Inosine in Biology and Disease
Abstract
1. Introduction
2. Detection and Quantification of Inosine
2.1. Chromatography-Based Methods
2.2. Reverse Transcription (RT)-Based Methods
2.3. Other Methods
3. Molecular Inosine in Metabolism and Signaling
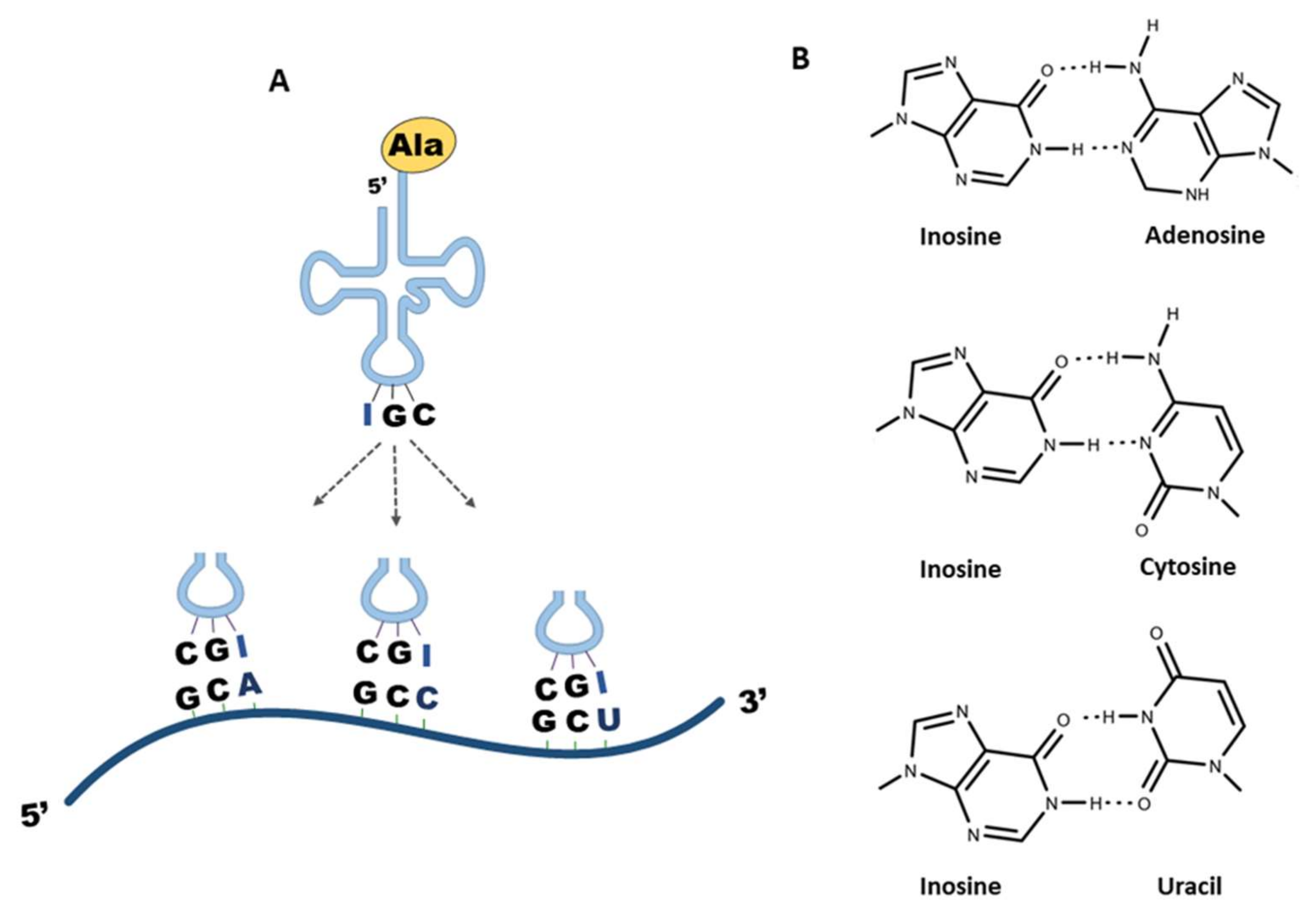
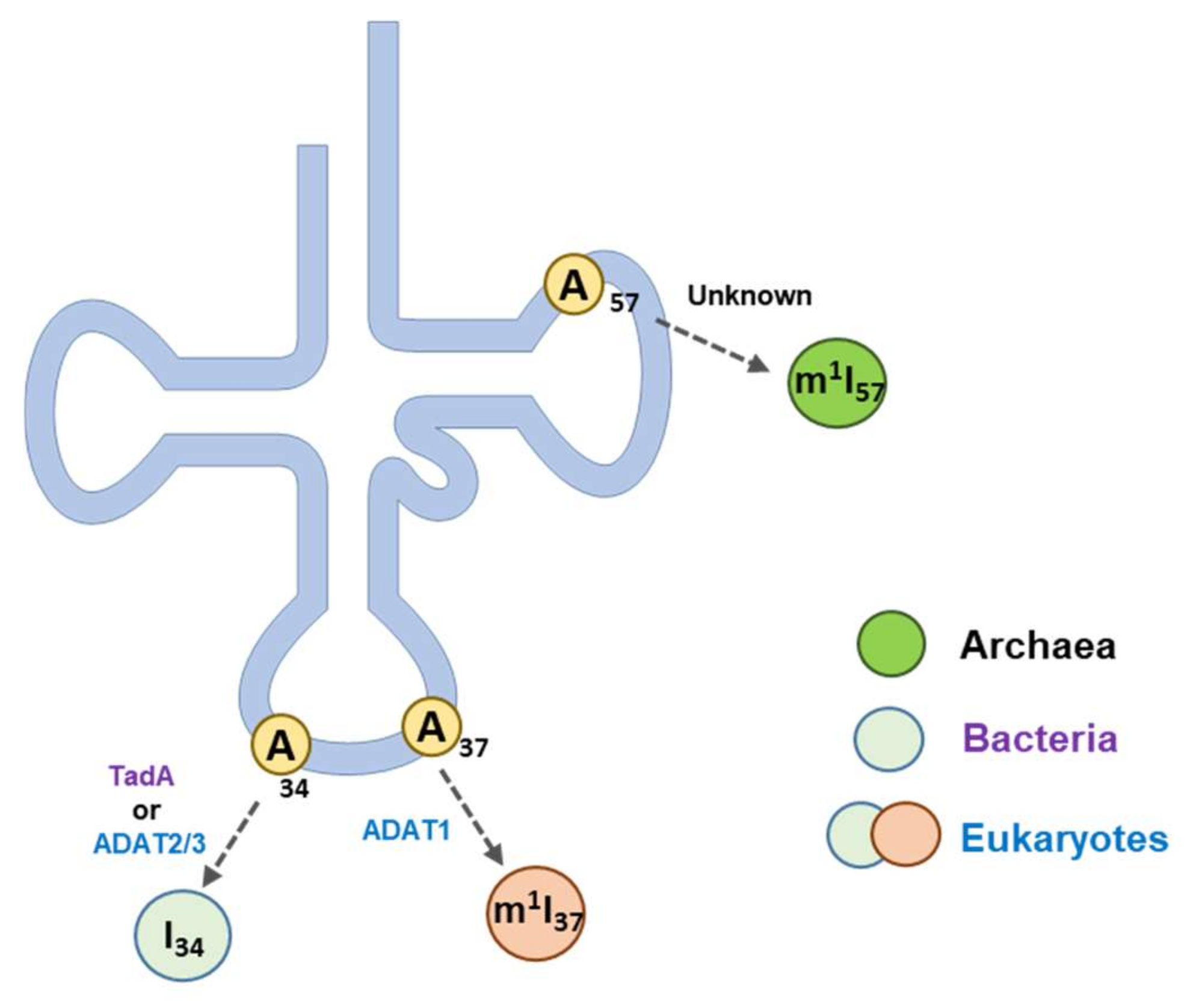
4. Inosine in tRNA
5. Inosine in mRNA
6. Inosine in MicroRNAs
7. Inosine in Viral RNAs and Mobile Elements
7.1. Inosine in Viral RNAs
7.2. Inosine in Mobile Elements
8. Inosine in Other Nucleic Acids
8.1. Inosine in Ribosomal RNAs
8.2. Inosine in DNA
9. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holley, R.W.; Apgar, J.; Everett, G.A.; Madison, J.T.; Marquisee, M.; Merrill, S.H.; Penswick, J.R.; Zamir, A. Structure of a Ribonucleic Acid. Science 1965, 147, 1462–1465. [Google Scholar] [CrossRef]
- Schoonen, M.A.; Xu, Y. Nitrogen reduction under hydrothermal vent conditions: Implications for the prebiotic synthesis of C-H-O-N compounds. Astrobiology 2001, 1, 133–142. [Google Scholar] [CrossRef]
- Pearce, B.K.D.; Pudritz, R.E.; Semenov, D.A.; Henning, T.K. Origin of the RNA world: The fate of nucleobases in warm little ponds. Proc. Natl. Acad. Sci. USA 2017, 114, 11327–11332. [Google Scholar] [CrossRef]
- Nutman, A.P.; Bennett, V.C.; Friend, C.R.; Van Kranendonk, M.J.; Chivas, A.R. Rapid emergence of life shown by discovery of 3,700-million-year-old microbial structures. Nature 2016, 537, 535–538. [Google Scholar] [CrossRef]
- Kasting, J.F. Earth’s early atmosphere. Science 1993, 259, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Nuevo, C.K.M.M.; Scott, A. Sandford. The photochemistry of pyrimidine in realistic astrophysical ices and the production of nucleobases. Astrophys. J. 2014, 793, 7. [Google Scholar] [CrossRef]
- Leu, K.; Obermayer, B.; Rajamani, S.; Gerland, U.; Chen, I.A. The prebiotic evolutionary advantage of transferring genetic information from RNA to DNA. Nucleic Acids Res. 2011, 39, 8135–8147. [Google Scholar] [CrossRef] [PubMed]
- Szostak, J.W. The Narrow Road to the Deep Past: In Search of the Chemistry of the Origin of Life. Angew. Chem. 2017, 56, 11037–11043. [Google Scholar] [CrossRef]
- Nagaswamy, U.; Voss, N.; Zhang, Z.; Fox, G.E. Database of non-canonical base pairs found in known RNA structures. Nucleic Acids Res. 2000, 28, 375–376. [Google Scholar] [CrossRef]
- Machnicka, M.A.; Milanowska, K.; Oglou, O.O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. MODOMICS: A database of RNA modification pathways--2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; O’Flaherty, D.K.; Zhou, L.; Lelyveld, V.S.; Szostak, J.W. Inosine, but none of the 8-oxo-purines, is a plausible component of a primordial version of RNA. Proc. Natl. Acad. Sci. USA 2018, 115, 13318–13323. [Google Scholar] [CrossRef]
- Stairs, S.; Nikmal, A.; Bucar, D.K.; Zheng, S.L.; Szostak, J.W.; Powner, M.W. Divergent prebiotic synthesis of pyrimidine and 8-oxo-purine ribonucleotides. Nat. Commun. 2017, 8, 15270. [Google Scholar] [CrossRef]
- Heuberger, B.D.; Pal, A.; Del Frate, F.; Topkar, V.V.; Szostak, J.W. Replacing uridine with 2-thiouridine enhances the rate and fidelity of nonenzymatic RNA primer extension. J. Am. Chem. Soc. 2015, 137, 2769–2775. [Google Scholar] [CrossRef]
- Wolf, J.; Gerber, A.P.; Keller, W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 2002, 21, 3841–3851. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef]
- Palladino, M.J.; Keegan, L.P.; O’Connell, M.A.; Reenan, R.A. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 2000, 102, 437–449. [Google Scholar] [CrossRef]
- Gu, T.; Buaas, F.W.; Simons, A.K.; Ackert-Bicknell, C.L.; Braun, R.E.; Hibbs, M.A. Canonical A-to-I and C-to-U RNA editing is enriched at 3’UTRs and microRNA target sites in multiple mouse tissues. PLoS ONE 2012, 7, e33720. [Google Scholar] [CrossRef] [PubMed]
- Wulff, T.F.; Arguello, R.J.; Jordan, M.M.; Frigole, H.R.; Hauquier, G.; Filonava, L.; Camacho, N.; Gatti, E.; Pierre, P.; de Pouplana, L.R.; et al. Detection of a Subset of Posttranscriptional Transfer RNA Modifications in Vivo with a Restriction Fragment Length Polymorphism-Based Method. Biochemistry 2017, 56, 4029–4038. [Google Scholar] [CrossRef]
- Paz, N.; Levanon, E.Y.; Amariglio, N.; Heimberger, A.B.; Ram, Z.; Constantini, S.; Barbash, Z.S.; Adamsky, K.; Safran, M.; Hirschberg, A.; et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007, 17, 1586–1595. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; An, O.; Hong, H.; Chan, T.H.M.; Song, Y.; Shen, H.; Tang, S.J.; Lin, J.S.; Ng, V.H.E.; Tay, D.J.T.; et al. Suppression of adenosine-to-inosine (A-to-I) RNA editome by death associated protein 3 (DAP3) promotes cancer progression. Sci. Adv. 2020, 6, eaba5136. [Google Scholar] [CrossRef]
- Torres, A.G.; Pineyro, D.; Rodriguez-Escriba, M.; Camacho, N.; Reina, O.; Saint-Leger, A.; Filonava, L.; Batlle, E.; de Pouplana, L.R. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015, 43, 5145–5157. [Google Scholar] [CrossRef]
