I. Extracellular vesicles or free circulating DNA: Where to search for BRAF and cKIT mutations? Nanomed. Nanotechnol. Biol. Med. 2018, 14, 875–882. [Google Scholar] [CrossRef]
- Helmig, S.; Frühbeis, C.; Krämer-Albers, E.-M.; Simon, P.; Tug, S. Release of bulk cell free DNA during physical exercise occurs independent of extracellular vesicles. Eur. J. Appl. Physiol. 2015, 115, 2271–2280. [Google Scholar] [CrossRef]
- Cai, J.; Guan, W.; Tan, X.; Chen, C.; Li, L.; Wang, N.; Zou, X.; Zhou, F.; Wang, J.; Pei, F.; et al. SRY gene transferred by extracellular vesicles accelerates atherosclerosis by promotion of leucocyte adherence to endothelial cells. Clin. Sci. 2015, 129, 259–269. [Google Scholar] [CrossRef]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monguió-Tortajada, M.; Gálvez-Montón, C.; Bayes-Genis, A.; Roura, S.; Borràs, F.E. Extracellular vesicle isolation methods: Rising impact of size-exclusion chromatography. Cell. Mol. Life Sci. 2019, 76, 2369–2382. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, J.B. What Are We Looking At? Extracellular Vesicles, Lipoproteins, or Both? Circ. Res. 2017, 121, 920–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2015; pp. 179–209. [Google Scholar]
- Brahmer, A.; Neuberger, E.W.I.; Simon, P.; Krämer-Albers, E.-M. Considerations for the analysis of small extracellular vesicles in physical exercise. Front. Physiol. 2020, 11, 1611. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, C. Rating of Perceived Exertion (RPE). J. Physiother. 2012, 58, 62. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, R.; Judicone, C.; Poncelet, P.; Robert, S.; Arnaud, L.; Sampol, J.; Dignat-George, F. Impact of pre-analytical parameters on the measurement of circulating microparticles: Towards standardization of protocol. J. Thromb. Haemost. 2012, 10, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, Ö.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; Bertier, L.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef]
- Neuberger, E.W.I.; Brahmer, A.; Ehlert, T.; Kluge, K.; Philippi, K.F.A.; Boedecker, S.C.; Weinmann-Menke, J.; Simon, P. Validating quantitative PCR assays for cell-free DNA detection without DNA extraction: Exercise induced kinetics in systemic lupus erythematosus patients. medRXiv 2021. [Google Scholar] [CrossRef]
- Sumenkova, D.V.; Polyakov, L.M.; Panin, L.E. Plasma lipoproteins as a transport form of extracellular DNA. Bull. Exp. Biol. Med. 2013, 154, 622–623. [Google Scholar] [CrossRef] [PubMed]
- Tug, S.; Mehdorn, M.; Helmig, S.; Breitbach, S.; Ehlert, T.; Simon, P. Exploring the potential of Cell-Free-DNA measurements after an exhaustive cycle-ergometer test as a marker for performance-related parameters. Int. J. Sports Physiol. Perform. 2017, 12, 597–604. [Google Scholar] [CrossRef]
- Napirei, M.; Wulf, S.; Eulitz, D.; Mannherz, H.G.; Kloeckl, T. Comparative characterization of rat deoxyribonuclease 1 (Dnase1) and murine deoxyribonuclease 1-like 3 (Dnase1l3). Biochem. J. 2005, 389, 355–364. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef] [PubMed]
- Möller, A.; Lobb, R.J. The evolving translational potential of small extracellular vesicles in cancer. Nat. Rev. Cancer 2020, 20, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef] [Green Version]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Tandia, B.-M.; Vandenbranden, M.; Wattiez, R.; Lakhdar, Z.; Ruysschaert, J.-M.; Elouahabi, A. Identification of human plasma proteins that bind to cationic lipid/DNA complex and analysis of their effects on transfection efficiency: Implications for intravenous gene transfer. Mol. Ther. 2003, 8, 264–273. [Google Scholar] [CrossRef]
- Qubit® dsDNA HS Assay Kits. Available online: https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FQubit_dsDNA_HS_Assay_UG.pdf&title=VXNlciBHdWlkZTogUXViaXQgZHNETkEgSFMgQXNzYXkgS2l0cw== (accessed on 27 January 2021).
- Vechetti, I.J.; Valentino, T.; Mobley, C.B.; McCarthy, J.J. The role of extracellular vesicles in skeletal muscle and systematic adaptation to exercise. J. Physiol. 2020, 599, 845–861. [Google Scholar] [CrossRef] [PubMed]
- Fuller, O.K.; Whitham, M.; Mathivanan, S.; Febbraio, M.A. The protective effect of exercise in neurodegenerative diseases: The potential role of extracellular vesicles. Cells 2020, 9, 2182. [Google Scholar] [CrossRef] [PubMed]
- Denham, J.; Spencer, S.J. Emerging roles of extracellular vesicles in the intercellular communication for exercise-induced adaptations. Am. J. Physiol. Metab. 2020, 319, E320–E329. [Google Scholar] [CrossRef] [PubMed]


| Subjects | Age (y) | BMI (kg/m2) | Exercise | Time Until Exhaustion (min) | Maximal Heart Rate (1/min) | RPE |
|---|---|---|---|---|---|---|
| n = 5 | 23.8 (±1.47) | 22.95 (±1.57) | Running | 22.96 (±2.56) | 193.4 (±2.87) | 19.6 (±0.49) |
| Cycling | 28.32 (±3.54) | 188.2 (±8.30) | 19.8 (±0.40) | |||
| n = 5 | 29.6 (±4.72) | 23.56 (±1.06) | Running | 21.10 (±1.56) | 189.8 (±7.08) | 18.8 (± 0.40) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neuberger, E.W.I.; Hillen, B.; Mayr, K.; Simon, P.; Krämer-Albers, E.-M.; Brahmer, A. Kinetics and Topology of DNA Associated with Circulating Extracellular Vesicles Released during Exercise. Genes 2021, 12, 522. https://doi.org/10.3390/genes12040522
Neuberger EWI, Hillen B, Mayr K, Simon P, Krämer-Albers E-M, Brahmer A. Kinetics and Topology of DNA Associated with Circulating Extracellular Vesicles Released during Exercise. Genes. 2021; 12(4):522. https://doi.org/10.3390/genes12040522
Chicago/Turabian StyleNeuberger, Elmo W. I., Barlo Hillen, Katharina Mayr, Perikles Simon, Eva-Maria Krämer-Albers, and Alexandra Brahmer. 2021. "Kinetics and Topology of DNA Associated with Circulating Extracellular Vesicles Released during Exercise" Genes 12, no. 4: 522. https://doi.org/10.3390/genes12040522
APA StyleNeuberger, E. W. I., Hillen, B., Mayr, K., Simon, P., Krämer-Albers, E. -M., & Brahmer, A. (2021). Kinetics and Topology of DNA Associated with Circulating Extracellular Vesicles Released during Exercise. Genes, 12(4), 522. https://doi.org/10.3390/genes12040522