- Sakurai, M.; Yano, T.; Kawabata, H.; Ueda, H.; Suzuki, T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol. 2010, 6, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Wulff, T.F.; Rodriguez-Escriba, M.; Camacho, N.; de Pouplana, L.R. Detection of Inosine on Transfer RNAs without a Reverse Transcription Reaction. Biochemistry 2018, 57, 5641–5647. [Google Scholar] [CrossRef] [PubMed]
- Morse, D.P.; Bass, B.L. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry 1997, 36, 8429–8434. [Google Scholar] [CrossRef] [PubMed]
- Saint-Leger, A.; Bello, C.; Dans, P.D.; Torres, A.G.; Novoa, E.M.; Camacho, N.; Orozco, M.; Kondrashov, F.A.; de Pouplana, L.R. Saturation of recognition elements blocks evolution of new tRNA identities. Sci. Adv. 2016, 2, e1501860. [Google Scholar] [CrossRef]
- Vik, E.S.; Nawaz, M.S.; Strom Andersen, P.; Fladeby, C.; Bjoras, M.; Dalhus, B.; Alseth, I. Endonuclease V cleaves at inosines in RNA. Nat. Commun. 2013, 4, 2271. [Google Scholar] [CrossRef]
- Morita, Y.; Shibutani, T.; Nakanishi, N.; Nishikura, K.; Iwai, S.; Kuraoka, I. Human endonuclease V is a ribonuclease specific for inosine-containing RNA. Nat. Commun. 2013, 4, 2273. [Google Scholar] [CrossRef]
- Workman, R.E.; Tang, A.D.; Tang, P.S.; Jain, M.; Tyson, J.R.; Razaghi, R.; Zuzarte, P.C.; Gilpatrick, T.; Payne, A.; Quick, J.; et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 2019, 16, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Purine bases can be synthesized de novo or recycled by salvage pathways. In Biochemistry; W H Freeman: New York, NY, USA, 2002. [Google Scholar]
- Goswami, M.T.; Chen, G.; Chakravarthi, B.V.; Pathi, S.S.; Anand, S.K.; Carskadon, S.L.; Giordano, T.J.; Chinnaiyan, A.M.; Thomas, D.G.; Palanisamy, N.; et al. Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer. Oncotarget 2015, 6, 23445–23461. [Google Scholar] [CrossRef] [PubMed]
- Barfeld, S.J.; Fazli, L.; Persson, M.; Marjavaara, L.; Urbanucci, A.; Kaukoniemi, K.M.; Rennie, P.S.; Ceder, Y.; Chabes, A.; Visakorpi, T.; et al. Myc-dependent purine biosynthesis affects nucleolar stress and therapy response in prostate cancer. Oncotarget 2015, 6, 12587–12602. [Google Scholar] [CrossRef]
- Bahreyni, A.; Samani, S.S.; Rahmani, F.; Behnam-Rassouli, R.; Khazaei, M.; Ryzhikov, M.; Parizadeh, M.R.; Avan, A.; Hassanian, S.M. Role of adenosine signaling in the pathogenesis of breast cancer. J. Cell. Physiol. 2018, 233, 1836–1843. [Google Scholar] [CrossRef] [PubMed]
- Di Virgilio, F. Purines, purinergic receptors, and cancer. Cancer Res. 2012, 72, 5441–5447. [Google Scholar] [CrossRef]
- Pellegatti, P.; Raffaghello, L.; Bianchi, G.; Piccardi, F.; Pistoia, V.; Di Virgilio, F. Increased level of extracellular ATP at tumor sites: In vivo imaging with plasma membrane luciferase. PLoS ONE 2008, 3, e2599. [Google Scholar] [CrossRef]
- Hagner, N.; Joerger, M. Cancer chemotherapy: Targeting folic acid synthesis. Cancer Manag. Res. 2010, 2, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.C.; Weber, G.; Morris, H.P. IMP dehydrogenase, an enzyme linked with proliferation and malignancy. Nature 1975, 256, 331–333. [Google Scholar] [CrossRef]
- Xu, Y.; Zheng, Z.; Gao, Y.; Duan, S.; Chen, C.; Rong, J.; Wang, K.; Yun, M.; Weng, H.; Ye, S.; et al. High expression of IMPDH2 is associated with aggressive features and poor prognosis of primary nasopharyngeal carcinoma. Sci. Rep. 2017, 7, 745. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Natsumeda, Y.; Yamaji, Y.; Jayaram, H.N.; Tricot, G.J.; Hoffman, R.; Weber, G. IMP dehydrogenase: Inhibition by the anti-leukemic drug, tiazofurin. Leuk. Res. 1989, 13, 179–184. [Google Scholar] [CrossRef]
- Elion, G.B. The purine path to chemotherapy. Science 1989, 244, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Teml, A.; Schaeffeler, E.; Herrlinger, K.R.; Klotz, U.; Schwab, M. Thiopurine treatment in inflammatory bowel disease: Clinical pharmacology and implication of pharmacogenetically guided dosing. Clin. Pharmacokinet. 2007, 46, 187–208. [Google Scholar] [CrossRef] [PubMed]
- Pedley, A.M.; Benkovic, S.J. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci. 2017, 42, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Parker, W.B. Enzymology of purine and pyrimidine antimetabolites used in the treatment of cancer. Chem. Rev. 2009, 109, 2880–2893. [Google Scholar] [CrossRef]
- Bierau, J.; Lindhout, M.; Bakker, J.A. Pharmacogenetic significance of inosine triphosphatase. Pharmacogenomics 2007, 8, 1221–1228. [Google Scholar] [CrossRef]
- Handley, M.T.; Reddy, K.; Wills, J.; Rosser, E.; Kamath, A.; Halachev, M.; Falkous, G.; Williams, D.; Cox, P.; Meynert, A.; et al. ITPase deficiency causes a Martsolf-like syndrome with a lethal infantile dilated cardiomyopathy. PLoS Genet. 2019, 15, e1007605. [Google Scholar] [CrossRef] [PubMed]
- Kevelam, S.H.; Bierau, J.; Salvarinova, R.; Agrawal, S.; Honzik, T.; Visser, D.; Weiss, M.M.; Salomons, G.S.; Abbink, T.E.; Waisfisz, Q.; et al. Recessive ITPA mutations cause an early infantile encephalopathy. Ann. Neurol. 2015, 78, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Burnstock, G. Purinergic signalling and disorders of the central nervous system. Nat. Rev. Drug Discov. 2008, 7, 575–590. [Google Scholar] [CrossRef]
- Abbracchio, M.P.; Burnstock, G.; Verkhratsky, A.; Zimmermann, H. Purinergic signalling in the nervous system: An overview. Trends Neurosci. 2009, 32, 19–29. [Google Scholar] [CrossRef]
- Nascimento, F.P.; Figueredo, S.M.; Marcon, R.; Martins, D.F.; Macedo, S.J., Jr.; Lima, D.A.; Almeida, R.C.; Ostroski, R.M.; Rodrigues, A.L.; Santos, A.R. Inosine reduces pain-related behavior in mice: Involvement of adenosine A1 and A2A receptor subtypes and protein kinase C pathways. J. Pharmacol. Exp. Ther. 2010, 334, 590–598. [Google Scholar] [CrossRef]
- Liu, F.; You, S.W.; Yao, L.P.; Liu, H.L.; Jiao, X.Y.; Shi, M.; Zhao, Q.B.; Ju, G. Secondary degeneration reduced by inosine after spinal cord injury in rats. Spinal Cord 2006, 44, 421–426. [Google Scholar] [CrossRef]
- Muto, J.; Lee, H.; Lee, H.; Uwaya, A.; Park, J.; Nakajima, S.; Nagata, K.; Ohno, M.; Ohsawa, I.; Mikami, T. Oral administration of inosine produces antidepressant-like effects in mice. Sci. Rep. 2014, 4, 4199. [Google Scholar] [CrossRef]
- Chen, P.; Goldberg, D.E.; Kolb, B.; Lanser, M.; Benowitz, L.I. Inosine induces axonal rewiring and improves behavioral outcome after stroke. Proc. Natl. Acad. Sci. USA 2002, 99, 9031–9036. [Google Scholar] [CrossRef] [PubMed]
- Benowitz, L.I.; Goldberg, D.E.; Madsen, J.R.; Soni, D.; Irwin, N. Inosine stimulates extensive axon collateral growth in the rat corticospinal tract after injury. Proc. Natl. Acad. Sci. USA 1999, 96, 13486–13490. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, C.E.; Spitsin, S.; Zimmerman, V.; Jacobs, D.; Udupa, J.K.; Hooper, D.C.; Koprowski, H. The treatment of multiple sclerosis with inosine. J. Altern. Complement. Med. 2009, 15, 619–625. [Google Scholar] [CrossRef]
- Nicholson, K.; Chan, J.; Macklin, E.A.; Levine-Weinberg, M.; Breen, C.; Bakshi, R.; Grasso, D.L.; Wills, A.M.; Jahandideh, S.; Taylor, A.A.; et al. Pilot trial of inosine to elevate urate levels in amyotrophic lateral sclerosis. Ann. Clin. Transl. Neurol. 2018, 5, 1522–1533. [Google Scholar] [CrossRef]
- Parkinson Study Group; Schwarzschild, M.A.; Ascherio, A.; Beal, M.F.; Cudkowicz, M.E.; Curhan, G.C.; Hare, J.M.; Hooper, D.C.; Kieburtz, K.D.; Macklin, E.A.; et al. Inosine to increase serum and cerebrospinal fluid urate in Parkinson disease: A randomized clinical trial. JAMA Neurol. 2014, 71, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Hung, A.Y.; Schwarzschild, M.A. Approaches to Disease Modification for Parkinson’s Disease: Clinical Trials and Lessons Learned. Neurotherapeutics 2020, 17, 1393–1405. [Google Scholar] [CrossRef] [PubMed]
- Sliva, J.; Pantzartzi, C.N.; Votava, M. Inosine Pranobex: A Key Player in the Game Against a Wide Range of Viral Infections and Non-Infectious Diseases. Adv. Ther. 2019, 36, 1878–1905. [Google Scholar] [CrossRef]
- Beran, J.; Spajdel, M.; Katzerova, V.; Holousova, A.; Malys, J.; Finger Rouskova, J.; Sliva, J. Inosine Pranobex Significantly Decreased the Case-Fatality Rate among PCR Positive Elderly with SARS-CoV-2 at Three Nursing Homes in the Czech Republic. Pathogens 2020, 9, 1055. [Google Scholar] [CrossRef]
- Klinge, S.; Woolford, J.L., Jr. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef]
- Kim, S.H.; Quigley, G.J.; Suddath, F.L.; McPherson, A.; Sneden, D.; Kim, J.J.; Weinzierl, J.; Rich, A. Three-dimensional structure of yeast phenylalanine transfer RNA: Folding of the polynucleotide chain. Science 1973, 179, 285–288. [Google Scholar] [CrossRef]
- Dixit, S.; Henderson, J.C.; Alfonzo, J.D. Multi-Substrate Specificity and the Evolutionary Basis for Interdependence in tRNA Editing and Methylation Enzymes. Front. Genet. 2019, 10, 104. [Google Scholar] [CrossRef]
- Torres, A.G.; Pineyro, D.; Filonava, L.; Stracker, T.H.; Batlle, E.; de Pouplana, L.R. A-to-I editing on tRNAs: Biochemical, biological and evolutionary implications. FEBS Lett. 2014, 588, 4279–4286. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Batlle, E.; de Pouplana, L.R. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014, 20, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Novoa, E.M.; de Pouplana, L.R. Speeding with control: Codon usage, tRNAs, and ribosomes. Trends Genet. 2012, 28, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Rafels-Ybern, A.; Torres, A.G.; Camacho, N.; Herencia-Ropero, A.; Frigole, H.R.; Wulff, T.F.; Raboteg, M.; Bordons, A.; Grau-Bove, X.; Ruiz-Trillo, I.; et al. The Expansion of Inosine at the Wobble Position of tRNAs, and Its Role in the Evolution of Proteomes. Mol. Biol. Evol. 2019, 36, 650–662. [Google Scholar] [CrossRef]
- Zhou, W.; Karcher, D.; Bock, R. Identification of enzymes for adenosine-to-inosine editing and discovery of cytidine-to-uridine editing in nucleus-encoded transfer RNAs of Arabidopsis. Plant Physiol. 2014, 166, 1985–1997. [Google Scholar] [CrossRef]
- Crick, F.H. Codon--anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966, 19, 548–555. [Google Scholar] [CrossRef]
- Novoa, E.M.; Pavon-Eternod, M.; Pan, T.; de Pouplana, L.R. A role for tRNA modifications in genome structure and codon usage. Cell 2012, 149, 202–213. [Google Scholar] [CrossRef]
- de Crecy-Lagard, V.; Marck, C.; Brochier-Armanet, C.; Grosjean, H. Comparative RNomics and modomics in Mollicutes: Prediction of gene function and evolutionary implications. IUBMB Life 2007, 59, 634–658. [Google Scholar] [CrossRef]
- Diwan, G.D.; Agashe, D. Wobbling Forth and Drifting Back: The Evolutionary History and Impact of Bacterial tRNA Modifications. Mol. Biol. Evol. 2018, 35, 2046–2059. [Google Scholar] [CrossRef]
- Yokobori, S.; Kitamura, A.; Grosjean, H.; Bessho, Y. Life without tRNAArg-adenosine deaminase TadA: Evolutionary consequences of decoding the four CGN codons as arginine in Mycoplasmas and other Mollicutes. Nucleic Acids Res. 2013, 41, 6531–6543. [Google Scholar] [CrossRef]
- Bar-Yaacov, D.; Mordret, E.; Towers, R.; Biniashvili, T.; Soyris, C.; Schwartz, S.; Dahan, O.; Pilpel, Y. RNA editing in bacteria recodes multiple proteins and regulates an evolutionarily conserved toxin-antitoxin system. Genome Res. 2017, 27, 1696–1703. [Google Scholar] [CrossRef]
- Gerber, A.P.; Keller, W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 1999, 286, 1146–1149. [Google Scholar] [CrossRef]
- Grosjean, H.; de Crecy-Lagard, V.; Marck, C. Deciphering synonymous codons in the three domains of life: Co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010, 584, 252–264. [Google Scholar] [CrossRef]
- Maraia, R.J.; Arimbasseri, A.G. Factors That Shape Eukaryotic tRNAomes: Processing, Modification and Anticodon-Codon Use. Biomolecules 2017, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- McKenney, K.M.; Rubio, M.A.T.; Alfonzo, J.D. The Evolution of Substrate Specificity by tRNA Modification Enzymes. Enzymes 2017, 41, 51–88. [Google Scholar] [CrossRef]
- Elias, Y.; Huang, R.H. Biochemical and structural studies of A-to-I editing by tRNA:A34 deaminases at the wobble position of transfer RNA. Biochemistry 2005, 44, 12057–12065. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, R.; Sun, Y.; Chen, R.; Zhou, J.; Tian, Q.; Tao, X.; Zhang, Z.; Luo, G.Z.; Xie, W. Crystal structure of the yeast heterodimeric ADAT2/3 deaminase. BMC Biol. 2020, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.A.; Pastar, I.; Gaston, K.W.; Ragone, F.L.; Janzen, C.J.; Cross, G.A.; Papavasiliou, F.N.; Alfonzo, J.D. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc. Natl. Acad. Sci. USA 2007, 104, 7821–7826. [Google Scholar] [CrossRef]
- Rubio, M.A.; Gaston, K.W.; McKenney, K.M.; Fleming, I.M.; Paris, Z.; Limbach, P.A.; Alfonzo, J.D. Editing and methylation at a single site by functionally interdependent activities. Nature 2017, 542, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Ragone, F.L.; Spears, J.L.; Wohlgamuth-Benedum, J.M.; Kreel, N.; Papavasiliou, F.N.; Alfonzo, J.D. The C-terminal end of the Trypanosoma brucei editing deaminase plays a critical role in tRNA binding. Rna 2011, 17, 1296–1306. [Google Scholar] [CrossRef][Green Version]
- Tsutsumi, S.; Sugiura, R.; Ma, Y.; Tokuoka, H.; Ohta, K.; Ohte, R.; Noma, A.; Suzuki, T.; Kuno, T. Wobble inosine tRNA modification is essential to cell cycle progression in G(1)/S and G(2)/M transitions in fission yeast. J. Biol. Chem. 2007, 282, 33459–33465. [Google Scholar] [CrossRef]
- Delannoy, E.; Le Ret, M.; Faivre-Nitschke, E.; Estavillo, G.M.; Bergdoll, M.; Taylor, N.L.; Pogson, B.J.; Small, I.; Imbault, P.; Gualberto, J.M. Arabidopsis tRNA adenosine deaminase arginine edits the wobble nucleotide of chloroplast tRNAArg(ACG) and is essential for efficient chloroplast translation. Plant Cell 2009, 21, 2058–2071. [Google Scholar] [CrossRef]
- Pernod, K.; Schaeffer, L.; Chicher, J.; Hok, E.; Rick, C.; Geslain, R.; Eriani, G.; Westhof, E.; Ryckelynck, M.; Martin, F. The nature of the purine at position 34 in tRNAs of 4-codon boxes is correlated with nucleotides at positions 32 and 38 to maintain decoding fidelity. Nucleic Acids Res. 2020, 48, 6170–6183. [Google Scholar] [CrossRef]
- Kubo, M.; Imanaka, T. mRNA secondary structure in an open reading frame reduces translation efficiency in Bacillus subtilis. J. Bacteriol. 1989, 171, 4080–4082. [Google Scholar] [CrossRef]
- Parmley, J.L.; Huynen, M.A. Clustering of codons with rare cognate tRNAs in human genes suggests an extra level of expression regulation. PLoS Genet. 2009, 5, e1000548. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Yang, Q.; Li, L.; Dang, Y.; Zhou, Z.; Chen, S.; Liu, Y. Adaptation of codon usage to tRNA I34 modification controls translation kinetics and proteome landscape. PLoS Genet. 2020, 16, e1008836. [Google Scholar] [CrossRef]
- Chan, C.T.; Deng, W.; Li, F.; DeMott, M.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Highly Predictive Reprogramming of tRNA Modifications Is Linked to Selective Expression of Codon-Biased Genes. Chem. Res. Toxicol. 2015, 28, 978–988. [Google Scholar] [CrossRef]
- Bornelov, S.; Selmi, T.; Flad, S.; Dietmann, S.; Frye, M. Codon usage optimization in pluripotent embryonic stem cells. Genome Biol. 2019, 20, 119. [Google Scholar] [CrossRef] [PubMed]
- Rafels-Ybern, A.; Torres, A.G.; Grau-Bove, X.; Ruiz-Trillo, I.; de Pouplana, L.R. Codon adaptation to tRNAs with Inosine modification at position 34 is widespread among Eukaryotes and present in two Bacterial phyla. RNA Biol. 2018, 15, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Rafels-Ybern, A.; Attolini, C.S.; de Pouplana, L.R. Distribution of ADAT-Dependent Codons in the Human Transcriptome. Int. J. Mol. Sci. 2015, 16, 17303–17314. [Google Scholar] [CrossRef] [PubMed]
- de Pouplana, L.R.; Torres, A.G.; Rafels-Ybern, A. What Froze the Genetic Code? Life 2017, 7, 14. [Google Scholar] [CrossRef]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef]
- Lyons, S.M.; Fay, M.M.; Ivanov, P. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett. 2018, 592, 2828–2844. [Google Scholar] [CrossRef] [PubMed]
- Molla-Herman, A.; Angelova, M.T.; Ginestet, M.; Carre, C.; Antoniewski, C.; Huynh, J.R. tRNA Fragments Populations Analysis in Mutants Affecting tRNAs Processing and tRNA Methylation. Front. Genet. 2020, 11, 518949. [Google Scholar] [CrossRef]
- Gerber, A.P.; Keller, W. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001, 26, 376–384. [Google Scholar] [CrossRef]
- Grosjean, H.; Auxilien, S.; Constantinesco, F.; Simon, C.; Corda, Y.; Becker, H.F.; Foiret, D.; Morin, A.; Jin, Y.X.; Fournier, M.; et al. Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: A review. Biochimie 1996, 78, 488–501. [Google Scholar] [CrossRef]
- Bjork, G.R.; Jacobsson, K.; Nilsson, K.; Johansson, M.J.; Bystrom, A.S.; Persson, O.P. A primordial tRNA modification required for the evolution of life? EMBO J. 2001, 20, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, C.G.; Crain, P.F.; Gupta, R.; Hashizume, T.; Hocart, C.H.; Kowalak, J.A.; Pomerantz, S.C.; Stetter, K.O.; McCloskey, J.A. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J. Bacteriol. 1991, 173, 3138–3148. [Google Scholar] [CrossRef]
- Grosjean, H.; Constantinesco, F.; Foiret, D.; Benachenhou, N. A novel enzymatic pathway leading to 1-methylinosine modification in Haloferax volcanii tRNA. Nucleic Acids Res. 1995, 23, 4312–4319. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Tomizawa, K. Human transfer RNA modopathies: Diseases caused by aberrations in transfer RNA modifications. FEBS J. 2021. [Google Scholar] [CrossRef]
- Bednarova, A.; Hanna, M.; Durham, I.; VanCleave, T.; England, A.; Chaudhuri, A.; Krishnan, N. Lost in Translation: Defects in Transfer RNA Modifications and Neurological Disorders. Front. Mol. Neurosci. 2017, 10, 135. [Google Scholar] [CrossRef]
- El-Hattab, A.W.; Saleh, M.A.; Hashem, A.; Al-Owain, M.; Asmari, A.A.; Rabei, H.; Abdelraouf, H.; Hashem, M.; Alazami, A.M.; Patel, N.; et al. ADAT3-related intellectual disability: Further delineation of the phenotype. Am. J. Med Genet. Part A 2016, 170A, 1142–1147. [Google Scholar] [CrossRef]
- Alazami, A.M.; Hijazi, H.; Al-Dosari, M.S.; Shaheen, R.; Hashem, A.; Aldahmesh, M.A.; Mohamed, J.Y.; Kentab, A.; Salih, M.A.; Awaji, A.; et al. Mutation in ADAT3, encoding adenosine deaminase acting on transfer RNA, causes intellectual disability and strabismus. J. Med. Genet. 2013, 50, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Sharkia, R.; Zalan, A.; Jabareen-Masri, A.; Zahalka, H.; Mahajnah, M. A new case confirming and expanding the phenotype spectrum of ADAT3-related intellectual disability syndrome. Eur. J. Med. Genet. 2019, 62, 103549. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.; Han, L.; Li, Y.; Hagelskamp, F.; Kellner, S.M.; Alkuraya, F.S.; Phizicky, E.M.; Fu, D. Formation of tRNA Wobble Inosine in Humans Is Disrupted by a Millennia-Old Mutation Causing Intellectual Disability. Mol. Cell Biol. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.; Lewis, A.M.; Yang, Y.; Chanprasert, S.; Potocki, L.; Scott, D.A. Novel Missense Variants in ADAT3 as a Cause of Syndromic Intellectual Disability. J. Pediatr. Genet. 2019, 8, 244–251. [Google Scholar] [CrossRef]
- Chaleshtori, A.R.S.; Miyake, N.; Ahmadvand, M.; Bashti, O.; Matsumoto, N.; Noruzinia, M. A novel 8-bp duplication in ADAT3 causes mild intellectual disability. Hum. Genome Var. 2018, 5, 7. [Google Scholar] [CrossRef]
- Bass, B.L.; Weintraub, H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 1988, 55, 1089–1098. [Google Scholar] [CrossRef]
- Li, J.B.; Levanon, E.Y.; Yoon, J.K.; Aach, J.; Xie, B.; Leproust, E.; Zhang, K.; Gao, Y.; Church, G.M. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 2009, 324, 1210–1213. [Google Scholar] [CrossRef]
- Kim, U.; Wang, Y.; Sanford, T.; Zeng, Y.; Nishikura, K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. USA 1994, 91, 11457–11461. [Google Scholar] [CrossRef]
- Chen, C.X.; Cho, D.S.; Wang, Q.; Lai, F.; Carter, K.C.; Nishikura, K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 2000, 6, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; McFaline, J.L.; Burgis, N.E.; Dong, M.; Taghizadeh, K.; Sullivan, M.R.; Elmquist, C.E.; Cunningham, R.P.; Dedon, P.C. Defects in purine nucleotide metabolism lead to substantial incorporation of xanthine and hypoxanthine into DNA and RNA. Proc. Natl. Acad. Sci. USA 2012, 109, 2319–2324. [Google Scholar] [CrossRef]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar] [CrossRef]
- Sommer, B.; Kohler, M.; Sprengel, R.; Seeburg, P.H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 1991, 67, 11–19. [Google Scholar] [CrossRef]
- Burns, C.M.; Chu, H.; Rueter, S.M.; Hutchinson, L.K.; Canton, H.; Sanders-Bush, E.; Emeson, R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 1997, 387, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Patton, D.E.; Silva, T.; Bezanilla, F. RNA editing generates a diverse array of transcripts encoding squid Kv2 K+ channels with altered functional properties. Neuron 1997, 19, 711–722. [Google Scholar] [CrossRef]
- Saccomanno, L.; Bass, B.L. A minor fraction of basic fibroblast growth factor mRNA is deaminated in Xenopus stage VI and matured oocytes. RNA 1999, 5, 39–48. [Google Scholar] [CrossRef][Green Version]
- Hanrahan, C.J.; Palladino, M.J.; Ganetzky, B.; Reenan, R.A. RNA editing of the Drosophila para Na(+) channel transcript. Evolutionary conservation and developmental regulation. Genetics 2000, 155, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef]
- Licht, K.; Hartl, M.; Amman, F.; Anrather, D.; Janisiw, M.P.; Jantsch, M.F. Inosine induces context-dependent recoding and translational stalling. Nucleic Acids Res. 2019, 47, 3–14. [Google Scholar] [CrossRef]
- Presnyak, V.; Alhusaini, N.; Chen, Y.H.; Martin, S.; Morris, N.; Kline, N.; Olson, S.; Weinberg, D.; Baker, K.E.; Graveley, B.R.; et al. Codon optimality is a major determinant of mRNA stability. Cell 2015, 160, 1111–1124. [Google Scholar] [CrossRef] [PubMed]
- Brachova, P.; Alvarez, N.S.; Hong, X.; Gunewardena, S.; Vincent, K.A.; Latham, K.E.; Christenson, L.K. Inosine RNA modifications are enriched at the codon wobble position in mouse oocytes and eggsdagger. Biol. Reprod. 2019, 101, 938–949. [Google Scholar] [CrossRef]
- Brachova, P.; Alvarez, N.S.; Christenson, L.K. Loss of Cnot6l Impairs Inosine RNA Modifications in Mouse Oocytes. Int. J. Mol. Sci. 2021, 22, 1191. [Google Scholar] [CrossRef]
- Feldmeyer, D.; Kask, K.; Brusa, R.; Kornau, H.C.; Kolhekar, R.; Rozov, A.; Burnashev, N.; Jensen, V.; Hvalby, O.; Sprengel, R.; et al. Neurological dysfunctions in mice expressing different levels of the Q/R site-unedited AMPAR subunit GluR-B. Nat. Neurosci. 1999, 2, 57–64. [Google Scholar] [CrossRef]
- Brusa, R.; Zimmermann, F.; Koh, D.S.; Feldmeyer, D.; Gass, P.; Seeburg, P.H.; Sprengel, R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 1995, 270, 1677–1680. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, I.; Tamir, H.; Arango, V.; Dwork, A.J.; Mann, J.J.; Schmauss, C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 2002, 34, 349–356. [Google Scholar] [CrossRef]
- Hundley, H.A.; Bass, B.L. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem. Sci. 2010, 35, 377–383. [Google Scholar] [CrossRef]
- Prasanth, K.V.; Prasanth, S.G.; Xuan, Z.; Hearn, S.; Freier, S.M.; Bennett, C.F.; Zhang, M.Q.; Spector, D.L. Regulating gene expression through RNA nuclear retention. Cell 2005, 123, 249–263. [Google Scholar] [CrossRef]
- Yang, C.C.; Chen, Y.T.; Chang, Y.F.; Liu, H.; Kuo, Y.P.; Shih, C.T.; Liao, W.C.; Chen, H.W.; Tsai, W.S.; Tan, B.C. ADAR1-mediated 3’ UTR editing and expression control of antiapoptosis genes fine-tunes cellular apoptosis response. Cell Death Dis. 2017, 8, e2833. [Google Scholar] [CrossRef]
- Hundley, H.A.; Krauchuk, A.A.; Bass, B.L.C. elegans and H. sapiens mRNAs with edited 3’ UTRs are present on polysomes. RNA 2008, 14, 2050–2060. [Google Scholar] [CrossRef]
- Benne, R.; Van den Burg, J.; Brakenhoff, J.P.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef]
- Rueter, S.M.; Dawson, T.R.; Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature 1999, 399, 75–80. [Google Scholar] [CrossRef]
- Licht, K.; Kapoor, U.; Mayrhofer, E.; Jantsch, M.F. Adenosine to Inosine editing frequency controlled by splicing efficiency. Nucleic Acids Res. 2016, 44, 6398–6408. [Google Scholar] [CrossRef]
- Tonkin, L.A.; Saccomanno, L.; Morse, D.P.; Brodigan, T.; Krause, M.; Bass, B.L. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002, 21, 6025–6035. [Google Scholar] [CrossRef] [PubMed]
- Liddicoat, B.J.; Piskol, R.; Chalk, A.M.; Ramaswami, G.; Higuchi, M.; Hartner, J.C.; Li, J.B.; Seeburg, P.H.; Walkley, C.R. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 2015, 349, 1115–1120. [Google Scholar] [CrossRef] [PubMed]
- Hartner, J.C.; Schmittwolf, C.; Kispert, A.; Muller, A.M.; Higuchi, M.; Seeburg, P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004, 279, 4894–4902. [Google Scholar] [CrossRef]
- XuFeng, R.; Boyer, M.J.; Shen, H.; Li, Y.; Yu, H.; Gao, Y.; Yang, Q.; Wang, Q.; Cheng, T. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc. Natl. Acad. Sci. USA 2009, 106, 17763–17768. [Google Scholar] [CrossRef] [PubMed]
- Hartner, J.C.; Walkley, C.R.; Lu, J.; Orkin, S.H. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 2009, 10, 109–115. [Google Scholar] [CrossRef]
- Picardi, E.; Manzari, C.; Mastropasqua, F.; Aiello, I.; D’Erchia, A.M.; Pesole, G. Profiling RNA editing in human tissues: Towards the inosinome Atlas. Sci. Rep. 2015, 5, 14941. [Google Scholar] [CrossRef]
- Peng, X.; Xu, X.; Wang, Y.; Hawke, D.H.; Yu, S.; Han, L.; Zhou, Z.; Mojumdar, K.; Jeong, K.J.; Labrie, M.; et al. A-to-I RNA Editing Contributes to Proteomic Diversity in Cancer. Cancer Cell 2018, 33, 817–828.817. [Google Scholar] [CrossRef]
- Fritzell, K.; Xu, L.D.; Lagergren, J.; Ohman, M. ADARs and editing: The role of A-to-I RNA modification in cancer progression. Semin. Cell. Dev. Biol. 2018, 79, 123–130. [Google Scholar] [CrossRef]
- Chan, T.H.; Qamra, A.; Tan, K.T.; Guo, J.; Yang, H.; Qi, L.; Lin, J.S.; Ng, V.H.; Song, Y.; Hong, H.; et al. ADAR-Mediated RNA Editing Predicts Progression and Prognosis of Gastric Cancer. Gastroenterology 2016, 151, 637–650.e610. [Google Scholar] [CrossRef]
- An, O.; Song, Y.; Ke, X.; So, J.B.; Sundar, R.; Yang, H.; Rha, S.Y.; Lee, M.H.; Tay, S.T.; Ong, X.; et al. “3G” Trial: An RNA Editing Signature to Guide Gastric Cancer Chemotherapy. Cancer Res. 2021. [Google Scholar] [CrossRef]
- Anadon, C.; Guil, S.; Simo-Riudalbas, L.; Moutinho, C.; Setien, F.; Martinez-Cardus, A.; Moran, S.; Villanueva, A.; Calaf, M.; Vidal, A.; et al. Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene 2016, 35, 4407–4413. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, J.J.; Manguso, R.T.; Cheruiyot, C.K.; Bi, K.; Panda, A.; Iracheta-Vellve, A.; Miller, B.C.; Du, P.P.; Yates, K.B.; Dubrot, J.; et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 2019, 565, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Rice, G.I.; Kasher, P.R.; Forte, G.M.; Mannion, N.M.; Greenwood, S.M.; Szynkiewicz, M.; Dickerson, J.E.; Bhaskar, S.S.; Zampini, M.; Briggs, T.A.; et al. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 2012, 44, 1243–1248. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Suzuki, T.; Inagaki, K.; Ito, S.; Kono, M.; Horikawa, T.; Fujiwara, S.; Ishiko, A.; Matsunaga, K.; Aoyama, Y.; et al. Ten novel mutations of the ADAR1 gene in Japanese patients with dyschromatosis symmetrica hereditaria. J. Invest. Dermatol. 2007, 127, 309–311. [Google Scholar] [CrossRef]
- Pedersen, K.; Gerdes, K. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 1999, 32, 1090–1102. [Google Scholar] [CrossRef]
- Verstraeten, N.; Knapen, W.J.; Kint, C.I.; Liebens, V.; Van den Bergh, B.; Dewachter, L.; Michiels, J.E.; Fu, Q.; David, C.C.; Fierro, A.C.; et al. Obg and Membrane Depolarization Are Part of a Microbial Bet-Hedging Strategy that Leads to Antibiotic Tolerance. Mol. Cell 2015, 59, 9–21. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Kawahara, Y.; Zinshteyn, B.; Sethupathy, P.; Iizasa, H.; Hatzigeorgiou, A.G.; Nishikura, K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 2007, 315, 1137–1140. [Google Scholar] [CrossRef]
- Ota, H.; Sakurai, M.; Gupta, R.; Valente, L.; Wulff, B.E.; Ariyoshi, K.; Iizasa, H.; Davuluri, R.V.; Nishikura, K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013, 153, 575–589. [Google Scholar] [CrossRef]
- Kawahara, Y.; Megraw, M.; Kreider, E.; Iizasa, H.; Valente, L.; Hatzigeorgiou, A.G.; Nishikura, K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008, 36, 5270–5280. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Sinha, A.N.; Ray, A.; Lal, M.; Nayak, S.; Sharma, A.; Mehani, B.; Mukherjee, D.; Laddha, S.V.; Suri, A.; et al. A-to-I editing in human miRNAs is enriched in seed sequence, influenced by sequence contexts and significantly hypoedited in glioblastoma multiforme. Sci. Rep. 2017, 7, 2466. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Yu, S.; Jeong, K.J.; Zhou, Z.; Han, L.; Tsang, Y.H.; Li, J.; Chen, H.; Mangala, L.S.; et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017, 27, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, Y.; Tay, F.C.; Lam, D.H.; Sandanaraj, E.; Tang, C.; Ang, B.T.; Wang, S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig. 2012, 122, 4059–4076. [Google Scholar] [CrossRef]
- Chiu, C.G.; St-Pierre, P.; Nabi, I.R.; Wiseman, S.M. Autocrine motility factor receptor: A clinical review. Expert Rev. Anticancer. Ther. 2008, 8, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, R.; Schmid, A.; Eschle, D.; Baczko, K.; ter Meulen, V.; Billeter, M.A. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 1988, 55, 255–265. [Google Scholar] [CrossRef]
- Cattaneo, R. Biased (A-->I) hypermutation of animal RNA virus genomes. Curr. Opin. Genet. Dev. 1994, 4, 895–900. [Google Scholar] [CrossRef]
- Zahn, R.C.; Schelp, I.; Utermohlen, O.; von Laer, D. A-to-G hypermutation in the genome of lymphocytic choriomeningitis virus. J. Virol. 2007, 81, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Mannion, N.M.; Greenwood, S.M.; Young, R.; Cox, S.; Brindle, J.; Read, D.; Nellaker, C.; Vesely, C.; Ponting, C.P.; McLaughlin, P.J.; et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014, 9, 1482–1494. [Google Scholar] [CrossRef]
- Lempp, F.A.; Ni, Y.; Urban, S. Hepatitis delta virus: Insights into a peculiar pathogen and novel treatment options. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.Y.; Chao, M.; Taylor, J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: Role of delta antigen. J. Virol. 1989, 63, 1945–1950. [Google Scholar] [CrossRef]
- Ryu, W.S.; Bayer, M.; Taylor, J. Assembly of hepatitis delta virus particles. J. Virol. 1992, 66, 2310–2315. [Google Scholar] [CrossRef] [PubMed]
- Jayan, G.C.; Casey, J.L. Effects of conserved RNA secondary structures on hepatitis delta virus genotype I RNA editing, replication, and virus production. J. Virol. 2005, 79, 11187–11193. [Google Scholar] [CrossRef] [PubMed]
- Krebs, J.E.; Goldstein, E.S.; Kilpatrick, S.T. Lewin’s Genes XII; Jones & Barlett: Burlington, MA, USA, 2018; p. 387. [Google Scholar]
- Ule, J. Alu elements: At the crossroads between disease and evolution. Biochem. Soc. Trans. 2013, 41, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R. The birth of new exons: Mechanisms and evolutionary consequences. Rna 2007, 13, 1603–1608. [Google Scholar] [CrossRef]
- Lev-Maor, G.; Sorek, R.; Levanon, E.Y.; Paz, N.; Eisenberg, E.; Ast, G. RNA-editing-mediated exon evolution. Genome Biol. 2007, 8, R29. [Google Scholar] [CrossRef] [PubMed]
- Bazak, L.; Haviv, A.; Barak, M.; Jacob-Hirsch, J.; Deng, P.; Zhang, R.; Isaacs, F.J.; Rechavi, G.; Li, J.B.; Eisenberg, E.; et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014, 24, 365–376. [Google Scholar] [CrossRef]
- Pullirsch, D.; Jantsch, M.F. Proteome diversification by adenosine to inosine RNA editing. RNA Biol. 2010, 7, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Fukami, T.; Gotoh, S.; Nakajima, M. A-to-I RNA Editing Up-regulates Human Dihydrofolate Reductase in Breast Cancer. J. Biol. Chem. 2017, 292, 4873–4884. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.W. O2’-Methylinosine, a constituent of the ribosomal RNA of Crithidia fasciculata. Nucleic Acids Res. 1976, 3, 977–988. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moreira, S.; Valach, M.; Aoulad-Aissa, M.; Otto, C.; Burger, G. Novel modes of RNA editing in mitochondria. Nucleic Acids Res. 2016, 44, 4907–4919. [Google Scholar] [CrossRef]
- Prestwich, E.G.; Mangerich, A.; Pang, B.; McFaline, J.L.; Lonkar, P.; Sullivan, M.R.; Trudel, L.J.; Taghizedeh, K.; Dedon, P.C. Increased levels of inosine in a mouse model of inflammation. Chem. Res. Toxicol. 2013, 26, 538–546. [Google Scholar] [CrossRef]
- Zheng, Y.; Lorenzo, C.; Beal, P.A. DNA editing in DNA/RNA hybrids by adenosine deaminases that act on RNA. Nucleic Acids Res. 2017, 45, 3369–3377. [Google Scholar] [CrossRef]
- Myrnes, B.; Guddal, P.H.; Krokan, H. Metabolism of dITP in HeLa cell extracts, incorporation into DNA by isolated nuclei and release of hypoxanthine from DNA by a hypoxanthine-DNA glycosylase activity. Nucleic Acids Res. 1982, 10, 3693–3701. [Google Scholar] [CrossRef][Green Version]
- Tsuruoka, N.; Arima, M.; Yoshida, N.; Okada, S.; Sakamoto, A.; Hatano, M.; Satake, H.; Arguni, E.; Wang, J.Y.; Yang, J.H.; et al. ADAR1 protein induces adenosine-targeted DNA mutations in senescent Bcl6 gene-deficient cells. J. Biol. Chem. 2013, 288, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Dalhus, B.; Arvai, A.S.; Rosnes, I.; Olsen, O.E.; Backe, P.H.; Alseth, I.; Gao, H.; Cao, W.; Tainer, J.A.; Bjoras, M. Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair. Nat. Struct. Mol. Biol. 2009, 16, 138–143. [Google Scholar] [CrossRef]
- Cao, W. Endonuclease V: An unusual enzyme for repair of DNA deamination. Cell Mol. Life Sci. CMLS 2013, 70, 3145–3156. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Wood, R.D. Quality control by DNA repair. Science 1999, 286, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srinivasan, S.; Torres, A.G.; Ribas de Pouplana, L. Inosine in Biology and Disease. Genes 2021, 12, 600. https://doi.org/10.3390/genes12040600
Srinivasan S, Torres AG, Ribas de Pouplana L. Inosine in Biology and Disease. Genes. 2021; 12(4):600. https://doi.org/10.3390/genes12040600
Chicago/Turabian StyleSrinivasan, Sundaramoorthy, Adrian Gabriel Torres, and Lluís Ribas de Pouplana. 2021. "Inosine in Biology and Disease" Genes 12, no. 4: 600. https://doi.org/10.3390/genes12040600
APA StyleSrinivasan, S., Torres, A. G., & Ribas de Pouplana, L. (2021). Inosine in Biology and Disease. Genes, 12(4), 600. https://doi.org/10.3390/genes12040600






